Phenolic Compounds Content and Genetic Diversity at Population Level across the Natural Distribution Range of Bearberry (Arctostaphylos uva-ursi, Ericaceae) in the Iberian Peninsula
Abstract
:1. Introduction
2. Results and Discussion
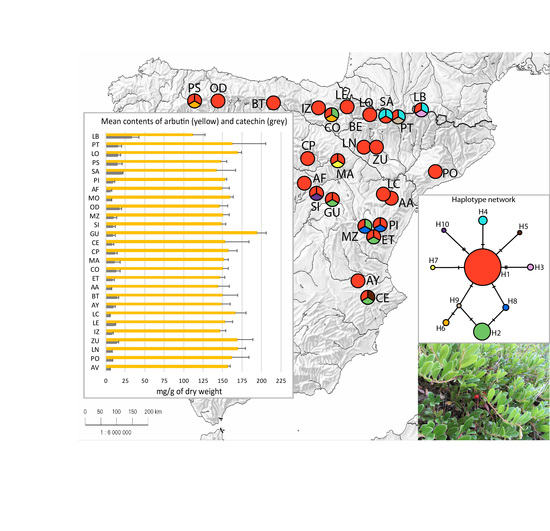
2.1. Chemical Diversity Analysis
2.2. Genome Size and Sequence Data Analysis
3. Materials and Methods
3.1. Plant Material
3.2. Preparation of Leaf Extracts and Total Phenols Determination
3.3. Arbutin and Other Phenolic Metabolites Determination
3.4. Genome Size Estimates and DNA Sequencing
3.5. Genome Size and Genetic Analysis
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Moore, B.D.; Andrew, R.L.; Külheim, C.; Foley, W.J. Explaining intraspecific diversity in plant secondary metabolites in an ecological context. New Phytol. 2014, 201, 733–750. [Google Scholar] [CrossRef] [PubMed]
- Cheynier, V.; Comte, G.; Davies, K.M.; Lattanzio, V. Plant phenolics: Recent advances on their biosynthesis, genetics and ecophysiology. Plant Physiol. Biochem. 2013, 72, 1–20. [Google Scholar] [CrossRef]
- Bernal, M.; Llorens, L.; Julkunen-Tiitto, R.; Badosa, J.; Verdaguer, D. Altitudinal and seasonal changes of phenolic compounds in Buxus sempervirens leaves and cuticles. Plant Physiol. Biochem. 2013, 70, 471–482. [Google Scholar] [CrossRef] [PubMed]
- Upton, R. (Ed.) Uva-ursi Leaf. Arctostaphylos uva-ursi (L.) Spreng. Standard of Analysis, Quality Control and Therapeutics; American Herbal Pharmacopoeia: Scotts Valley, CA, USA, 2008; 30p. [Google Scholar]
- European Medicines Agency. Assessment Report on Arctostaphylos uva-ursi (L.) Spreng. Folium. EMA/HMPC/750266/2016; European Medicines Agency: Amsterdam, The Netherlands, 2018.
- Kanlayavattanakul, M.; Lourith, N. Plants and natural products for the treatment of skin hyperpigmentation—A review. Planta Med. 2018, 84, 988–1006. [Google Scholar] [CrossRef] [Green Version]
- Migas, P.; Krauze-Baranowska, M. The significance of arbutin and its derivatives in therapy and cosmetics. Phytochem. Lett. 2015, 13, 35–40. [Google Scholar] [CrossRef]
- Seo, D.H.; Jung, J.H.; Lee, J.E.; Jeon, E.J.; Kim, W.; Park, C.S. Biotechnological production of arbutins (α- and β-arbutins), skin-lightening agents, and their derivatives. Appl. Microbiol. Biotechnol. 2012, 95, 1417–1425. [Google Scholar] [CrossRef]
- Zhu, X.; Tian, Y.; Zhang, W.; Zhang, T.; Guang, C.; Mu, W. Recent progress on biological production of α-arbutin. Appl. Microbiol. Biotechnol. 2018, 102, 8145–8152. [Google Scholar] [CrossRef] [PubMed]
- Davey, M. Secondary metabolism in plant cell cultures. In Encyclopedia of Applied Plant Sciences, 2nd ed.; Thomas, B., Murphy, D.J., Murray, B.G., Eds.; Elsevier: London, UK, 2017; Volume 2, pp. 462–467. [Google Scholar] [CrossRef]
- Kurkin, V.A.; Ryazanova, T.K.; Daeva, E.D.; Kadentsev, V.I. Constituents of Arctostaphylos uva-ursi leaves. Chem. Nat. Compd. 2018, 54, 278–280. [Google Scholar] [CrossRef]
- Panusa, A.; Petrucci, R.; Marrosu, G.; Multari, G.; Romana-Gallo, F. HPLC-PDAESI-TOF/MS metabolic profiling of Arctostaphylos pungens and Arctostaphylos uva-ursi: A comparative study of phenolic compounds from leaf metabolic extract. Phytochemistry 2015, 115, 79–88. [Google Scholar] [CrossRef]
