Plasma Proteomics Elucidated a Protein Signature in COVID-19 Patients with Comorbidities and Early-Diagnosis Biomarkers
Abstract
1. Introduction
2. Materials and Methods
2.1. Study Cohort
2.2. Sample Preparation for Mass Spectrometry
2.3. Mass Spectrometry Analysis
2.4. Mass Spectrometry Data Analysis
2.5. Proteomics Data and Statistical Analysis
3. Results
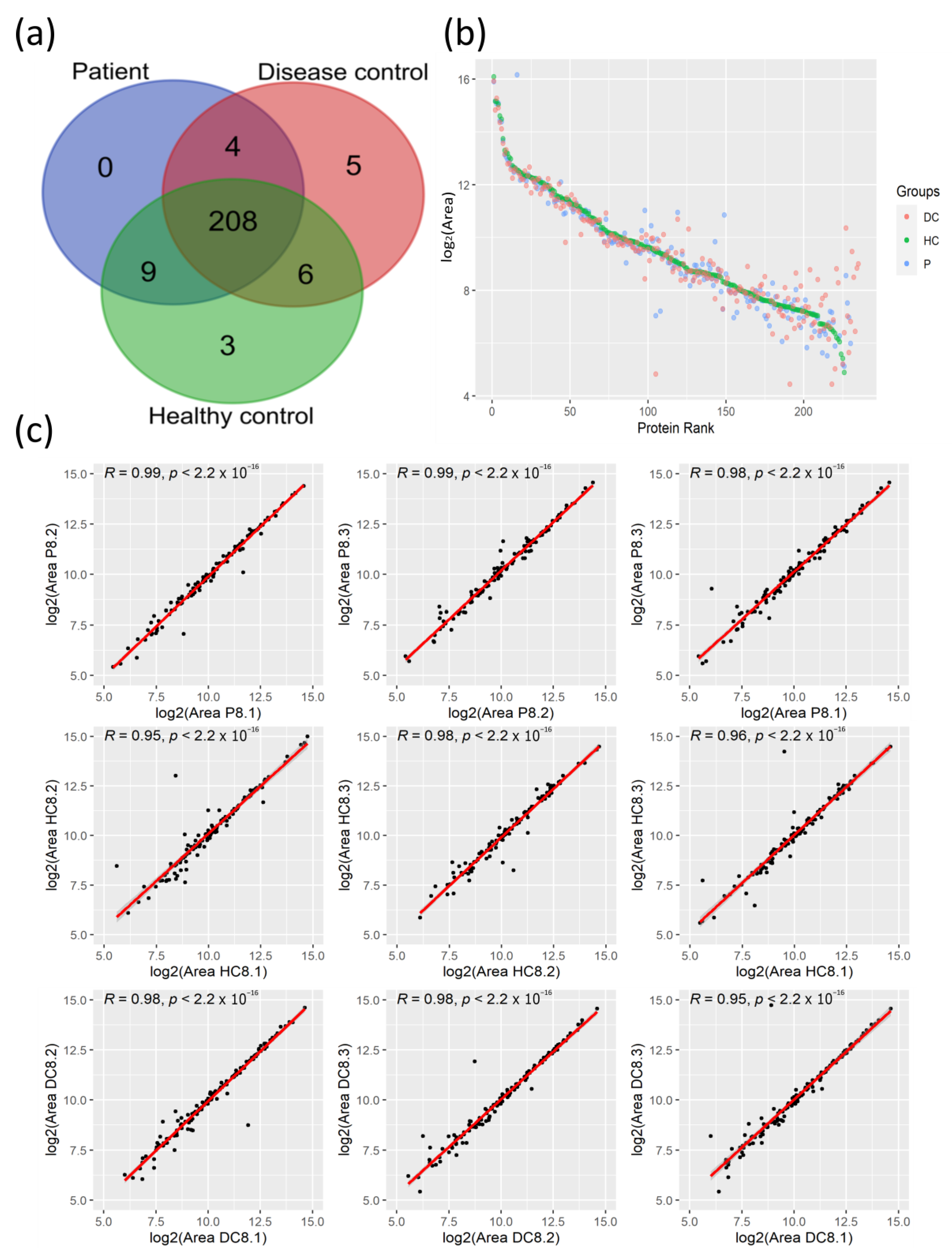
3.1. Proteomic Profiles of Plasma from COVID-19 Patients and Their Controls
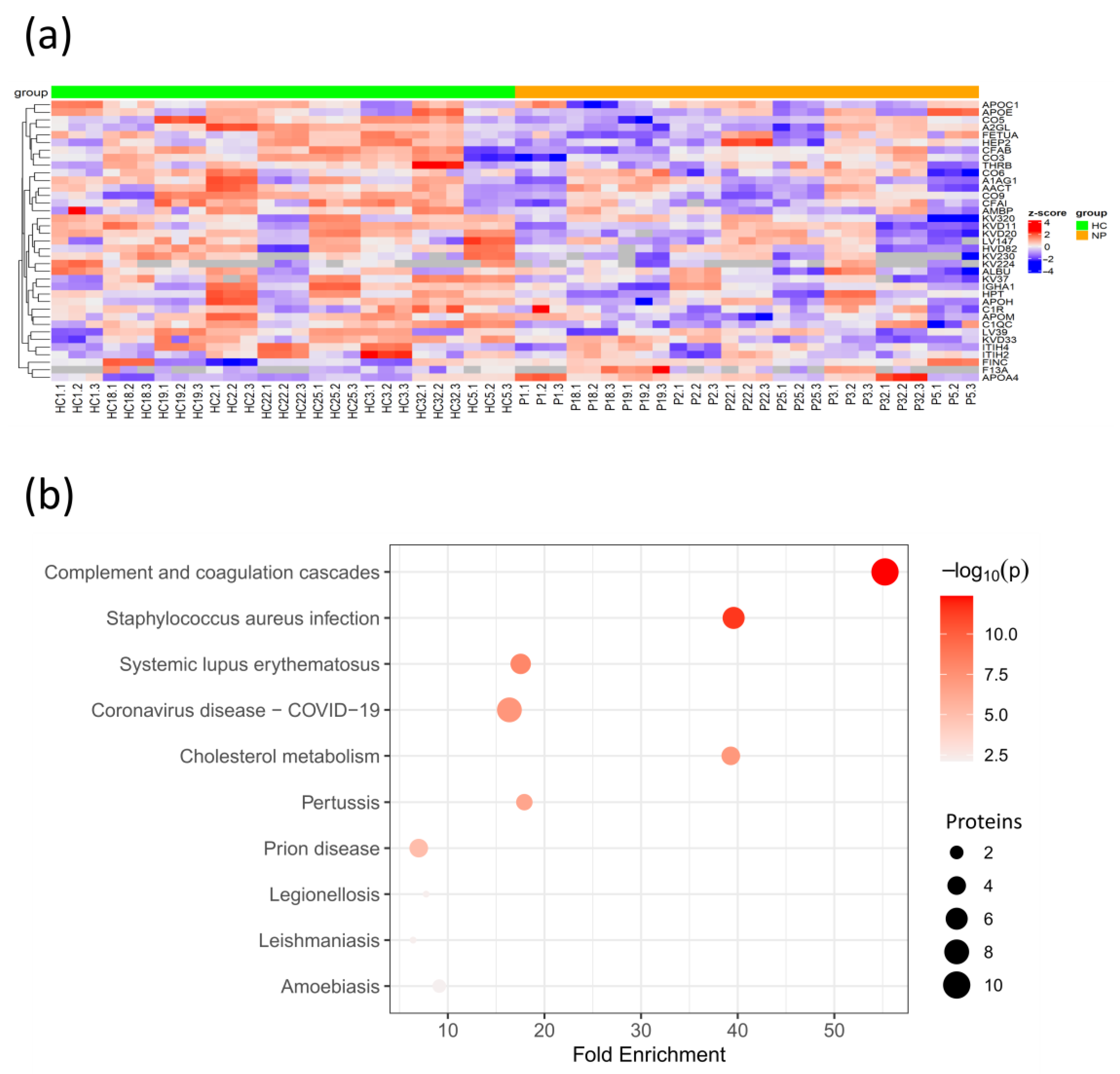
3.2. COVID-19 Patients with Comorbidities Share a Common Plasma Protein Signature
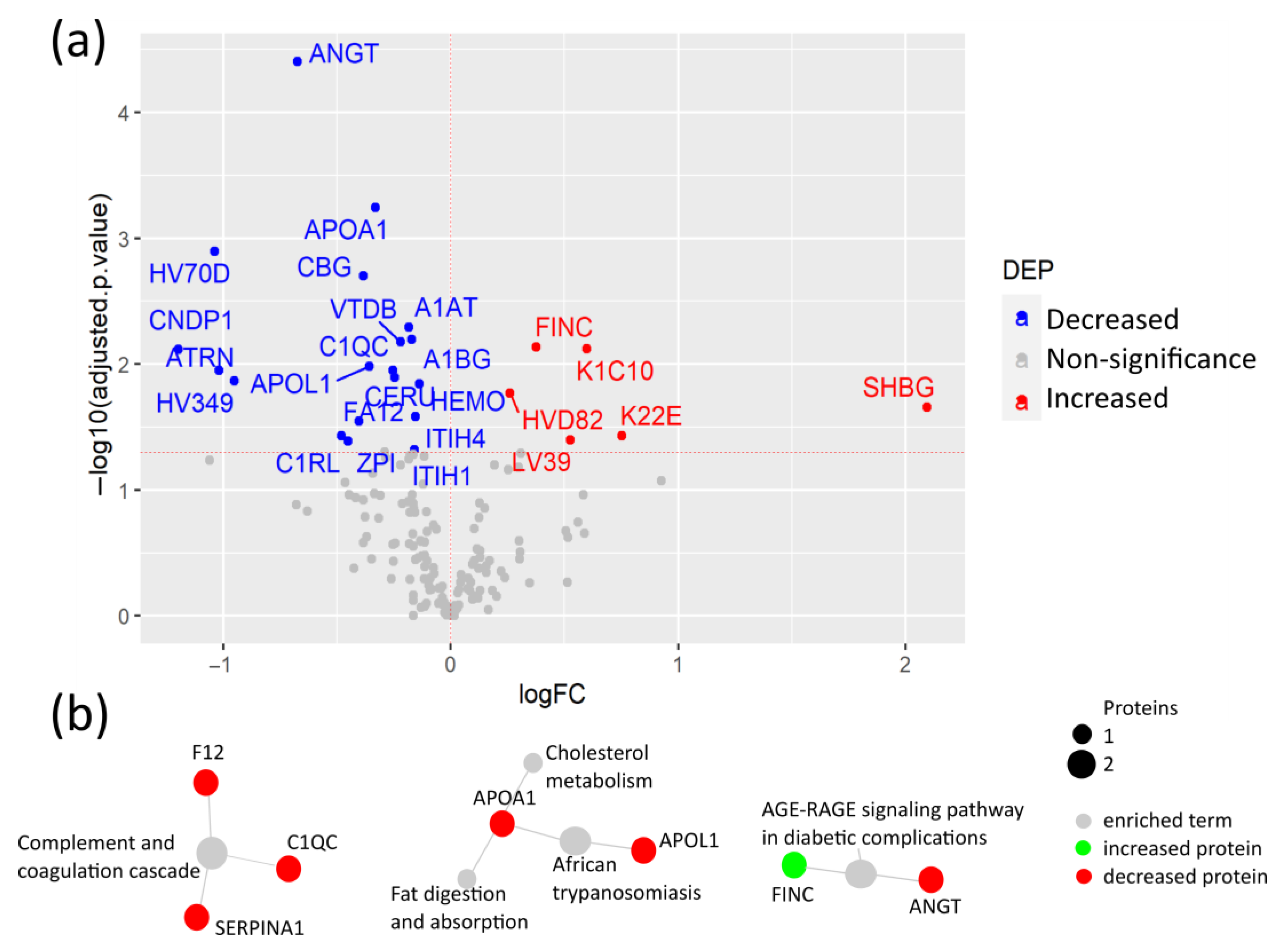
