Furfural Biodegradation in a Moving Bed Biofilm Reactor Using Native Bacteria and Agroforestry Waste as Supports
Abstract
1. Introduction
2. Materials and Methods
2.1. Microorganisms
2.2. Biofilm Formation Assays
2.2.1. Inoculum Preparation
2.2.2. Plate Assay by Crystal Violet Dying Technique
2.2.3. Biofilm Development on Wastes
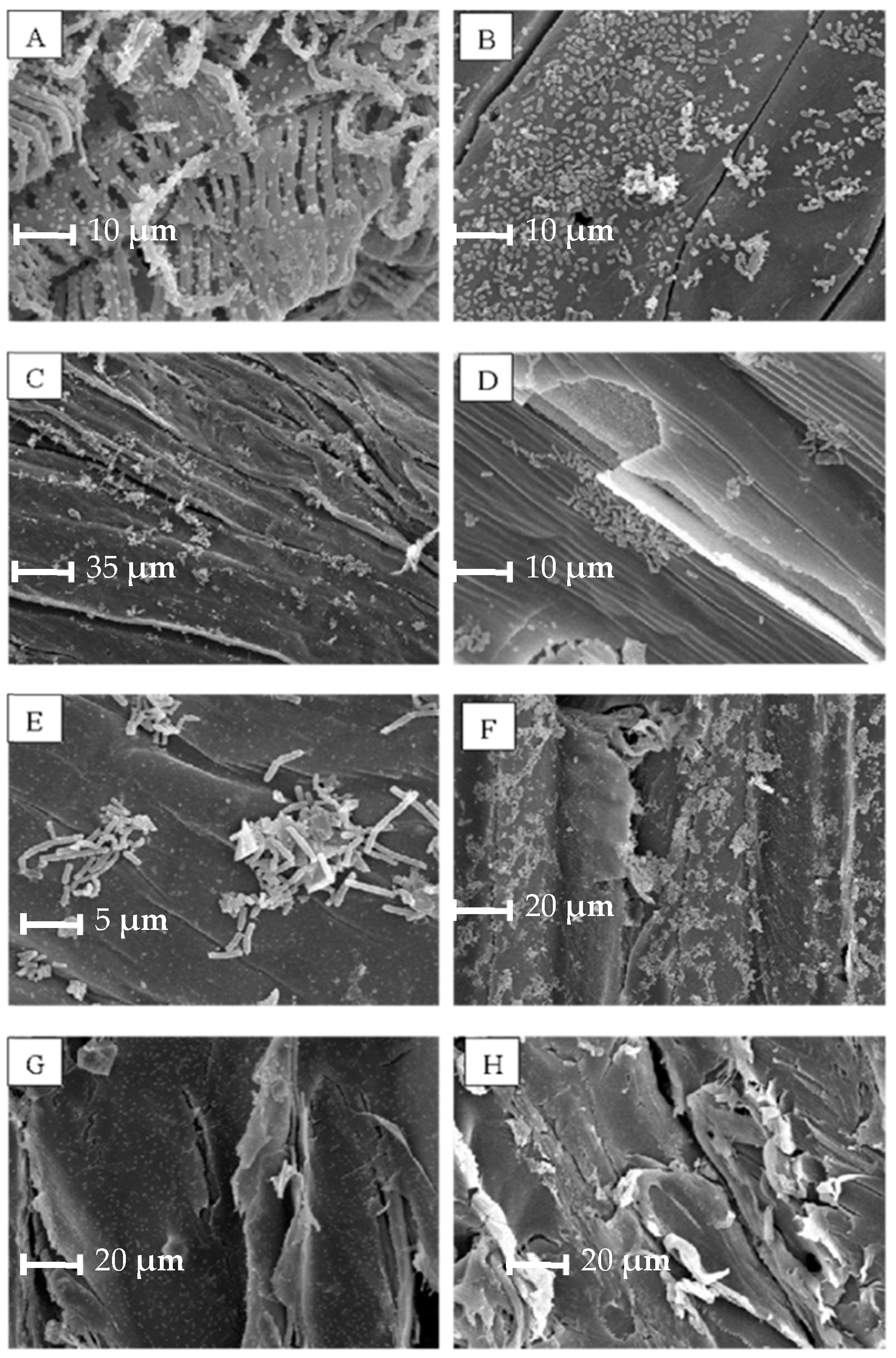
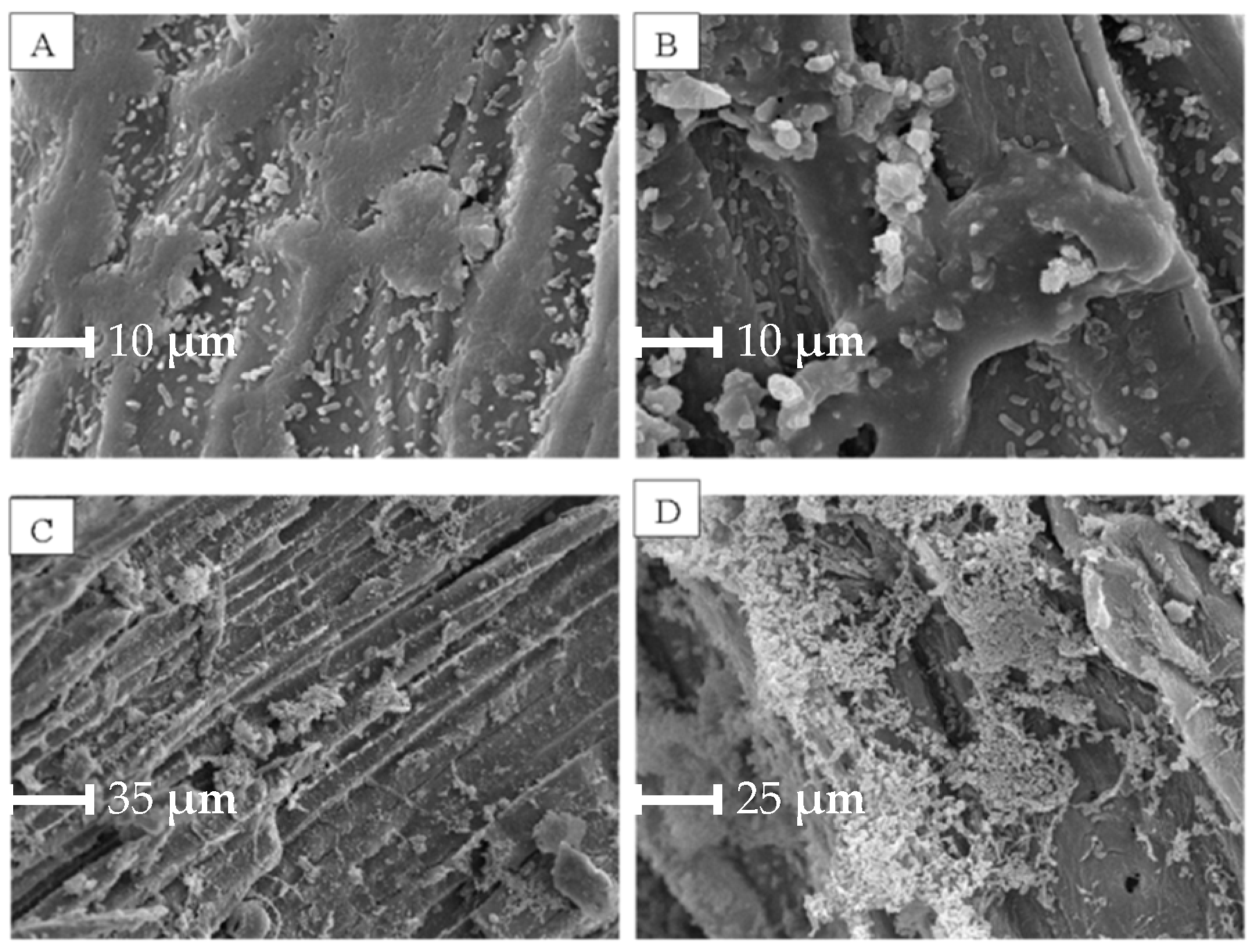
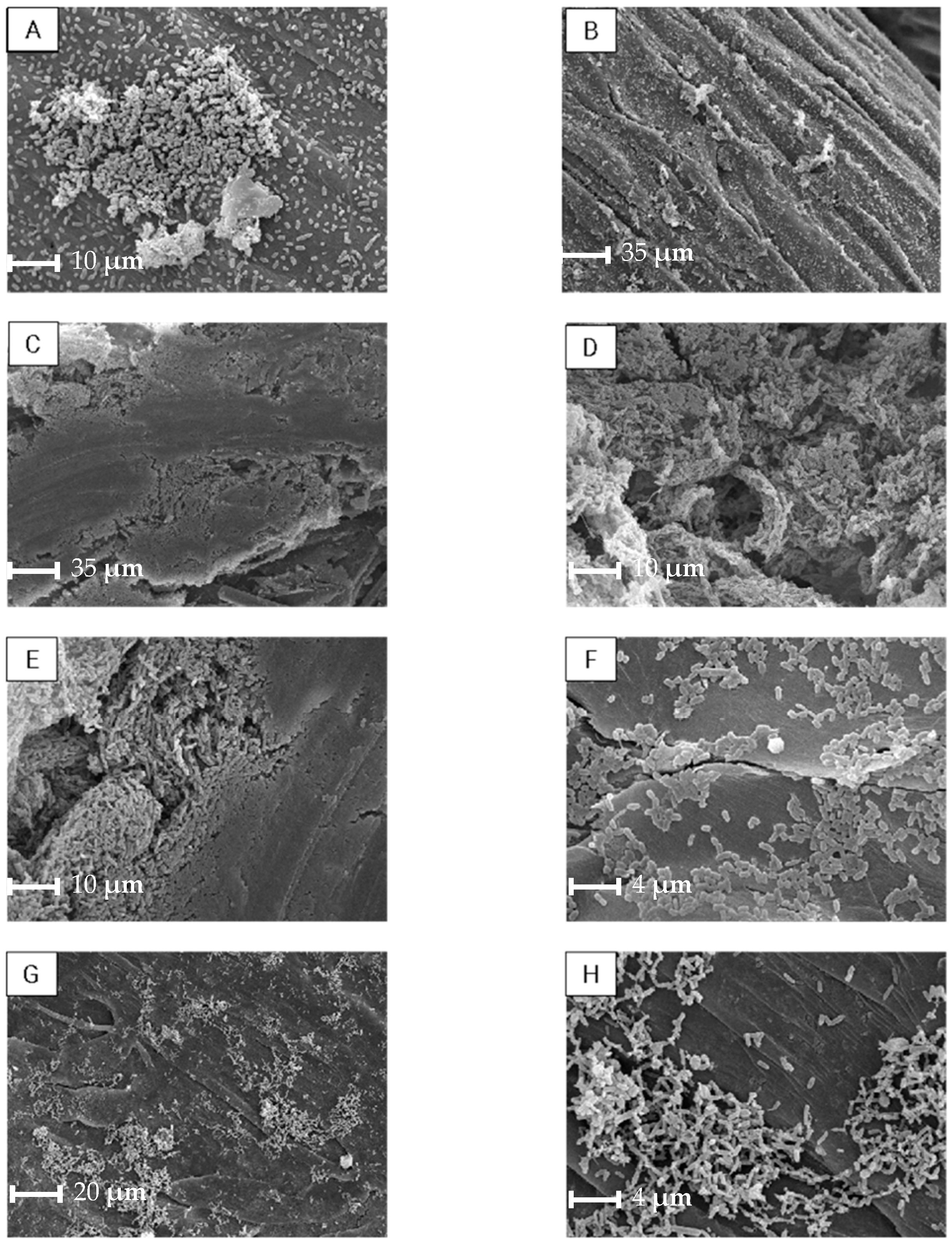
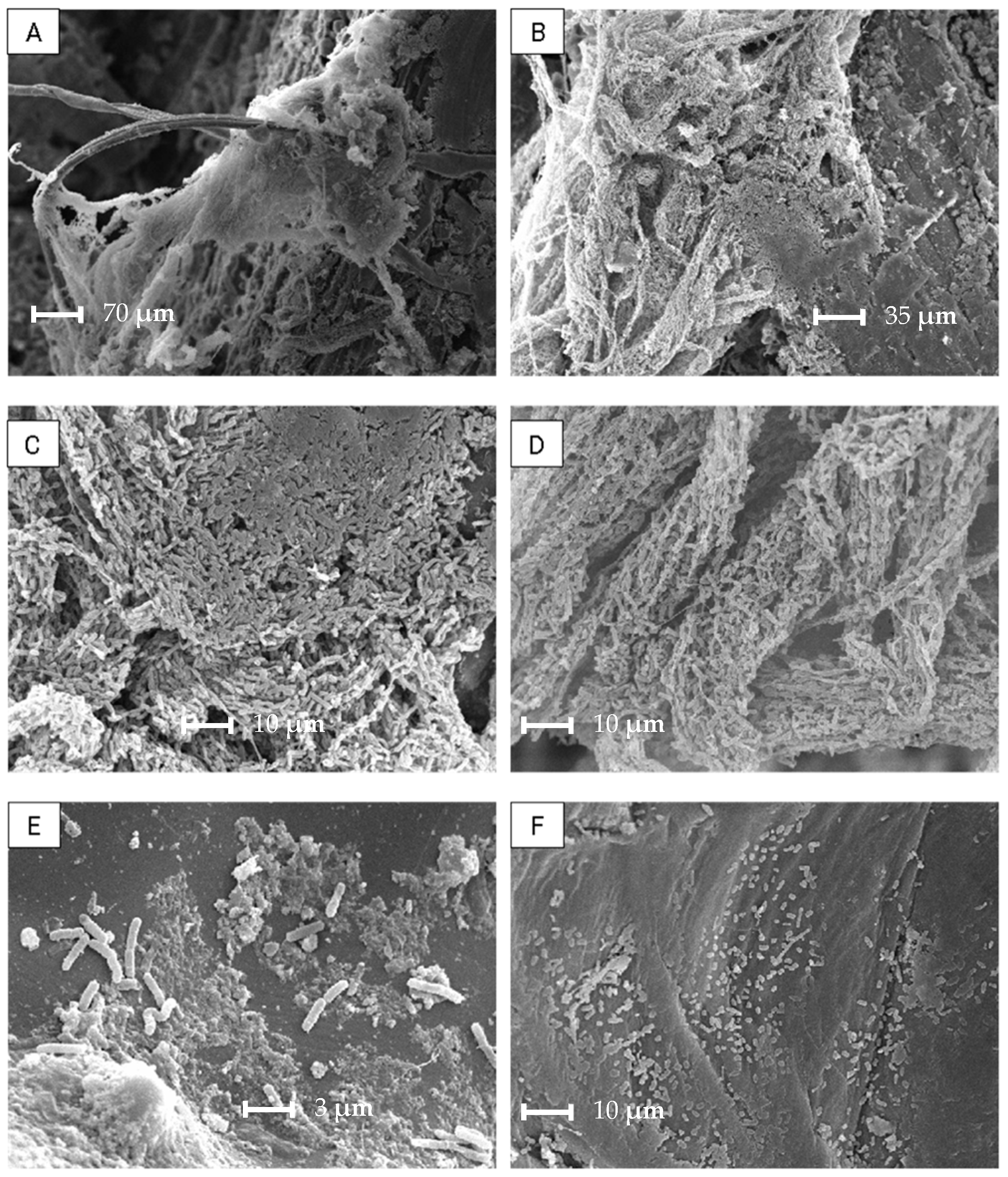
2.2.4. Biofilm Evaluation by Scanning Electron Microscopy
2.3. MBBR Setup
2.4. MBBR Degradation Tests
2.5. Analytical Determination of FUR
2.6. Statistical Evaluation
3. Results and Discussion
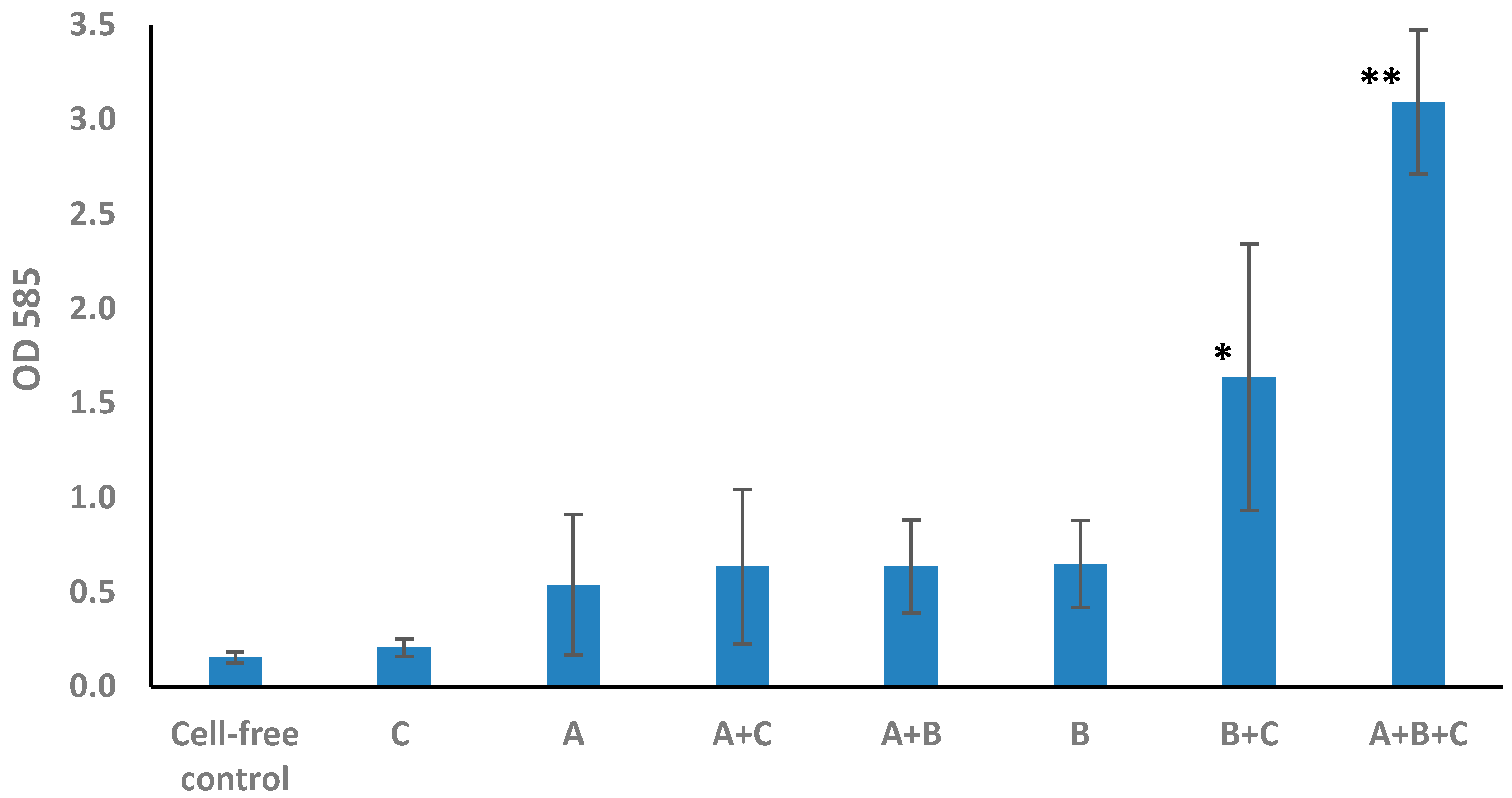
3.1. Determination of Biofilm Formation by Plate Assay
- The presence of biofilm in any of the individual cultures cannot be statistically confirmed.
- Regarding double combinations, in those that contain Brevundimonas sp. GISTAQ1, the presence of biofilm cannot be confirmed.
- The only double combination that could develop biofilm was Microbacterium sp. GISTAQ2 + Bacillus licheniformis GTQ1.
- The highest biofilm production was observed with the triple combination Brevundimonas sp. GISTAQ1 + Microbacterium sp. GISTAQ2 + Bacillus licheniformis GTQ1.
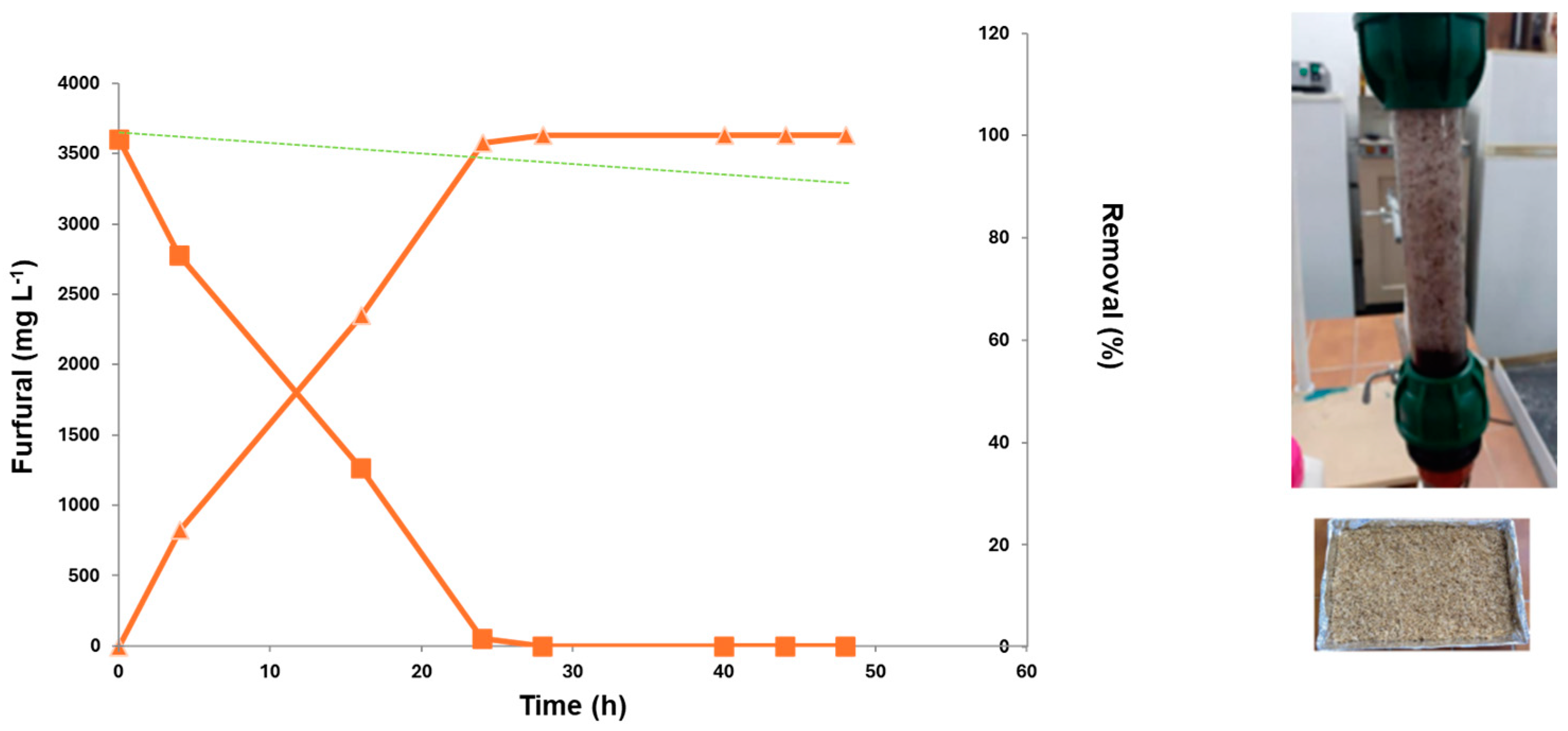
3.2. FUR Degradation in MBBR
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| FUR | Furfural |
| MBBRs | Moving bed biofilm reactors |
| FOL | Furfuryl alcohol |
| EPSs | Extracellular polymeric substances |
| MBRs | Membrane bioreactors |
| OD610 nm | Optical Density at 610 nm |
| SEM | Scanning Electron Microscope |
| FA | Furoic acid |
| HRT | Hydraulic retention time |
| HMF | 5-Hidroximetylfurfural |
References
- Mariscal, R.; Maireles-Torres, P.; Ojeda, M.; Sádaba, I.; Granados, M.L. Furfural: A renewable and versatile platform molecule for the synthesis of chemicals and fuels. Energy Environ. Sci. 2016, 9, 1144–1189. [Google Scholar] [CrossRef]
- SCCS. Opinion on Furfural. Eur. Comm. 2012, 1–26. [Google Scholar]
- Ran, H.; Zhang, J.; Gao, Q.; Lin, Z.; Bao, J. Analysis of biodegradation performance of furfural and 5-hydroxymethylfurfural by Amorphotheca resinae ZN1. Biotechnol. Biofuels 2014, 7, 51. [Google Scholar] [CrossRef]
