A High Temperature Environment Regulates the Olive Oil Biosynthesis Network
Abstract
1. Introduction
1.1. Heat-Shock Proteins
1.2. Olive Oil Biosynthesis
1.3. Transcription Factors Regulate Olive Oil Biosynthesis
2. Materials and Methods
2.1. Experimental Design and Plant Material
2.2. Transcriptome Analysis
2.3. RT-PCR
2.4. Data Analysis
2.5. Statistical Analysis
3. Results
3.1. Gene Expression Regulation
3.2. Heat-Shock Protein Genes are Upregulated in the HT Environment
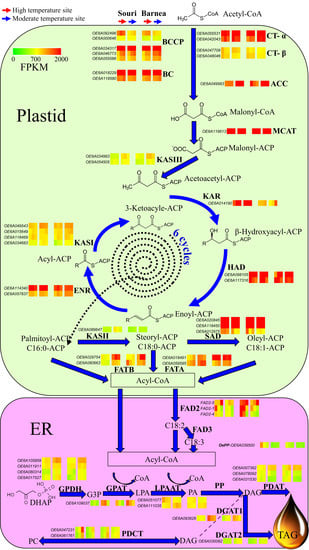
3.3. The Effect of High Summer Temperatures on the Olive Oil Biosynthesis Pathway
3.4. Transcription Factors Regulating Olive Oil Biosynthesis Pathway
4. Discussion
4.1. HT Environment Induced A Heat Stress Response in Both Cultivars
4.2. High Summer Temperatures Repress Genes Involved in the Olive Oil Biosynthesis Pathway
4.3. WRI1 and Dof4 Are the Main Transcription Factors Regulating Olive Oil Biosynthesis Pathway
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Maestri, E.; Klueva, N.; Perrotta, C.; Gulli, M.; Nguyen, H.T.; Marmiroli, N. Molecular genetics of heat tolerance and heat shock proteins in cereals. Plant Mol. Biol. 2002, 48, 667–681. [Google Scholar] [CrossRef]
- Wilhelm, E.P.; Mullen, R.; Keeling, P.; Singletary, G. Heat stress during grain filling in maize: Effects on kernel growth and metabolism. Crop Sci. 1999, 39, 1733–1741. [Google Scholar] [CrossRef]
- Barnabas, B.; Jager, K.; Feher, A. The effect of drought and heat stress on reproductive processes in cereals. Plant Cell Environ. 2008, 31, 11–38. [Google Scholar] [CrossRef] [PubMed]
- Bita, C.E.; Gerats, T. Plant tolerance to high temperature in a changing environment: Scientific fundamentals and production of heat stress-tolerant crops. Front. Plant Sci. 2013, 4, 273. [Google Scholar] [CrossRef] [PubMed]
- Bray, E.A.; Bailey-Serres, J.; Weretilnyk, E. Biochemistry and Molecular Biology of Plants, Responses to Abiotic Stresses; Gruissem, W., Jones, R., Eds.; American Society of Plant Biologists: Rockville, MD, USA, 2000; pp. 1158–1203. [Google Scholar]
- Guy, C. Molecular responses of plants to cold shock and cold acclimation. J. Mol. Microbiol. Biotechnol. 1999, 1, 231–242. [Google Scholar]
- Weis, E.; Berry, J.A. Plants and high temperature stress. Symp. Soc. Exp. Biol. 1988, 42, 329–346. [Google Scholar]
- Fahad, S.; Bajwa, A.A.; Nazir, U.; Anjum, S.A.; Farooq, A.; Zohaib, A.; Sadia, S.; Nasim, W.; Adkins, S.; Saud, S.; et al. Crop production under drought and heat stress: Plant responses and management options. Front. Plant Sci. 2017, 8. [Google Scholar] [CrossRef]
- Camejo, D.; Rodríguez, P.; Angeles Morales, M.; Miguel Dell’Amico, J.; Torrecillas, A.; Alarcón, J.J. High temperature effects on photosynthetic activity of two tomato cultivars with different heat susceptibility. J. Plant Physiol. 2005, 162, 281–289. [Google Scholar] [CrossRef]
- Fahad, S.; Hussain, S.; Saud, S.; Hassan, S.; Tanveer, M.; Ihsan, M.Z.; Shah, A.N.; Ullah, A.; Nasrullah; Khan, F.; et al. A combined application of biochar and phosphorus alleviates heat-induced adversities on physiological, agronomical and quality attributes of rice. Plant Physiol. Biochem. 2016, 103, 191–198. [Google Scholar] [CrossRef]
- Ferris, R.; Ellis, R.H.; Wheeler, T.R.; Hadley, P. Effect of high temperature stress at anthesis on grain yield and biomass of field-grown crops of wheat. Ann. Bot. 1998, 82, 631–639. [Google Scholar] [CrossRef]
- Rainey, K.M.; Griffiths, P.D. Evaluation of Phaseolus acutifolius A. Gray plant introductions under high temperatures in a controlled environment. Genet. Resour. Crop Evol. 2005, 52, 117–120. [Google Scholar] [CrossRef]