- Olennikov, D.N.; Chekhirova, G.V. 6′-Galloylpicein and other phenolic compounds from Arctostaphylos uva-ursi. Chem. Nat. Compd. 2013, 49, 1–7. [Google Scholar] [CrossRef]
- Brewer, M.S. Natural antioxidants: Sources, compounds, mechanisms of action, and potential applications. Compr. Rev. Food Sci. Food Saf. 2011, 10, 221–247. [Google Scholar] [CrossRef]
- Amarowicz, R.; Pegg, R.B.; Rahimi-Moghaddam, P.; Barl, B.; Weil, J.A. Free-radical scavenging capacity and antioxidant activity of selected plant species from the Canadian prairies. Food Chem. 2004, 84, 551–562. [Google Scholar] [CrossRef]
- Carpenter, R.; O’Grady, M.N.; O’Callaghan, Y.C.; O’Brien, N.M.; Kerry, J.P. Evaluation of the antioxidant potential of grape seed and bearberry extracts in raw and cooked pork. Meat Sci. 2007, 76, 604–610. [Google Scholar] [CrossRef] [PubMed]
- Mekinić, I.G.; Skroza, D.; Ljubenkov, I.; Katalinić, V.; Šimat, V. Antioxidant and antimicrobial potential of phenolic metabolites from traditionally used Mediterranean herbs and spices. Foods 2019, 8, 579. [Google Scholar] [CrossRef] [Green Version]
- Mohd Azman, N.A.; Gallego, M.G.; Segovia, F.; Abdullah, S.; Shaarani, S.M.; Almajano, M.P. Study of the properties of bearberry leaf extract as a natural antioxidant in model foods. Antioxidants 2016, 5, 11. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pegg, R.B.; Amarowicz, R.; Naczk, M. Antioxidant activity of polyphenolics from bearberry leaf (Arctostaphylos uva-ursi L. Sprengel) extract in meat systems. In Phenolic Compounds in Foods and Natural Health Products; Shahidi, F., Ho, C.T., Eds.; American Chemical Society: Washington, DC, USA, 2005; pp. 67–82. [Google Scholar] [CrossRef]
- Wrona, M.; Blasco, S.; Becerril, R.; Nerín, C.; Sales, E.; Asensio, E. Antioxidant and antimicrobial markers by UPLC®–ESI-Q-TOF-MSE of a new multilayer active packaging based on Arctostaphylos uva-ursi. Talanta 2019, 196, 498–509. [Google Scholar] [CrossRef]
- Maier, M.; Oelbermann, A.L.; Renner, M.; Weidner, E. Screening of European medicinal herbs on their tannin content—New potential tanning agents for the leather industry. Ind. Crop. Prod. 2017, 99, 19–26. [Google Scholar] [CrossRef]
- Boros, B.; Jakabová, S.; Madarász, T.; Molnár, R.; Galambosi, B.; Kilár, F.; Felinger, A.; Farkas, A. Validated HPLC method for simultaneous quantitation of bergenin, arbutin and gallic acid in leaves of different Bergenia species. Chromatographia 2014, 77, 1129–1135. [Google Scholar] [CrossRef]
- Miaw-Ling, C.; Chur-Min, C. Simultaneous HPLC determination of hydrophilic whitening agent in cosmetic product. J. Pharm. Biomed. Anal. 2003, 33, 617–626. [Google Scholar] [CrossRef]
- Parejo, I.; Viladomat, F.; Bastida, J.; Codina, C. A single extraction step in the quantitative analysis of arbutin in bearberry (Arctostaphylos uva-ursi) leaves by high performance liquid chromatography. Phytochem. Anal. 2001, 12, 336–339. [Google Scholar] [CrossRef]
- POWO. Plants of the World Online. Available online: http://www.plantsoftheworldonline.org/ (accessed on 29 April 2020).