3.3. Coagulation and Cholesterol Metabolism Are Altered in COVID-19 Patients without Comorbidities
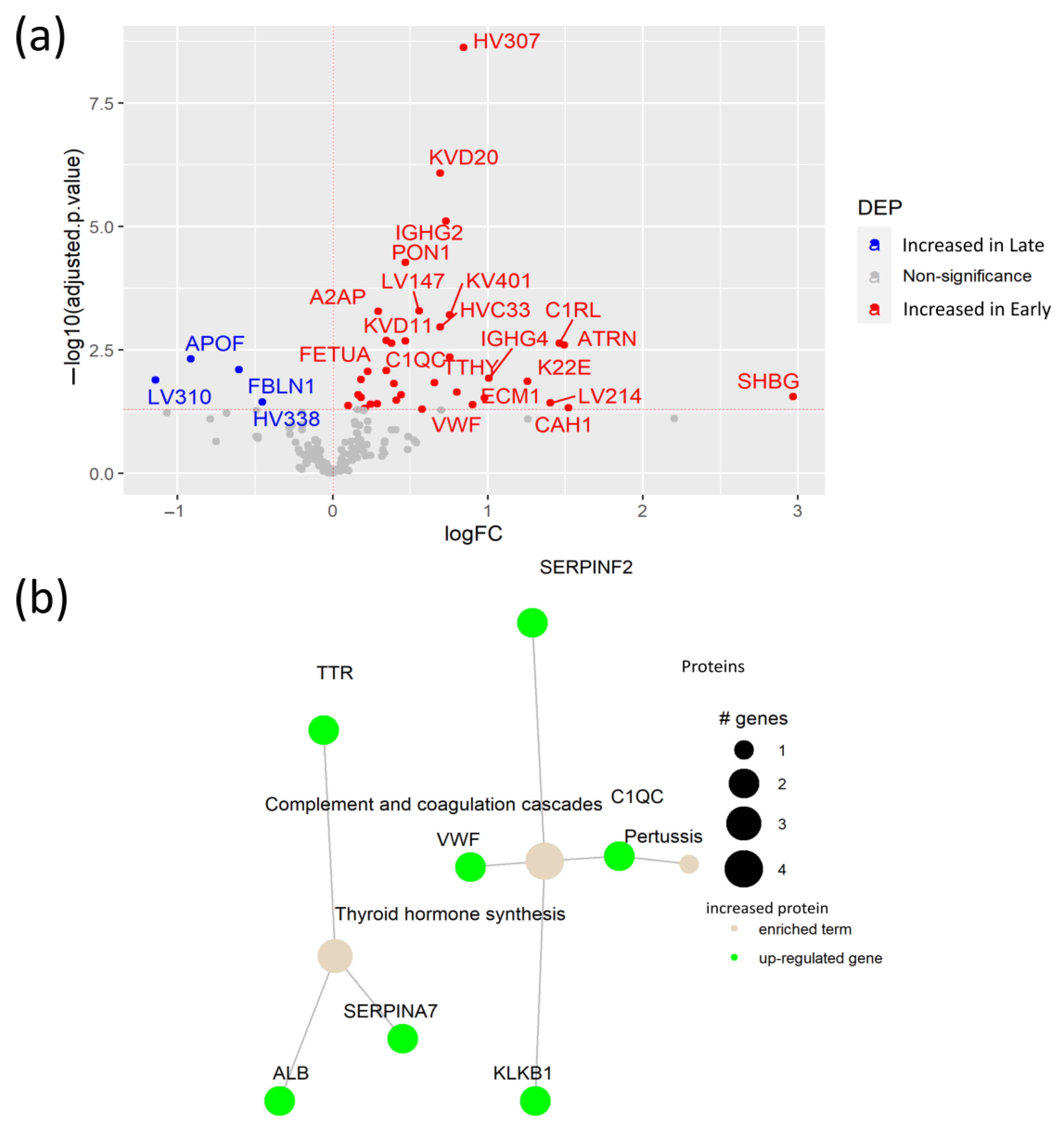
3.4. Early SARS-CoV-2 Infection Is Associated with Immune Protein Changes and Tissue Remodeling
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Parasher, A. COVID-19: Current understanding of its Pathophysiology, Clinical presentation and Treatment. Postgrad. Med. J. 2021, 97, 312–320. [Google Scholar] [CrossRef] [PubMed]
- Merad, M.; Blish, C.A.; Sallusto, F.; Iwasaki, A. The immunology and immunopathology of COVID-19. Science 2022, 375, 1122–1127. [Google Scholar] [CrossRef] [PubMed]