- Park, H.S.; Um, Y.; Sim, S.J.; Lee, S.Y.; Woo, H.M. Transcriptomic analysis of Corynebacterium glutamicum in the response to the toxicity of furfural present in lignocellulosic hydrolysates. Process Biochem. 2015, 50, 347–356. [Google Scholar] [CrossRef]
- Choi, S.Y.; Gong, G.; Park, H.-S.; Um, Y.; Sim, S.J.; Woo, H.M. Extreme furfural tolerance of a soil bacterium Enterobacter cloacae GGT036. J. Biotechnol. 2015, 193, 11–13. [Google Scholar] [CrossRef]
- Seo, H.M.; Jeon, J.M.; Lee, J.H.; Song, H.-S.; Joo, H.B.; Park, S.H.; Choi, K.Y.; Kim, Y.H.; Park, K.; Ahn, J.; et al. Combinatorial application of two aldehyde oxidoreductases on isobutanol production in the presence of furfural. J. Ind. Microbiol. Biotechnol. 2016, 43, 37–44. [Google Scholar] [CrossRef] [PubMed]
- Ren, J.; Zhang, M.; Guo, X.; Zhou, X.; Ding, N.; Lei, C.; Jia, C.; Wang, Y.; Zhao, J.; Dong, Z.; et al. Furfural tolerance of mutant Saccharomyces cerevisiae selected via ionizing radiation combined with adaptive laboratory evolution. Biotechnol. Biofuels Bioprod. 2024, 17, 117. [Google Scholar] [CrossRef] [PubMed]
- Li, B.; Liu, N.; Zhao, X. Response mechanisms of Saccharomyces cerevisiae to the stress factors present in lignocellulose hydrolysate and strategies for constructing robust strains. Biotechnol. Biofuels Bioprod. 2022, 15, 28. [Google Scholar] [CrossRef]
- Cho, H.Y.; Nam, M.S.; Hong, H.J.; Song, W.S.; Yoon, S.-I. Structural and Biochemical Analysis of the Furan Aldehyde Reductase YugJ from Bacillus subtilis. Int. J. Mol. Sci. 2022, 23, 1882. [Google Scholar] [CrossRef]
- Kim, J.; Son, H.F.; Hwang, S.; Gong, G.; Ko, J.K.; Um, Y.; Han, S.O.; Lee, S. Improving Lipid Production of Yarrowia lipolytica by the Aldehyde Dehydrogenase-Mediated Furfural Detoxification. Int. J. Mol. Sci. 2022, 23, 4761. [Google Scholar] [CrossRef]
- Becerra, M.L.; Lizarazo, L.M.; Rojas, H.A.; Prieto, G.A.; Martinez, J.J. Biotransformation of 5-hydroxymethylfurfural and furfural with bacteria of Bacillus genus. Biocatal. Agric. Biotechnol. 2022, 39, 102281. [Google Scholar] [CrossRef]
- Feldman, D.; Kowbel, D.J.; Cohen, A.; Glass, N.L.; Hadar, Y.; Yarden, O. Identification and manipulation of Neurospora crassa genes involved in sensitivity to furfural. Biotechnol. Biofuels 2019, 12, 210. [Google Scholar] [CrossRef]