- Saleem, A.; Harris, C.S.; Asim, M.; Cuerrier, A.; Martineau, L.; Haddad, P.S.; Arnasona, J.T. RP-HPLC-DAD-APCI/MSD method for the characterisation of medicinal Ericaceae used by the Eeyou Istchee Cree First Nations. Phytochem. Anal. 2010, 21, 328–339. [Google Scholar] [CrossRef] [PubMed]
- Stefanescu, B.E.; Szabo, K.; Mocan, A.; Crisan, G. Phenolic compounds from five Ericaceae species leaves and their related bioavailability and health benefits. Molecules 2019, 24, 2046. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Onda, Y.; Mochida, K. Exploring genetic diversity in plants using high-throughput sequencing techniques. Curr. Genom. 2016, 17, 358–367. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Iannicelli, J.; Guariniello, J.; Tossib, V.E.; Regalado, J.J.; Di Ciaccioa, L.; van Barene, C.M.; Pitta Álvarez, S.I.; Escandón, A.S. The “polyploid effect” in the breeding of aromatic and medicinal species. Sci. Hortic. 2020, 260, 108854. [Google Scholar] [CrossRef]
- Leitch, I.J.; Leitch, A.R. Genome size diversity and evolution in land plants. In Plant Genome Diversity Volume 2, Physical Structure, Behaviour and Evolution of Plant Genomes; Leitch, I.J., Greilhuber, J., Doležel, J., Wendel, J.F., Eds.; Springer: Vienna, Austria, 2013; pp. 307–322. [Google Scholar] [CrossRef]
- Bennett, M.D.; Leitch, I.J. Nuclear DNA amounts in angiosperms—Progress, problems and prospects. Ann. Bot. 2005, 95, 45–90. [Google Scholar] [CrossRef] [Green Version]
- Gregory, T.R. (Ed.) The Evolution of the Genome; Elsevier: San Diego, CA, USA, 2005. [Google Scholar]
- Hoang, P.N.; Schubert, V.; Meister, A.; Fuchs, J.; Shubert, I. Variation in genome size, cell and nucleus volume, chromosome number and rDNA loci among duckweeds. Sci. Rep. 2019, 9, 3234. [Google Scholar] [CrossRef]
- Knight, C.A.; Molinari, N.; Petrov, D.A. The large genome constraint hypothesis: Evolution, ecology and phenotype. Ann. Bot. 2005, 95, 177–190. [Google Scholar] [CrossRef] [Green Version]
- Pellicer, J.; Hidalgo, O.; Dodsworth, S.; Leitch, I.J. Genome size diversity and its impact on the evolution of land plants. Genes 2018, 9, 88. [Google Scholar] [CrossRef] [Green Version]
- Carev, I.; Ruščić, M.; Skočibušić, M.; Maravić, A.; Siljak-Yakovlev, S.; Politeo, O. Phytochemical and cytogenetic characterization of Centaurea solstitialis (Asteraceae) from Croatia. Chem. Biodivers. 2017, 14, e1600213. [Google Scholar] [CrossRef]