- Ejaz, H.; Alsrhani, A.; Zafar, A.; Javed, H.; Junaid, K.; Abdalla, A.E.; Abosalif, K.O.A.; Ahmed, Z.; Younas, S. COVID-19 and comorbidities: Deleterious impact on infected patients. J. Infect. Public Health 2020, 13, 1833–1839. [Google Scholar] [CrossRef] [PubMed]
- Belchior-Bezerra, M.; Lima, R.S.; Medeiros, N.I.; Gomes, J.A.S. COVID-19, obesity, and immune response 2 years after the pandemic: A timeline of scientific advances. Obes. Rev. 2022, 23, e13496. [Google Scholar] [CrossRef] [PubMed]
- Yu, S.; Li, X.; Xin, Z.; Sun, L.; Shi, J. Proteomic insights into SARS-CoV-2 infection mechanisms, diagnosis, therapies and prognostic monitoring methods. Front. Immunol. 2022, 13, 923387. [Google Scholar] [CrossRef] [PubMed]
- Wiśniewski, J.R. Filter Aided Sample Preparation—A tutorial. Anal. Chim. Acta 2019, 1090, 23–30. [Google Scholar] [CrossRef] [PubMed]
- Rappsilber, J.; Mann, M.; Ishihama, Y. Protocol for micro-purification, enrichment, pre-fractionation and storage of peptides for proteomics using StageTips. Nat. Protoc. 2007, 2, 1896–1906. [Google Scholar] [CrossRef] [PubMed]
- R Core Team. A Language and Environment for Statistical Computing; R Foundation for Statistical Computing: Vienna, Austria, 2022; Available online: https://www.R-project.org/ (accessed on 12 January 2024).
- RStudio Team. RStudio: Integrated Development Environment for R; RStudio, PBC: Boston, MA, USA, 2022; Available online: http://www.rstudio.com/ (accessed on 12 January 2024).