- Sárvári Horváth, I.; Franzén, C.J.; Taherzadeh, M.J.; Niklasson, C.; Lidén, G. Effects of furfural on the respiratory metabolism of Saccharomyces cerevisiae in glucose-limited chemostats. Appl. Environ. Microbiol. 2003, 69, 4076–4086. [Google Scholar] [CrossRef]
- Igeño, M.I.; Macias, D.; Blasco, R. A case of adaptive laboratory evolution (ALE): Biodegradation of furfural by Pseudomonas pseudoalcaligenes CECT 5344. Genes 2019, 10, 499. [Google Scholar] [CrossRef] [PubMed]
- Ishii, J.; Yoshimura, K.; Hasunuma, T.; Kondo, A. Reduction of furan derivatives by overexpressing NADH-dependent Adh1 improves ethanol fermentation using xylose as sole carbon source with Saccharomyces cerevisiae harboring XR-XDH pathway. Appl. Microbiol. Biotechnol. 2013, 97, 2597–2607. [Google Scholar] [CrossRef] [PubMed]
- Rodríguez, A.; Rache, L.Y.; Brijaldo, M.H.; Romanelli, G.P.; Luque, R.; Martinez, J.J. Biocatalytic transformation of furfural into furfuryl alcohol using resting cells of Bacillus cereus. Catal. Today 2021, 372, 220–225. [Google Scholar] [CrossRef]
- Zheng, Z.; Xu, Q.; Tan, H.; Zhou, F.; Ouyang, J. Selective Biosynthesis of Furoic Acid From Furfural by Pseudomonas Putida and Identification of Molybdate Transporter Involvement in Furfural Oxidation. Front. Chem. 2020, 8, 587456. [Google Scholar] [CrossRef]
- Flemming, H.C.; Wuertz, S. Bacteria and archaea on Earth and their abundance in biofilms. Nat. Rev. Microbiol. 2019, 17, 247–260. [Google Scholar] [CrossRef]
- Dos Santos, A.L.S.; Galdino, A.C.M.; de Mello, T.P.; Ramos, L.d.S.; Branquinha, M.H.; Bolognese, A.M.; Neto, J.C.; Roudbary, M. What are the advantages of living in a community? A microbial biofilm perspective! Memórias Do Inst. Oswaldo Cruz 2018, 113, e180212. [Google Scholar] [CrossRef]
- Koo, H.; Allan, R.N.; Howlin, R.P.; Stoodley, P.; Hall-Stoodley, L. Targeting microbial biofilms: Current and prospective therapeutic strategies. Nat. Rev. Microbiol. 2017, 15, 740–755. [Google Scholar] [CrossRef]
- Rabin, N.; Zheng, Y.; Opoku-Temeng, C.; Du, Y.; Bonsu, E.; Sintim, H.O. Biofilm formation mechanisms and targets for developing antibiofilm agents. Future Med. Chem. 2015, 7, 493–512. [Google Scholar] [CrossRef]
- Saini, S.; Tewari, S.; Dwivedi, J.; Sharma, V. Biofilm-mediated wastewater treatment: A comprehensive review. Mater. Adv. 2023, 4, 1415–1443. [Google Scholar] [CrossRef]