- Cole, I.B.; Cao, J.; Alan, R.A.; Saxena, P.K.; Murch, S.J. Comparisons of Scutellaria baicalensis, Scutellaria lateriflora and Scutellaria racemosa: Genome size, antioxidant potential and phytochemistry. Planta Med. 2008, 74, 474–481. [Google Scholar] [CrossRef]
- Carvalho, Y.G.S.; Vitorino, L.C.; de Souza, U.J.B.; Bessa, L.A. Recent trends in research on the genetic diversity of plants: Implications for conservation. Diversity 2019, 11, 62. [Google Scholar] [CrossRef] [Green Version]
- Wei, S.; Yang, W.; Wang, X.; Hou, Y. High genetic diversity in an endangered medicinal plant, Saussurea involucrata (Saussurea, Asteraceae), in western Tianshan Mountains, China. Conserv. Genet. 2017, 18, 1435–1447. [Google Scholar] [CrossRef]
- Chang, Y.J.; Cao, Y.F.; Zhang, J.M.; Tian, L.M.; Dong, X.G.; Zhang, Y.; Qi, D.; Zhang, X.S. Study on chloroplast DNA diversity of cultivated and wild pears (Pyrus L.) in Northern China. Tree Genet. Genomes 2017, 13, 44. [Google Scholar] [CrossRef]
- Parejo, I.; Viladomat, F.; Bastida, J.; Codina, C. Variation of the arbutin content in different wild populations of Arctostaphylos uva-ursi in Catalonia, Spain. J. Herbs Spices Med. Plants 2002, 9, 329–333. [Google Scholar] [CrossRef]
- Sonnenschein, M.; Tegtmeier, M. Experiments on the domestication of bearberry (Arctostaphylos uva-ursi (L.) Spreng.). J. Med. Spices Plants 2012, 17, 124–128. [Google Scholar]
- Chen, H.Y.; Lin, Y.C.; Hsieh, C.L. Evaluation of antioxidant activity of aqueous extract of some selected nutraceutical herbs. Food Chem. 2008, 104, 1418–1424. [Google Scholar] [CrossRef]
- Del Valle, J.C.; Buide, M.L.; Casimiro-Soriguer, I.; Whittall, J.B.; Narbona, E. On flavonoid accumulation indifferent plant parts: Variation patterns among individuals and populations in the shore campion (Silene littorea). Front. Plant Sci. 2015, 6, 939. [Google Scholar] [CrossRef] [Green Version]
- Martínez-Cadenas, C.; López, S.; Ribas, G.; Flores, C.; García, O.; Sevilla, A.; Smith-Zubiaga, I.; Ibarrola-Villaba, M.; Pino-Yanes, M.M.; Gardeazabal, J.; et al. Simultaneous purifying selection on the ancestral MC1R allele and positive selection on the melanoma-risk allele v60l in south Europeans. Mol. Biol. Evol. 2013, 30, 2654–2665. [Google Scholar] [CrossRef] [Green Version]