- Sticker, A.; Goeminne, L.; Martens, L.; Clement, L. Robust Summarization and Inference in Proteome-wide Label-free Quantification. Mol. Cell. Proteom. 2020, 19, 1209–1219. [Google Scholar] [CrossRef] [PubMed]
- Benjamini, Y.; Hochberg, Y. Controlling the False Discovery Rate: A Practical and Powerful Approach to Multiple Testing. J. R. Stat. Soc. Ser. B 1995, 57, 289–300. [Google Scholar] [CrossRef]
- Ulgen, E.; Ozisik, O.; Sezerman, O.U. PathfindR: An R package for comprehensive identification of enriched pathways in omics data through active subnetworks. Front. Genet. 2019, 10, 858. [Google Scholar] [CrossRef]
- Overmyer, K.A.; Shishkova, E.; Miller, I.J.; Balnis, J.; Bernstein, M.N.; Peters-Clarke, T.M.; Meyer, J.G.; Quan, Q.; Muehlbauer, L.K.; Trujillo, E.A.; et al. Large-Scale Multi-omic Analysis of COVID-19 Severity. Cell Syst. 2021, 12, 23–40.e7. [Google Scholar] [CrossRef] [PubMed]
- Huang, J.-J.; Wang, C.-W.; Liu, Y.; Zhang, Y.-Y.; Yang, N.-B.; Yu, Y.-C.; Jiang, Q.; Song, Q.-F.; Qian, G.-Q. Role of the extracellular matrix in COVID-19. World J. Clin. Cases 2023, 11, 73–83. [Google Scholar] [CrossRef] [PubMed]
- Yeung, M.L.; Teng, J.L.L.; Jia, L.; Zhang, C.; Huang, C.; Cai, J.-P.; Zhou, R.; Chan, K.-H.; Zhao, H.; Zhu, L.; et al. Soluble ACE2-mediated cell entry of SARS-CoV-2 via interaction with proteins related to the renin-angiotensin system. Cell 2021, 184, 2212–2228.e12. [Google Scholar] [CrossRef] [PubMed]
- Razanakolona, M.; Adam, F.; Bianchini, E.; Saller, F.; de Carvalho, A.; Diehl, J.-L.; Denis, C.V.; Meziani, F.; Borgel, D.; Helms, J.; et al. Anti-inflammatory Activity of the Protein Z-Dependent Protease Inhibitor. TH Open 2021, 05, e220–e229. [Google Scholar] [CrossRef]
- Hill, L.A.; Bodnar, T.S.; Weinberg, J.; Hammond, G.L. Corticosteroid-Binding Globulin is a biomarker of inflammation onset and severity in female rats. J. Endocrinol. 2016, 230, 215–225. [Google Scholar] [CrossRef] [PubMed]
- Hilser, J.R.; Han, Y.; Biswas, S.; Gukasyan, J.; Cai, Z.; Zhu, R.; Tang, W.H.W.; Deb, A.; Lusis, A.J.; Hartiala, J.A.; et al. Association of serum HDL-cholesterol and apolipoprotein A1 levels with risk of severe SARS-CoV-2 infection. J. Lipid Res. 2021, 62, 100061. [Google Scholar] [CrossRef] [PubMed]
- Wu, J.; Ma, Z.; Raman, A.; Beckerman, P.; Dhillon, P.; Mukhi, D.; Palmer, M.; Chen, H.C.; Cohen, C.R.; Dunn, T.; et al. APOL1 risk variants in individuals of African genetic ancestry drive endothelial cell defects that exacerbate sepsis. Immunity 2021, 54, 2632–2649.e6. [Google Scholar] [CrossRef] [PubMed]
- Oczypok, E.A.; Perkins, T.N.; Oury, T.D. All the “RAGE” in lung disease: The receptor for advanced glycation endproducts (RAGE) is a major mediator of pulmonary inflammatory responses. Paediatr. Respir. Rev. 2017, 23, 40–49. [Google Scholar] [CrossRef] [PubMed]
- Bierhaus, A.; Schiekofer, S.; Schwaninger, M.; Andrassy, M.; Humpert, P.M.; Chen, J.; Hong, M.; Luther, T.; Henle, T.; Klöting, I.; et al. Diabetes-associated sustained activation of the transcription factor nuclear factor-κB. Diabetes 2001, 50, 2792–2808. [Google Scholar] [CrossRef]
- Cavezzi, A.; Troiani, E.; Corrao, S. COVID-19: Hemoglobin, iron, and hypoxia beyond inflammation. A narrative review. Clin. Pract. 2020, 10, 1271. [Google Scholar] [CrossRef]
- Camps, J.; Iftimie, S.; García-Heredia, A.; Castro, A.; Joven, J. Paraoxonases and infectious diseases. Clin. Biochem. 2017, 50, 804–811. [Google Scholar] [CrossRef] [PubMed]