- Kilic, T. Factors Affecting Biofilm Formation and the Effects of These Factors on Bacteria. In Exploring Bacterial Biofilms; Dincer, S., Ed.; IntechOpen: Rijeka, Croatia, 2025; p. 17. [Google Scholar]
- Mehrotra, T.; Dev, S.; Banerjee, A.; Chatterjee, A.; Singh, R.; Aggarwal, S. Use of immobilized bacteria for environmental bioremediation: A review. J. Environ. Chem. Eng. 2021, 9, 105920. [Google Scholar] [CrossRef]
- Dzionek, A.; Wojcieszyńska, D.; Guzik, U. Natural carriers in bioremediation: A review. Electron. J. Biotechnol. 2016, 23, 28–36. [Google Scholar] [CrossRef]
- Partovinia, A.; Rasekh, B. Review of the immobilized microbial cell systems for bioremediation of petroleum hydrocarbons polluted environments. Crit. Rev. Environ. Sci. Technol. 2018, 48, 1–38. [Google Scholar] [CrossRef]
- Asri, M.; Elabed, S.; Ibnsouda Koraichi, S.; El Ghachtouli, N. Biofilm-Based Systems for Industrial Wastewater Treatment. In Handbook of Environmental Materials Management; Hussain, C.M., Ed.; Springer: Berlin/Heidelberg, Germany, 2018; pp. 1–21. [Google Scholar]
- Mazioti, A.A.; Koutsokeras, L.E.; Constantinides, G.; Vyrides, I. Untapped potential of moving bed biofilm reactors with different biocarrier types for bilge water treatment: A laboratory-scale study. Water 2021, 13, 1810. [Google Scholar] [CrossRef]
- Rehman, F.; Thebo, K.H.; Aamir, M.; Akhtar, J. Nanomembranes for water treatment. In Nanotechnology in the Beverage Industry: Fundamentals and Applications; Amrane, A., Rajendran, S., Nguyen, T.A., Assadi, A.A., Sharoba, A.M., Eds.; Elsevier: Amsterdam, The Netherlands, 2020; pp. 207–240. [Google Scholar]
- Moktadir, M.A.; Maliha, M.; Tujjohra, F.; Munmun, S.A.; Alam, M.S.; Islam, M.A.; Rahman, M.M. Treatment of tannery wastewater by different membrane bioreactors: A critical review. Environ. Adv. 2024, 15, 100478. [Google Scholar] [CrossRef]
- Bhushan, S.; Gogoi, M.; Bora, A.; Ghosh, S.; Barman, S.; Biswas, T.; Sudarshan, M.; Thakur, A.R.; Mukherjee, I.; Dey, S.K.; et al. Understanding bacterial biofilm stimulation using different methods—A criterion for selecting epiphytes by plants. Microbiol. Biotechnol. Lett. 2019, 47, 303–309. [Google Scholar] [CrossRef]
- Gogoi, M.; Mukherjee, I.; Ray Chaudhuri, S. Characterization of ammonia remover Bacillus albus (ASSF01) in terms of biofilm-forming ability with application in aquaculture effluent treatment. Environ. Sci. Pollut. Res. 2022, 29, 61838–61855. [Google Scholar] [CrossRef]