- Jakaaola, L.; Hohtola, A. Effect of latitude on flavonoid biosynthesis in plants. Plant Cell Environ. 2010, 33, 1239–1247. [Google Scholar] [CrossRef]
- Martz, F.; Peltola, R.; Fontanay, S.; Duval, R.E.; Julkunen-Tiitto, R.; Stark, S. Effect of latitude and altitude on the terpenoid and soluble phenolic composition of juniper (Juniperus communis) needles and evaluation of their antibacterial activity in the boreal zone. J. Agric. Food Chem. 2009, 57, 9575–9584. [Google Scholar] [CrossRef]
- Yang, B.; Zheng, J.; Laaksonen, O.; Tahvonen, R.; Kallio, H. Effects of latitude and weather conditions on phenolic compounds in currant (Ribes spp.) cultivars. J. Agric. Food Chem. 2013, 61, 3517–3532. [Google Scholar] [CrossRef] [PubMed]
- Lätti, A.K.; Jaakola, L.; Riihinen, K.R.; Kainulainen, P.S. Anthocyanin and flavonol variation in bogbil berries (Vaccinium uliginosum L.) in Finland. J. Agric. Food Chem. 2010, 58, 427–433. [Google Scholar] [CrossRef] [PubMed]
- Lätti, A.K.; Riihinen, K.R.; Kainulainen, P.S. Analysis of anthocyanin variation in wild populations of bilberry (Vaccinium myrtillus L.) in Finland. J. Agric. Food Chem. 2008, 56, 190–196. [Google Scholar] [CrossRef]
- Monschein, M.; Iglesias, J.; Kunert, O.; Bucar, F. Phytochemistry of heather (Calluna vulgaris (L.) Hull) and its altitudinal alteration. Phytochem. Rev. 2010, 9, 205–215. [Google Scholar] [CrossRef] [Green Version]
- Perry, N.B.; Burgess, E.J.; Rodríguez Guitián, M.A.; Romero Franco, R.; López Mosquera, E.; Smallfield, B.M.; Joyce, N.I.; Littlejohn, R.P. Sesquiterpene lactones in Arnica montana: Helenalin and dihydrohelenalin chemotypes in Spain. Planta Med. 2009, 75, 660–666. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Abdala-Roberts, L.; Rasmann, S.; Berny-Mier, Y.; Terán, J.C.; Covelo, F.; Glauser, G.; Moreira, X. Biotic and abiotic factors associated with altitudinal variation in plant traits and herbivory in a dominant oak species. Am. J. Bot. 2016, 103, 2070–2078. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bentley, J.; Moore, J.P.; Farrant, J.M. Metabolomics as a complement to phylogenetics for assessing intraspecific boundaries in the desiccation-tolerant medicinal shrub Myrothamnus flabellifolia (Myrothamnaceae). Phytochemistry 2019, 159, 127–136. [Google Scholar] [CrossRef] [PubMed]