- Vara Prasad, P.V.; Craufurd, P.Q.; Summerfield, R.J. Fruit Number in Relation to Pollen Production and Viability in Groundnut Exposed to Short Episodes of Heat Stress. Ann. Bot. 1999, 84, 381–386. [Google Scholar] [CrossRef]
- Bellincontro, A.; Caruso, G.; Mencarelli, F.; Gucci, R. Oil accumulation in intact olive fruits measured by near infrared spectroscopy-acousto-optically tunable filter. J. Sci. Food Agric. 2013, 93, 1259–1265. [Google Scholar] [CrossRef]
- Conde, C.; Delrot, S.; Gerós, H. Physiological, biochemical and molecular changes occurring during olive development and ripening. J. Plant Physiol. 2008, 165, 1545–1562. [Google Scholar] [CrossRef]
- Servili, M. Olive oil processing technologies and investments. In Present and Future of the Mediterranean Olive Sector; Caballero, J., D’Andria, R., Fernández, M., Garrido, A., Rallo, L., Arroyo López, F.N., Arcas, N., Fernandez Escobar, R., López-Miranda, J., Msallem, M., et al., Eds.; CIHEAM IOC: Zaragoza, Spain, 2013; Volume 106, pp. 55–66. [Google Scholar]
- García-Inza, G.P.; Castro, D.N.; Hall, A.J.; Rousseaux, M.C. Responses to temperature of fruit dry weight, oil concentration, and oil fatty acid composition in olive (Olea europaea L. var. ‘Arauco’). Eur. J. Agron. 2014, 54, 107–115. [Google Scholar] [CrossRef]
- Miserere, A.; Searles, P.S.; García-Inza, G.P.; Rousseaux, M.C. Elevated temperature affects vegetative growth and fruit oil concentration in olive trees (Olea europaea). Acta Hortic. 2018, 1199, 523–528. [Google Scholar] [CrossRef]
- Rondanini, D.P.; Castro, D.N.; Searles, P.S.; Rousseaux, M.C. Contrasting patterns of fatty acid composition and oil accumulation during fruit growth in several olive varieties and locations in a non-Mediterranean region. Eur. J. Agron. 2014, 52, 237–246. [Google Scholar] [CrossRef]
- Trentacoste, E.R.; Puertas, C.M.; Sadras, V.O. Modelling the intraspecific variation in the dynamics of fruit growth, oil and water concentration in olive (Olea europaea L.). Eur. J. Agron. 2012, 38, 83–93. [Google Scholar] [CrossRef]
- García-Inza, G.P.; Castro, D.N.; Hall, A.J.; Rousseaux, M.C. Opposite oleic acid responses to temperature in oils from the seed and mesocarp of the olive fruit. Eur. J. Agron. 2016, 76, 138–147. [Google Scholar] [CrossRef]
- García-Inza, G.P.; Hall, A.J.; Rousseaux, M.C. Proportion of oleic acid in olive oil as influenced by the dimensions of the daily temperature oscillation. Sci. Hortic. 2018, 227, 305–312. [Google Scholar] [CrossRef]
- Lombardo, N.; Marone, E.; Alessandrino, M.; Godino, G.; Madeo, A.; Fiorino, P. Influence of growing season temperatures in the fatty acids (FAs) of triacilglycerols (TAGs) composition in Italian cultivars of Olea europaea. Adv. Hortic. Sci. 2008, 22, 49–53. [Google Scholar]
- Mailer, R.J.; Ayton, J.; Graham, K. The Influence of Growing Region, Cultivar and Harvest Timing on the Diversity of Australian Olive Oil. J. Am. Oil Chem. Soc. 2010, 87, 877–884. [Google Scholar] [CrossRef]
- Al-Whaibi, M.H. Plant heat-shock proteins: A mini review. J. King Saud Univ. Sci. 2011, 23, 139–150. [Google Scholar] [CrossRef]
- Ul Haq, S.; Khan, A.; Ali, M.; Khattak, A.M.; Gai, W.-X.; Zhang, H.-X.; Wei, A.-M.; Gong, Z.-H. Heat shock proteins: Dynamic biomolecules to counter plant biotic and abiotic stresses. Int. J. Mol. Sci. 2019, 20, 5321. [Google Scholar] [CrossRef] [PubMed]
- Janni, M.; Gullì, M.; Maestri, E.; Marmiroli, M.; Valliyodan, B.; Nguyen, H.T.; Marmiroli, N. Molecular and genetic bases of heat stress responses in crop plants and breeding for increased resilience and productivity. J. Exp. Bot. 2020, 71, 3780–3802. [Google Scholar] [CrossRef]