- Schaller, J.; Gerber, S.S. The plasmin–antiplasmin system: Structural and functional aspects. Cell. Mol. Life Sci. 2011, 68, 785–801. [Google Scholar] [CrossRef] [PubMed]
- Java, A.; Apicelli, A.J.; Kathryn Liszewski, M.; Coler-Reilly, A.; Atkinson, J.P.; Kim, A.H.; Kulkarni, H.S. The complement system in COVID-19: Friend and foe? JCI Insight 2020, 5, e14071. [Google Scholar] [CrossRef] [PubMed]
- Henry, B.M.; Szergyuk, I.; de Oliveira, M.H.S.; Lippi, G.; Benoit, J.L.; Vikse, J.; Benoit, S.W. Complement levels at admission as a reflection of coronavirus disease 2019 (COVID-19) severity state. J. Med. Virol. 2021, 93, 5515–5522. [Google Scholar] [CrossRef] [PubMed]
- Marfia, G.; Navone, S.; Guarnaccia, L.; Campanella, R.; Mondoni, M.; Locatelli, M.; Barassi, A.; Fontana, L.; Palumbo, F.; Garzia, E.; et al. Decreased serum level of sphingosine-1-phosphate: A novel predictor of clinical severity in COVID-19. EMBO Mol. Med. 2021, 13, e13424. [Google Scholar] [CrossRef]
- Kurt, N.; Ozgeris, F.B.; Kocak, O.F.; Yuce, N.; Bayraktutan, Z.; Parlak, E.; Coban, T.A.; Bakan, E. Evaluation of fetuin-A, CRP, and CRP/fetuin-A values in COVID-19 patients. Int. J. Med. Biochem. 2022, 5, 125–131. [Google Scholar] [CrossRef]
- Corneo, E.; Garbelotto, R.; Prestes, G.; Girardi, C.S.; Santos, L.; Moreira, J.C.F.; Gelain, D.P.; Westphal, G.A.; Kupek, E.; Walz, R.; et al. Coagulation biomarkers and coronavirus disease 2019 phenotyping: A prospective cohort study. Thromb. J. 2023, 21, 80. [Google Scholar] [CrossRef] [PubMed]
- Lee, J.-S.; Han, D.; Kim, S.Y.; Hong, K.H.; Jang, M.-J.; Kim, M.J.; Kim, Y.-G.; Park, J.H.; Cho, S.I.; Park, W.B.; et al. Longitudinal proteomic profiling provides insights into host response and proteome dynamics in COVID-19 progression. Proteomics 2021, 21, e2000278. [Google Scholar] [CrossRef] [PubMed]
- Feingold, K.R. The bidirectional interaction of COVID-19 infections and lipoproteins. Best Pract. Res. Clin. Endocrinol. Metab. 2023, 37, 101751. [Google Scholar] [CrossRef] [PubMed]
- Duke-Cohan, J.S.; Gu, J.; Mclaughlin, D.F.; Xu, Y.; Freeman, G.J.; Schlossman, S.F. Attractin (DPPT-L), a member of the CUB family of cell adhesion and guidance proteins, is secreted by activated human T lymphocytes and modulates immune cell interactions. Proc. Natl. Acad. Sci. USA 1998, 95, 11336–11341. [Google Scholar] [CrossRef]
- He, L.; Gu, W.; Wang, M.; Chang, X.; Sun, X.; Zhang, Y.; Lin, X.; Yan, C.; Fan, W.; Su, P.; et al. Extracellular matrix protein 1 promotes follicular helper T cell differentiation and antibody production. Proc. Natl. Acad. Sci. USA 2018, 115, 8621–8626. [Google Scholar] [CrossRef] [PubMed]
- Rostami, M.; Mansouritorghabeh, H.; Parsa-Kondelaji, M. High levels of Von Willebrand factor markers in COVID-19: A systematic review and meta-analysis. Clin. Exp. Med. 2022, 22, 347–357. [Google Scholar] [CrossRef] [PubMed]
- Shu, T.; Ning, W.; Wu, D.; Xu, J.; Han, Q.; Huang, M.; Zou, X.; Yang, Q.; Yuan, Y.; Bie, Y.; et al. Plasma Proteomics Identify Biomarkers and Pathogenesis of COVID-19. Immunity 2020, 53, 1108–1122.e5. [Google Scholar] [CrossRef] [PubMed]
- D’Alessandro, A.; Thomas, T.; Dzieciatkowska, M.; Hill, R.C.; Francis, R.O.; Hudson, K.E.; Zimring, J.C.; Hod, E.A.; Spitalnik, S.L.; Hansen, K.C. Serum Proteomics in COVID-19 Patients: Altered Coagulation and Complement Status as a Function of IL-6 Level. J. Proteome Res. 2020, 19, 4417–4427. [Google Scholar] [CrossRef] [PubMed]