- Siljak-Yakovlev, S.; Pustahija, F.; Šolić, E.M.; Bogunić, F.; Muratović, E.; Bašić, N.; Catrice, O.; Brown, C.S. Towards a database of genome size and chromosome number of Balkan flora: C-values in 343 taxa with novel values for 252. Adv. Sci. Lett. 2010, 3, 190–213. [Google Scholar] [CrossRef]
- Garrido, J.L.; Alcántara, J.M.; Rey, P.J.; Medrano, M.; Guitián, J.; Castellanos, M.C.; Bastida, J.M.; Jaime, R.; Herrera, C.M. Geographic genetic structure of Iberian columbines (gen. Aquilegia). Plant Syst. Evol. 2017, 303, 1145–1160. [Google Scholar] [CrossRef]
- Listl, D.; Poschlod, P.; Reisch, C. Phylogeography of a tough rock survivor in European dry grasslands. PLoS ONE 2017, 12, e0179961. [Google Scholar] [CrossRef]
- Olalde, M.; Herrán, A.; Espinel, S.; Goicoechea, P. White oaks phylogeography in the Iberian Peninsula. Ecol. Manag. 2002, 156, 89–102. [Google Scholar] [CrossRef] [Green Version]
- Barrett, S.C.H. Influences of clonality on plant sexual reproduction. Proc. Natl. Acad. Sci. USA 2015, 112, 8859–8866. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Skowyra, M.; Falguera, V.; Gallego, G.; Peiro, S.; Almajano, M.P. The effect of Perilla frutescens extract on the oxidative stability of model food emulsions. Antioxidants 2014, 3, 38–54. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pascoal, A.; Quirantes-Piné, R.; Fernando, A.L.; Alexopoulou, E.; Segura-Carretero, A. Phenolic composition and antioxidant activity of kenaf leaves. Ind. Crop. Prod. 2015, 78, 116–123. [Google Scholar] [CrossRef]
- Pellicer, J.; Garnatje, T.; Molero, J.; Pustahija, F.; Siljak-Yakovlev, S.; Vallès, J. Origin and evolution of South American Artemisia species (Asteraceae); evidence from molecular phylogeny, ribosomal DNA and genome size data. Aust. J. Bot. 2010, 58, 605–616. [Google Scholar] [CrossRef]
- Doležel, J. Flow cytometric analysis of nuclear DNA content in higher plants. Phytochem. Anal. 1991, 2, 143–154. [Google Scholar] [CrossRef]
- Doyle, J.J.; Doyle, J.L. A rapid DNA isolation procedure for small quantities of fresh leaf tissue. Phytochem. Bull. Bot. Soc. Am. 1987, 19, 11–15. [Google Scholar]
- Vitales, D.; Feliner, G.N.; Vallès, J.; Garnatje, T.; Firat, M.; Álvarez, I. A new circumscription of the Mediterranean genus Anacyclus (Anthemideae, Asteraceae) based on plastid and nuclear DNA markers. Phytotaxa 2018, 349, 1–17. [Google Scholar] [CrossRef]
- Hall, T.A. BioEdit: A user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symp. Ser. 1999, 41, 95–98. [Google Scholar]