- Montero-Barrientos, M.; Hermosa, R.; Nicolás, C.; Cardoza, R.E.; Gutiérrez, S.; Monte, E. Overexpression of a Trichoderma HSP70 gene increases fungal resistance to heat and other abiotic stresses. Fungal Genet. Biol. 2008, 45, 1506–1513. [Google Scholar] [CrossRef]
- Qi, Y.; Wang, H.; Zou, Y.; Liu, C.; Liu, Y.; Wang, Y.; Zhang, W. Over-expression of mitochondrial heat shock protein 70 suppresses programmed cell death in rice. FEBS Lett. 2011, 585, 231–239. [Google Scholar] [CrossRef]
- Wang, X.; Yan, B.; Shi, M.; Zhou, W.; Zekria, D.; Wang, H.; Kai, G. Overexpression of a Brassica campestris HSP70 in tobacco confers enhanced tolerance to heat stress. Protoplasma 2016, 253, 637–645. [Google Scholar] [CrossRef]
- He, M.; Qin, C.-X.; Wang, X.; Ding, N.-Z. Plant unsaturated fatty acids: Biosynthesis and regulation. Front. Plant Sci. 2020, 11. [Google Scholar] [CrossRef]
- Salas, J.; Harwood, J.; Martínez Force, E. Lipid metabolism in olive: Biosynthesis of triacylglycerols and aroma components. In Handbook of Olive Oil, 2nd ed.; Aparicio, R., Harwood, J., Eds.; Springer: New York, NY, USA, 2013. [Google Scholar] [CrossRef]
- Hu, Z.; Ren, Z.; Lu, C. The phosphatidylcholine diacylglycerol cholinephosphotransferase is required for efficient hydroxy fatty acid accumulation in transgenic Arabidopsis. Plant Physiol. 2012, 158, 1944–1954. [Google Scholar] [CrossRef]
- Manan, S.; Chen, B.; She, G.; Wan, X.; Zhao, J. Transport and transcriptional regulation of oil production in plants. Crit. Rev. Biotechnol. 2017, 37, 641–655. [Google Scholar] [CrossRef] [PubMed]
- Kong, Q.; Yuan, L.; Ma, W. WRINKLED1, a “Master Regulator” in transcriptional control of plant oil biosynthesis. Plants 2019, 8, 238. [Google Scholar] [CrossRef] [PubMed]
- Nissim, Y.; Shloberg, M.; Biton, I.; Many, Y.; Doron-Faigenboim, A.; Zemach, H.; Hovav, R.; Kerem, Z.; Avidan, B.; Ben-Ari, G. High temperature environment reduces olive oil yield and quality. PLoS ONE 2020, 15, e0231956. [Google Scholar] [CrossRef] [PubMed]
- Grabherr, M.G.; Haas, B.J.; Yassour, M.; Levin, J.Z.; Thompson, D.A.; Amit, I.; Adiconis, X.; Fan, L.; Raychowdhury, R.; Zeng, Q.; et al. Full-length transcriptome assembly from RNA-Seq data without a reference genome. Nat. Biotechnol. 2011, 29, 644–652. [Google Scholar] [CrossRef]
- Bolger, A.M.; Lohse, M.; Usadel, B. Trimmomatic: A flexible trimmer for Illumina sequence data. Bioinformatics 2014, 30. [Google Scholar] [CrossRef]
- Langmead, B.; Trapnell, C.; Pop, M.; Salzberg, S.L. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 2009, 10, R25. [Google Scholar] [CrossRef]
- Li, B.; Dewey, C.N. RSEM: Accurate transcript quantification from RNA-Seq data with or without a reference genome. BMC Bioinform. 2011, 12, 323. [Google Scholar] [CrossRef]
- Rozen, S.; Skaletsky, H. Primer3 on the WWW for general users and for biologist programmers. Methods Mol. Biol. 2000, 132, 365–386. [Google Scholar]
- Robinson, M.D.; McCarthy, D.J.; Smyth, G.K. edgeR: A Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 2010, 26, 139–140. [Google Scholar] [CrossRef]
- Benjamini, Y.; Hochberg, Y. Controlling the False Discovery Rate: A practical and powerful approach to multiple testing. J. R. Stat. Soc. 1995, 57, 289–300. [Google Scholar] [CrossRef]
- Gentleman, R.C.; Carey, V.J.; Bates, D.M.; Bolstad, B.; Dettling, M.; Dudoit, S.; Ellis, B.; Gautier, L.; Ge, Y.; Gentry, J.; et al. Bioconductor: Open software development for computational biology and bioinformatics. Genome Biol. 2004, 5, R80. [Google Scholar] [CrossRef] [PubMed]
- SAS Institute JMP® Version 7 User’s Guide; SAS Institute Inc.: Cary, NC, USA, 2007.