- Biswas, K.; Taylor, M.W.; Turner, S.J. Successional development of biofilms in moving bed biofilm reactor (MBBR) systems treating municipal wastewater. Appl. Microbiol. Biotechnol. 2014, 98, 1429–1440. [Google Scholar] [CrossRef]
- Liang, C.; de Jonge, N.; Carvalho, P.N.; Nielsen, J.L.; Bester, K. Biodegradation kinetics of organic micropollutants and microbial community dynamics in a moving bed biofilm reactor. Chem. Eng. J. 2021, 415, 128963. [Google Scholar] [CrossRef]
- Lago, A.; Rocha, V.; Barros, O.; Silva, B.; Tavares, T. Bacterial biofilm attachment to sustainable carriers as a clean-up strategy for wastewater treatment: A review. J. Water Process Eng. 2024, 63, 105368. [Google Scholar] [CrossRef]
- Zhang, Y.; Zhao, L.; Song, T.; Cheng, Y.; Bao, M.; Li, Y. Simultaneous nitrification and denitrification in an aerobic biofilm biosystem with loofah sponges as carriers for biodegrading hydrolyzed polyacrylamide-containing wastewater. Bioprocess Biosyst. Eng. 2020, 43, 529–540. [Google Scholar] [CrossRef]
- Wang, W.; Wu, Y.; Zhang, C. High-density natural luffa sponge as anaerobic microorganisms carrier for degrading 1,1,1-TCA in groundwater. Bioprocess Biosyst. Eng. 2016, 40, 383–393. [Google Scholar] [CrossRef] [PubMed]
- FAO. Food and Agriculture Organization of the United Nations. Agricultural Market Information System (AMIS). Mark. Monit. 2024, 115, 1–19. [Google Scholar]
- Peanparkdee, M.; Iwamoto, S. Bioactive compounds from by-products of rice cultivation and rice processing: Extraction and application in the food and pharmaceutical industries. Trends Food Sci. Technol. 2019, 86, 109–117. [Google Scholar] [CrossRef]
- Nadaleti, W.C. Utilization of residues from rice parboiling industries in southern Brazil for biogas and hydrogen-syngas generation: Heat, electricity and energy planning. Renew. Energy 2019, 131, 55–72. [Google Scholar] [CrossRef]
- Rawindran, H.; Leong, W.H.; Suparmaniam, U.; Liew, C.S.; Raksasat, R.; Kiatkittipong, W.; Mohamad, M.; Ghani, N.A.; Abdelfattah, E.A.; Lam, M.K.; et al. Residual palm kernel expeller as the support material and alimentation provider in enhancing attached microalgal growth for quality biodiesel production. J. Environ. Manag. 2022, 316, 357–363. [Google Scholar] [CrossRef]