- Geyer, P.E.; Arend, F.M.; Doll, S.; Louiset, M.-L.; Virreira Winter, S.; Müller-Reif, J.B.; Torun, F.M.; Weigand, M.; Eichhorn, P.; Bruegel, M.; et al. High-resolution serum proteome trajectories in COVID-19 reveal patient-specific seroconversion. EMBO Mol. Med. 2021, 13, e14167. [Google Scholar] [CrossRef] [PubMed]
- Filbin, M.R.; Mehta, A.; Schneider, A.M.; Kays, K.R.; Guess, J.R.; Gentili, M.; Fenyves, B.G.; Charland, N.C.; Gonye, A.L.K.; Gushterova, I.; et al. Longitudinal proteomic analysis of severe COVID-19 reveals survival-associated signatures, tissue-specific cell death, and cell-cell interactions. Cell Rep. Med. 2021, 2, 100287. [Google Scholar] [CrossRef]
- Keur, N.; Saridaki, M.; Ricaño-Ponce, I.; Netea, M.G.; Giamarellos-Bourboulis, E.J.; Kumar, V. Analysis of inflammatory protein profiles in the circulation of COVID-19 patients identifies patients with severe disease phenotypes. Respir. Med. 2023, 217, 107331. [Google Scholar] [CrossRef] [PubMed]
- Rahnavard, A.; Mann, B.; Giri, A.; Chatterjee, R.; Crandall, K.A. Metabolite, protein, and tissue dysfunction associated with COVID-19 disease severity. Sci. Rep. 2022, 12, 12204. [Google Scholar] [CrossRef] [PubMed]
- Zhang, N.; Wang, S.; Wong, C.C.L. Proteomics research of SARS-CoV-2 and COVID-19 disease. Med. Rev. 2022, 2, 427–445. [Google Scholar] [CrossRef] [PubMed]
- Alosaimi, B.; Mubarak, A.; Hamed, M.E.; Almutairi, A.Z.; Alrashed, A.A.; AlJuryyan, A.; Enani, M.; Alenzi, F.Q.; Alturaiki, W. Complement Anaphylatoxins and Inflammatory Cytokines as Prognostic Markers for COVID-19 Severity and In-Hospital Mortality. Front. Immunol. 2021, 12, 668725. [Google Scholar] [CrossRef] [PubMed]
- Risitano, A.M.; Mastellos, D.C.; Huber-Lang, M.; Yancopoulou, D.; Garlanda, C.; Ciceri, F.; Lambris, J.D. Complement as a target in COVID-19? Nat. Rev. Immunol. 2020, 20, 343–344. [Google Scholar] [CrossRef]
- Pretorius, E.; Vlok, M.; Venter, C.; Bezuidenhout, J.A.; Laubscher, G.J.; Steenkamp, J.; Kell, D.B. Persistent clotting protein pathology in Long COVID/Post-Acute Sequelae of COVID-19 (PASC) is accompanied by increased levels of antiplasmin. Cardiovasc. Diabetol. 2021, 20, 172. [Google Scholar] [CrossRef] [PubMed]
- Sierko, E.; Zabrocka, E.; Ostrowska-Cichocka, K.; Tokajuk, P.; Zimnoch, L.; Wojtukiewicz, M.Z. Co-localization of Coagulation Factor X and its Inhibitory System, PZ/ZPI, in Human Endometrial Cancer Tissue. In Vivo 2019, 33, 771–776. [Google Scholar] [CrossRef] [PubMed]
- Toomer, K.H.; Gerber, G.F.; Zhang, Y.; Daou, L.; Tushek, M.; Hooper, J.E.; Francischetti, I.M.B. SARS-CoV-2 infection results in upregulation of Plasminogen Activator Inhibitor-1 and Neuroserpin in the lungs, and an increase in fibrinolysis inhibitors associated with disease severity. eJHaem 2023, 4, 324–338. [Google Scholar] [CrossRef] [PubMed]
- van Lier, D.; Kox, M.; Pickkers, P. Commentary: Plasma angiotensin II is increased in critical coronavirus disease 2019. Front. Cardiovasc. Med. 2022, 9, 1012452. [Google Scholar] [CrossRef] [PubMed]
- van Lier, D.; Kox, M.; Santos, K.; van der Hoeven, H.; Pillay, J.; Pickkers, P. Increased blood angiotensin converting enzyme 2 activity in critically ill COVID-19 patients. ERJ Open Res. 2021, 7, 00848-2020. [Google Scholar] [CrossRef] [PubMed]
- Urbiola-Salvador, V.; Lima de Souza, S.; Grešner, P.; Qureshi, T.; Chen, Z. Plasma Proteomics Unveil Novel Immune Signatures and Biomarkers upon SARS-CoV-2 Infection. Int. J. Mol. Sci. 2023, 24, 6276. [Google Scholar] [CrossRef] [PubMed]