- Thompson, J.D.; Higgins, D.G.; Gibson, T.J. CLUSTAL W: Improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994, 22, 4673–4680. [Google Scholar] [CrossRef] [Green Version]
- Rozas, J.; Ferrer-Mata, A.; Sánchez-DelBarrio, J.C.; Guirao-Rico, S.; Librado, P.; Ramos-Onsins, S.E.; Sánchez-Gracia, A. DnaSP 6: DNA Sequence Polymorphism Analysis of Large Datasets. Mol. Biol. Evol. 2017, 34, 3299–3302. [Google Scholar] [CrossRef] [PubMed]
- Borchsenius, F. FastGap, Version 1.2; Department of Biosciences, Aarhus University: Aarhus, Denmark, 2009; Available online: http://www.aubot.dk/FastGap_home.htm (accessed on 1 July 2019).
- Clement, M.; Posada, D.C.K.A.; Crandall, K.A. TCS: A computer program to estimate gene genealogies. Mol. Ecol. 2000, 9, 1657–1659. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Leigh, J.W.; Bryant, D. Popart: Full-feature software for haplotype network construction. Methods Ecol. Evol. 2015, 6, 1110–1116. [Google Scholar] [CrossRef]
- Fox, J. Getting started with the R commander: A basic-statistics graphical user interface to R. J. Stat. Softw. 2005, 14, 1–42. [Google Scholar] [CrossRef] [Green Version]
- Oksanen, J.; Blanchet, F.G.; Kindt, R.; Legendre, P.; O’hara, R.B.; Simpson, G.L.; Solymos, P.; Stevens, M.; Wagner, H. Vegan: Community Ecology Package. R Package Version 1.17-4. 2010. Available online: http://CRAN.R-project.org/package=vegan (accessed on 19 September 2010).
- Malinowska, H. The selects of bearberry (Arctostaphylos uva-ursi (L.) Spreng.) from natural populations in Poland. Acta Soc. Bot. Pol. 1995, 64, 91–96. [Google Scholar] [CrossRef] [Green Version]
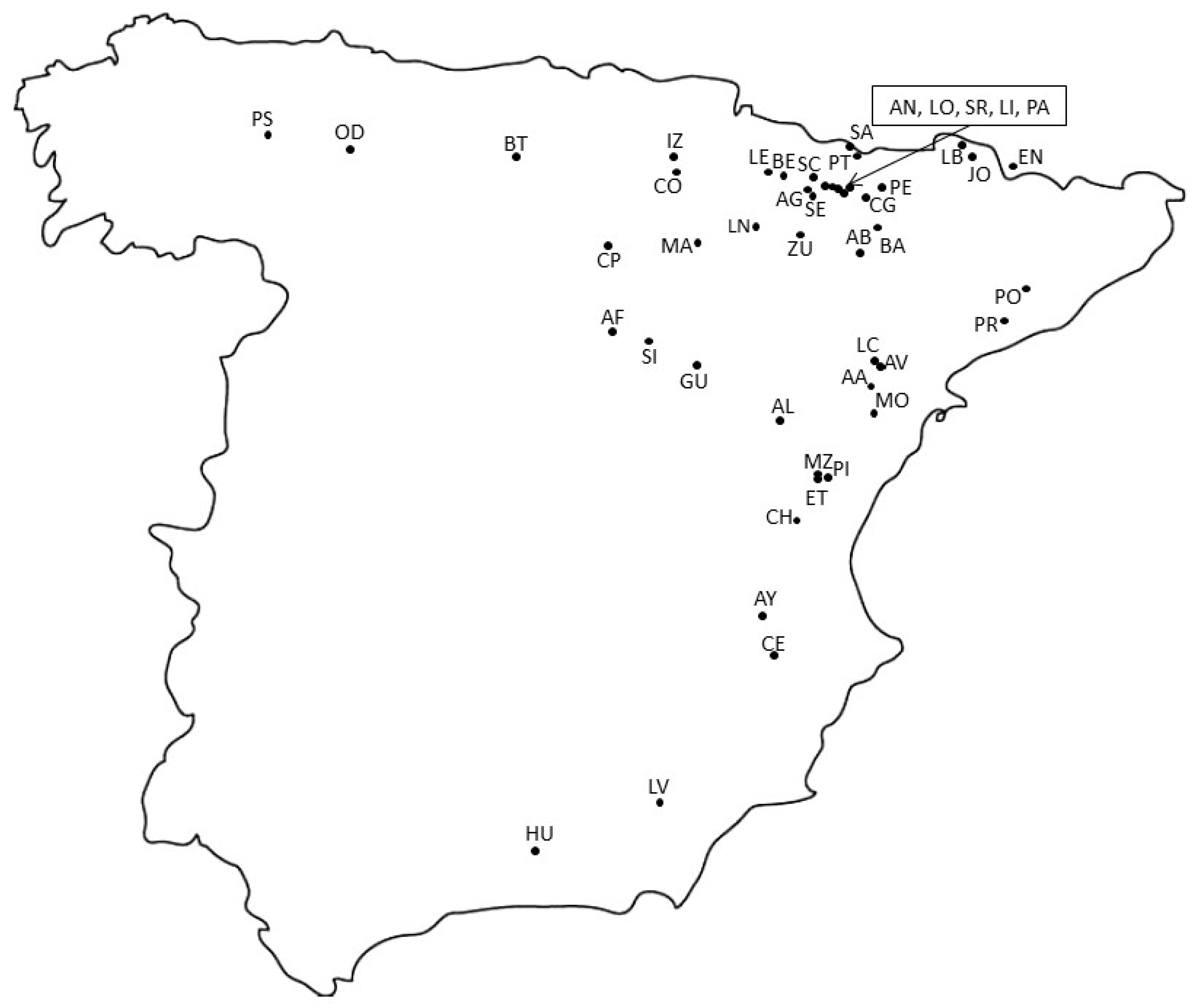


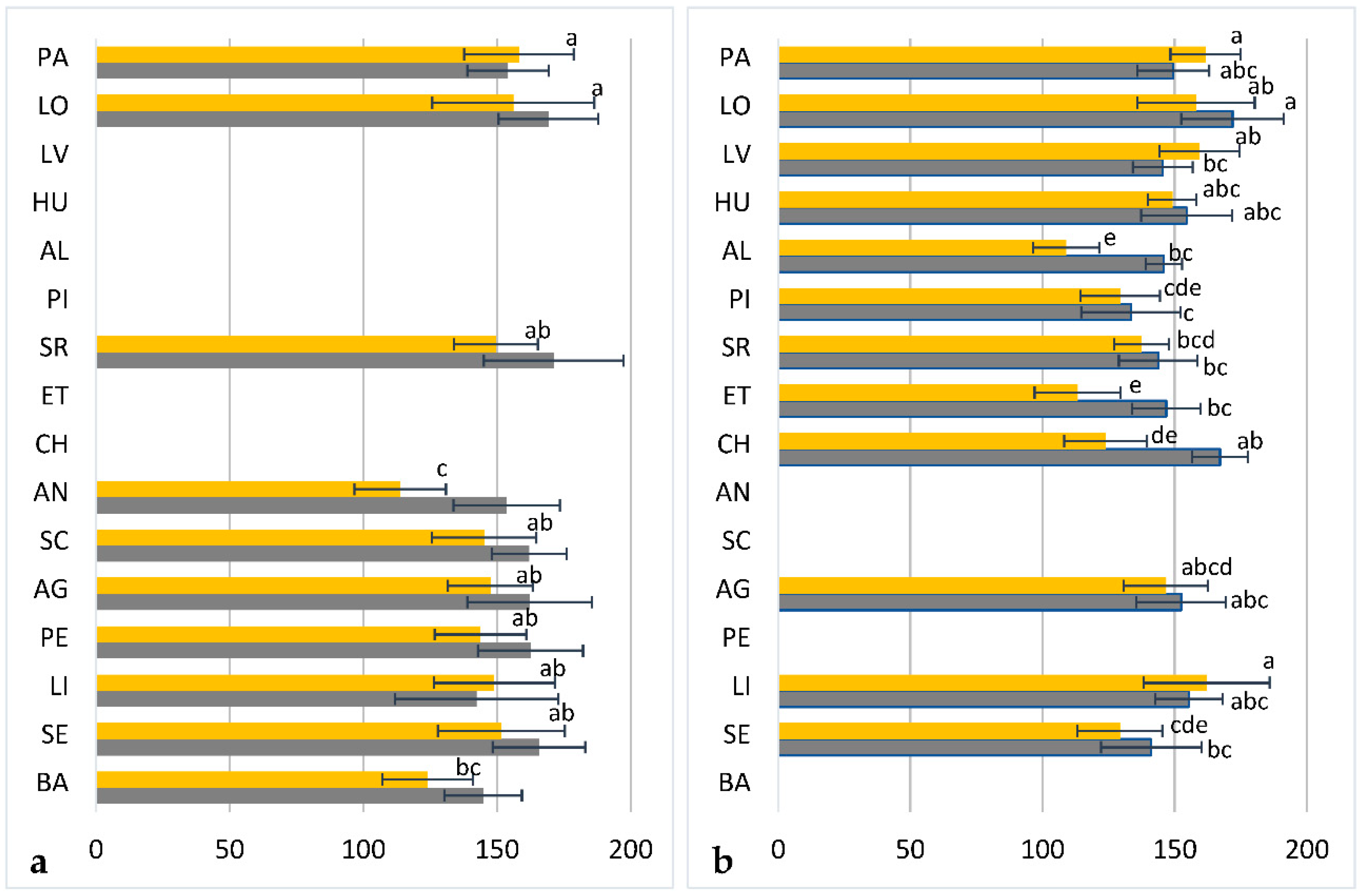

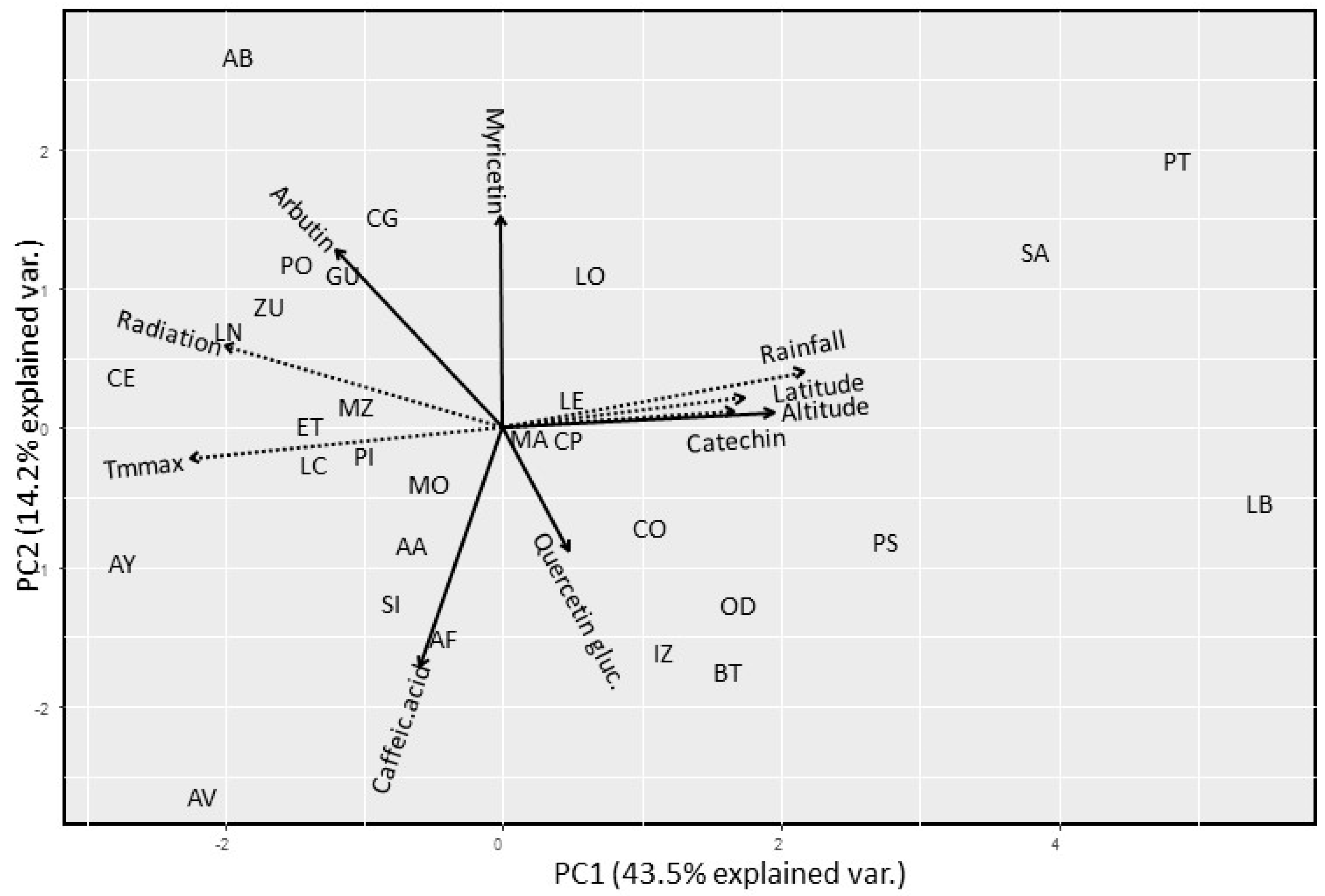

| Population Code | N (GS) | GS ± SD | N (H) | Haplotypes |
|---|---|---|---|---|
| AA | 5 | 2.66 ± 0.28 | 3 | H1 |
| AF | 5 | 2.87 ± 0.10 | 3 | H1 |
| AL | 5 | 2.53 ± 0.08 | 3 | H1, H2 |
| AV | 1 | 2.87 | - | - |
| AY | 5 | 2.66 ± 0.07 | 3 | H1 |
| BE | 5 | 2.70 ± 0.05 | 3 | H1, H3 |
| BT | 5 | 2.92 ± 0.08 | 3 | H1 |
| CE | 5 | 2.89 ± 0.27 | 3 | H1, H2, H5 |
| CH | 5 | 2.85 ± 0.10 | 3 | H1 |
| CO | 6 | 2.79 ± 0.09 | 3 | H1, H2, H6 |
| CP | 6 | 2.87 ± 0.09 | 3 | H1 |
| EN | 5 | 2.67 ± 0.09 | 3 | H1 |
| ET | 5 | 2.81 ± 0.12 | 3 | H1, H2 |