- Zakaria, Z.A.; Zakaria, Z.; Surif, S.; Ahmad, W.A. Biological detoxification of Cr(VI) using wood-husk immobilized Acinetobacter haemolyticus. J. Hazard. Mater. 2007, 148, 164–171. [Google Scholar] [CrossRef]
- Zhang, Q.; Yu, Z.; Jin, S.; Zhu, L.; Liu, C.; Zheng, H.; Zhou, T.; Liu, Y.; Ruan, R. Lignocellulosic residue as bio-carrier for algal biofilm growth: Effects of carrier physicochemical proprieties and toxicity on algal biomass production and composition. Bioresour. Technol. 2019, 293, 122091. [Google Scholar] [CrossRef]
- Yan, H.; Zhang, Q.; Wang, Y.; Cui, X.; Liu, Y.; Yu, Z.; Xu, S.; Ruan, R. Rice straw as microalgal biofilm bio-carrier: Effects of indigenous microorganisms on rice straw and microalgal biomass production. J. Environ. Manag. 2023, 341, 118075. [Google Scholar] [CrossRef] [PubMed]
- Shao, L.; Xu, Z.X.; Yin, H.L.; Chu, H. Rice husk as carbon source and biofilm carrier for water denitrification. J. Biotechnol. 2008, 18, S662. [Google Scholar] [CrossRef]
- Alitaleshi, F.; Daghbandan, A.; Alireza, P. Performance of rice husk biocarrier on ammonia nitrogen removal in the MBBR treating aquaculture wastewater using biological attached growth process: Performance and kinetic study. J. Environ. Chem. Eng. 2024, 12, 111446. [Google Scholar] [CrossRef]
- Zainab, A.; Meraj, S.; Liaquat, R. Study on Natural Organic Materials as Biofilm Carriers for the Optimization of Anaerobic Digestion. Waste Biomass Valorization 2020, 11, 2521–2531. [Google Scholar] [CrossRef]
- Al-Amshawee, S.K.A.; Yunus, M.Y.B.M.; Alalwan, H.A.; Lee, W.H.; Dai, F. Experimental investigation of biofilm carriers of varying shapes, sizes, and materials for wastewater treatment in fixed bed biofilm reactor: A qualitative study of biocarrier performance. J. Chem. Technol. Biotechnol. 2022, 97, 2592–2606. [Google Scholar] [CrossRef]
- Bu, C.Y.; Yan, Y.X.; Zou, L.H.; Zheng, Z.J.; Ouyang, J. One-pot biosynthesis of furfuryl alcohol and lactic acid via a glucose coupled biphasic system using single Bacillus coagulans NL01. Bioresour. Technol. 2020, 313, 123705. [Google Scholar] [CrossRef] [PubMed]
- Wierckx, N.; Koopman, F.; Ruijssenaars, H.J.; De Winde, J.H. Microbial degradation of furanic compounds: Biochemistry, genetics, and impact. Appl. Microbiol. Biotechnol. 2011, 92, 1095–1105. [Google Scholar] [CrossRef]