- Hoch, N.E.; Guzik, T.J.; Chen, W.; Deans, T.; Maalouf, S.A.; Gratze, P.; Weyand, C.; Harrison, D.G. Regulation of T-cell function by endogenously produced angiotensin II. Am. J. Physiol. Regul. Integr. Comp. Physiol. 2009, 296, R208–R216. [Google Scholar] [CrossRef] [PubMed]
- Gabaldó, X.; Juanpere, M.; Castañé, H.; Rodríguez-Tomàs, E.; López-Azcona, A.F.; Baiges-Gaya, G.; Castro, L.; Valverde-Díaz, E.; Muñoz-Blázquez, A.; Giménez-Cuenca, L.; et al. Usefulness of the Measurement of Serum Paraoxonase-1 Arylesterase Activity in the Diagnoses of COVID-19. Biomolecules 2022, 12, 879. [Google Scholar] [CrossRef] [PubMed]
- Muhammad, Y.; Kani, Y.A.; Iliya, S.; Muhammad, J.B.; Binji, A.; El-Fulaty Ahmad, A.; Kabir, M.B.; Umar Bindawa, K.; Ahmed, A. Deficiency of antioxidants and increased oxidative stress in COVID-19 patients: A cross-sectional comparative study in Jigawa, Northwestern Nigeria. SAGE Open Med. 2021, 9, 2050312121991246. [Google Scholar] [CrossRef]
- Dworzański, J.; Strycharz-Dudziak, M.; Kliszczewska, E.; Kiełczykowska, M.; Dworzańska, A.; Drop, B.; Polz-Dacewicz, M. Glutathione peroxidase (GPx) and superoxide dismutase (SOD) activity in patients with diabetes mellitus type 2 infected with Epstein-Barr virus. PLoS ONE 2020, 15, e0230374. [Google Scholar] [CrossRef] [PubMed]
- Vollbracht, C.; Kraft, K. Oxidative Stress and Hyper-Inflammation as Major Drivers of Severe COVID-19 and Long COVID: Implications for the Benefit of High-Dose Intravenous Vitamin C. Front. Pharmacol. 2022, 13, 899198. [Google Scholar] [CrossRef] [PubMed]
- Yang, Q.; Bai, S.-Y.; Li, L.-F.; Li, S.; Zhang, Y.; Munir, M.; Qiu, H.-J. Human Hemoglobin Subunit Beta Functions as a Pleiotropic Regulator of RIG-I/MDA5-Mediated Antiviral Innate Immune Responses. J. Virol. 2019, 93, e00718-19. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Z.; Lin, F.; Liu, F.; Li, Q.; Li, Y.; Zhu, Z.; Guo, H.; Liu, L.; Liu, X.; Liu, W.; et al. Proteomic profiling reveals a distinctive molecular signature for critically ill COVID-19 patients compared with asthma and chronic obstructive pulmonary disease. Int. J. Infect. Dis. 2022, 116, 258–267. [Google Scholar] [CrossRef] [PubMed]

Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Urbiola-Salvador, V.; Lima de Souza, S.; Macur, K.; Czaplewska, P.; Chen, Z. Plasma Proteomics Elucidated a Protein Signature in COVID-19 Patients with Comorbidities and Early-Diagnosis Biomarkers. Biomedicines 2024, 12, 840. https://doi.org/10.3390/biomedicines12040840
Urbiola-Salvador V, Lima de Souza S, Macur K, Czaplewska P, Chen Z. Plasma Proteomics Elucidated a Protein Signature in COVID-19 Patients with Comorbidities and Early-Diagnosis Biomarkers. Biomedicines. 2024; 12(4):840. https://doi.org/10.3390/biomedicines12040840
Chicago/Turabian StyleUrbiola-Salvador, Víctor, Suiane Lima de Souza, Katarzyna Macur, Paulina Czaplewska, and Zhi Chen. 2024. "Plasma Proteomics Elucidated a Protein Signature in COVID-19 Patients with Comorbidities and Early-Diagnosis Biomarkers" Biomedicines 12, no. 4: 840. https://doi.org/10.3390/biomedicines12040840
APA StyleUrbiola-Salvador, V., Lima de Souza, S., Macur, K., Czaplewska, P., & Chen, Z. (2024). Plasma Proteomics Elucidated a Protein Signature in COVID-19 Patients with Comorbidities and Early-Diagnosis Biomarkers. Biomedicines, 12(4), 840. https://doi.org/10.3390/biomedicines12040840