| GU | 5 | 2.83 ± 0.06 | 3 | H1, H2 |
| HU | 5 | 2.74 ± 0.04 | 3 | H2 |
| IZ | 5 | 2.78 ± 0.03 | 3 | H1 |
| JO | 5 | 2.61 ± 0.16 | 3 | H1, H2 |
| LB | 5 | 2.76 ± 0.07 | 3 | H1, H3, H4 |
| LC | 5 | 2.74 ± 0.25 | 3 | H1 |
| LE | 5 | 2.50 ± 0.16 | 3 | H1 |
| LN | 5 | 3.09 ± 0.35 | 3 | H1 |
| LO | 5 | 2.81 ± 0.11 | 3 | H1 |
| LV | 5 | 2.68 ± 0.11 | 3 | H2 |
| MA | 6 | 2.86 ± 0.05 | 3 | H1, H7 |
| MO | 1 | 2.83 | - | - |
| MZ | 5 | 2.87 ± 0.17 | 3 | H1, H2, H8 |
| OD | 5 | 2.82 ± 0.10 | 3 | H1 |
| PI | 5 | 2.86 ± 0.12 | 3 | H1, H8 |
| PO | 5 | 2.88 ± 0.35 | 3 | H1 |
| PR | 5 | 2.79 ± 0.18 | 3 | H2 |
| PS | 5 | 2.82 ± 0.08 | 3 | H1, H6 |
| PT | 5 | 2.72 ± 0.18 | 3 | H1, H4, H9 |
| SA | 2 | 3.13 ± 0.53 | 3 | H1, H4 |
| SE | 5 | 2.53 ± 0.11 | 3 | H1 |
| SI | 5 | 3.15 ± 0.33 | 3 | H1, H10 |
| SR | 5 | 2.66 ± 0.07 | 3 | H1 |
| ZU | 6 | 2.84 ± 0.19 | 3 | H1 |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Asensio, E.; Vitales, D.; Pérez, I.; Peralba, L.; Viruel, J.; Montaner, C.; Vallès, J.; Garnatje, T.; Sales, E. Phenolic Compounds Content and Genetic Diversity at Population Level across the Natural Distribution Range of Bearberry (Arctostaphylos uva-ursi, Ericaceae) in the Iberian Peninsula. Plants 2020, 9, 1250. https://doi.org/10.3390/plants9091250
Asensio E, Vitales D, Pérez I, Peralba L, Viruel J, Montaner C, Vallès J, Garnatje T, Sales E. Phenolic Compounds Content and Genetic Diversity at Population Level across the Natural Distribution Range of Bearberry (Arctostaphylos uva-ursi, Ericaceae) in the Iberian Peninsula. Plants. 2020; 9(9):1250. https://doi.org/10.3390/plants9091250
Chicago/Turabian StyleAsensio, Esther, Daniel Vitales, Iván Pérez, Laia Peralba, Juan Viruel, Celia Montaner, Joan Vallès, Teresa Garnatje, and Ester Sales. 2020. "Phenolic Compounds Content and Genetic Diversity at Population Level across the Natural Distribution Range of Bearberry (Arctostaphylos uva-ursi, Ericaceae) in the Iberian Peninsula" Plants 9, no. 9: 1250. https://doi.org/10.3390/plants9091250
APA StyleAsensio, E., Vitales, D., Pérez, I., Peralba, L., Viruel, J., Montaner, C., Vallès, J., Garnatje, T., & Sales, E. (2020). Phenolic Compounds Content and Genetic Diversity at Population Level across the Natural Distribution Range of Bearberry (Arctostaphylos uva-ursi, Ericaceae) in the Iberian Peninsula. Plants, 9(9), 1250. https://doi.org/10.3390/plants9091250