- Qin, L.Z.; He, Y.C. Chemoenzymatic Synthesis of Furfuryl Alcohol from Biomass in Tandem Reaction System. Appl. Biochem. Biotechnol. 2020, 190, 1289–1303. [Google Scholar] [CrossRef]
- Yan, Y.; Bu, C.; He, Q.; Zheng, Z.; Ouyang, J. Efficient bioconversion of furfural to furfuryl alcohol by Bacillus coagulans NL01. RSC Adv. 2018, 8, 26720–26727. [Google Scholar] [CrossRef]
- Wang, Z.W.; Gong, C.J.; He, Y.C. Improved biosynthesis of 5-hydroxymethyl-2-furancarboxylic acid and furoic acid from biomass-derived furans with high substrate tolerance of recombinant Escherichia coli HMFOMUT whole-cells. Bioresour. Technol. 2020, 303, 122930. [Google Scholar] [CrossRef]
- Koenig, K.; Andreesen, J.R. Xanthine dehydrogenase and 2-furoyl-coenzyme A dehydrogenase from Pseudomonas putida Fu1: Two molybdenum-containing dehydrogenases of novel structural composition. J. Bacteriol. 1990, 172, 5999–6600. [Google Scholar] [CrossRef]
- Wang, X.; Gao, Q.; Bao, J. Transcriptional analysis of Amorphotheca resinae ZN1 on biological degradation of furfural and 5-hydroxymethylfurfural derived from lignocellulose pretreatment. Biotechnol. Biofuels 2015, 8, 1–13. [Google Scholar] [CrossRef]
- Farías, A.; Echeverría, M.; Utgés, E.; Fontana, G.; Cuadra, P. Furfural Biodegradation in Consortium through Bacillus licheniformis, Microbacterium sp. and Brevundimonas sp. J. Sustain. Dev. Energy Water Environ. Syst. 2022, 10, 1–10. [Google Scholar] [CrossRef]
- Fernández Linares, L.C.; Rojas Avelizapa, N.G.; Roldán Carrillo, T.G.; Ramírez Islas, M.E.; Zegarra Martínez, H.G.; Uribe Hernández, R. Manual de Técnicas de Análisis de Suelos Aplicadas a la Remediación. 2006. Available online: https://es.scribd.com/doc/121876082/Manual-de-Analisis-de-Suelos (accessed on 15 April 2022).
- Boopathy, R.; Daniels, L. Isolation and characterization of a furfural degrading sulfate-reducing bacterium from an anaerobic digester. Curr. Microbiol. 1991, 23, 327–332. [Google Scholar] [CrossRef]
- Hareland, W.A.; Crawford, R.L.; Chapman, P.J.; Dagley, S. Metabolic function and properties of 4-hydroxyphenylacetic acid 1-hydroxylase from Pseudomonas acidovorans. J. Bacteriol. 1975, 121, 272–285. [Google Scholar] [CrossRef]
- Merritt, J.H.; Kadouri, D.E.; O’Toole, G.A. Growing and analyzing static biofilms. Curr. Protoc. Microbiol. 2011, 22, 1B-1.1–1B-1.18. [Google Scholar] [CrossRef]