- Cruz, F.; Julca, I.; Gómez-Garrido, J.; Loska, D.; Marcet-Houben, M.; Cano, E.; Galán, B.; Frias, L.; Ribeca, P.; Derdak, S.; et al. Genome sequence of the olive tree, Olea europaea. GigaScience 2016, 5, 29. [Google Scholar] [CrossRef] [PubMed]
- Huang, L.-J.; Cheng, G.-X.; Khan, A.; Wei, A.-M.; Yu, Q.-H.; Yang, S.-B.; Luo, D.-X.; Gong, Z.-H. CaHSP16.4, a small heat shock protein gene in pepper, is involved in heat and drought tolerance. Protoplasma 2019, 256, 39–51. [Google Scholar] [CrossRef] [PubMed]
- Xu, Y.; Zhan, C.; Huang, B. Heat shock proteins in association with heat tolerance in grasses. Int. J. Proteom. 2011, 2011, 529648. [Google Scholar] [CrossRef]
- Fernandez-Moya, R.; Da Silva, N.A. Engineering Saccharomyces cerevisiae for high-level synthesis of fatty acids and derived products. FEMS Yeast Res. 2017, 17. [Google Scholar] [CrossRef]
- Gomma, A.E.; Lee, S.-K.; Sun, S.M.; Yang, S.H.; Chung, G. Improvement in oil production by increasing Malonyl-CoA and Glycerol-3-Phosphate pools in scenedesmus quadricauda. Indian J. Microbiol. 2015, 55, 447–455. [Google Scholar] [CrossRef]
- Shin, G.-H.; Veen, M.; Stahl, U.; Lang, C. Overexpression of genes of the fatty acid biosynthetic pathway leads to accumulation of sterols in Saccharomyces cerevisiae. Yeast 2012, 29, 371–383. [Google Scholar] [CrossRef]
- Liu, Q.; Siloto, R.M.P.; Lehner, R.; Stone, S.J.; Weselake, R.J. Acyl-CoA:diacylglycerol acyltransferase: Molecular biology, biochemistry and biotechnology. Prog. Lipid Res. 2012, 51, 350–377. [Google Scholar] [CrossRef]
- Li, Q.; Zheng, Q.; Shen, W.; Cram, D.; Fowler, D.B.; Wei, Y.; Zou, J. Understanding the biochemical basis of temperature-Induced lipid pathway adjustments in plants. Plant Cell 2015, 27, 86–103. [Google Scholar] [CrossRef]
- Luisa Hernández, M.; Dolores Sicardo, M.; Arjona, P.M.; Martínez-Rivas, J.M. Specialized functions of olive FAD2 gene family members related to fruit development and the abiotic stress response. Plant Cell Physiol. 2019, 61, 427–441. [Google Scholar] [CrossRef]




© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Nissim, Y.; Shlosberg, M.; Biton, I.; Many, Y.; Doron-Faigenboim, A.; Hovav, R.; Kerem, Z.; Avidan, B.; Ben-Ari, G. A High Temperature Environment Regulates the Olive Oil Biosynthesis Network. Plants 2020, 9, 1135. https://doi.org/10.3390/plants9091135
Nissim Y, Shlosberg M, Biton I, Many Y, Doron-Faigenboim A, Hovav R, Kerem Z, Avidan B, Ben-Ari G. A High Temperature Environment Regulates the Olive Oil Biosynthesis Network. Plants. 2020; 9(9):1135. https://doi.org/10.3390/plants9091135
Chicago/Turabian StyleNissim, Yael, Maya Shlosberg, Iris Biton, Yair Many, Adi Doron-Faigenboim, Ran Hovav, Zohar Kerem, Benjamin Avidan, and Giora Ben-Ari. 2020. "A High Temperature Environment Regulates the Olive Oil Biosynthesis Network" Plants 9, no. 9: 1135. https://doi.org/10.3390/plants9091135
APA StyleNissim, Y., Shlosberg, M., Biton, I., Many, Y., Doron-Faigenboim, A., Hovav, R., Kerem, Z., Avidan, B., & Ben-Ari, G. (2020). A High Temperature Environment Regulates the Olive Oil Biosynthesis Network. Plants, 9(9), 1135. https://doi.org/10.3390/plants9091135