- Burmølle, M.; Webb, J.S.; Rao, D.; Hansen, L.H.; Sørensen, S.J.; Kjelleberg, S. Enhanced biofilm formation and increased resistance to antimicrobial agents and bacterial invasion are caused by synergistic interactions in multispecies biofilms. Appl. Environ. Microbiol. 2006, 72, 3916–3923. [Google Scholar] [CrossRef] [PubMed]
- Sanabria Cubillos, A.; Pacheco Ojeda, J.D. Diseño y Evaluación de un Reactor Biológico de Lecho Móvil de Cargas Secuenciales Como Alternativa de Tratamiento Para un Vertimiento Procedente de una Industria Farmacéutica. 2019. Available online: https://ciencia.lasalle.edu.co/items/d4d2886a-661a-4a60-a769-7bfc58f70b3d/full (accessed on 23 September 2023).
- D5837-99; A Standard Test Method for Furanic Compounds in Electrical Insulating Liquids by High- Performance Liquid Chromatography (HPLC). In ASTM Volume 10.03: Electrical Insulating Liquids and Gases. ASTM: West Conshohocken, PA, USA, 2005.
- Zhu, Z.; Shan, L.; Li, X.; Hu, F.; Yuan, Y.; Zhong, D.; Zhang, J. Effects of interspecific interactions on biofilm formation potential and chlorine resistance: Evaluation of dual-species biofilm observed in drinking water distribution systems. J. Water Process. Eng. 2020, 38, 101564. [Google Scholar] [CrossRef]
- Laganenka, L.; Sourjik, V. Autoinducer 2-dependent Escherichia coli biofilm formation is enhanced in a dual-species coculture. Appl. Environ. Microbiol. 2018, 84, e02638-17. [Google Scholar] [CrossRef]
- Liu, W.; Russel, J.; Røder, H.; Madsen, J.; Burmølle, M.; Sørensen, S.J. Low-abundant species facilitates specific spatial organisation that promotes multispecies biofilm formation. Environ. Microbiol. 2017, 19, 2893–2905. [Google Scholar] [CrossRef]
- Relucenti, M.; Familiari, G.; Donfrancesco, O.; Taurino, M.; Li, X.; Chen, R.; Artini, M.; Papa, R.; Selan, L. Microscopy methods for biofilm imaging: Focus on sem and VP-SEM pros and cons. Biology 2021, 10, 51. [Google Scholar] [CrossRef]
- Zheng, D.; Bao, J.; Lu, J.; Gao, C. Isolation and characterization of a furfural-degrading bacterium Bacillus cereus sp. strain DS1. Curr. Microbiol. 2015, 70, 199–205. [Google Scholar] [CrossRef] [PubMed]
- Maity, J.P.; Chandra Samal, A.; Rajnish, K.; Singha, S.; Sahoo, T.R.; Chakraborty, S.; Bhattacharya, P.; Chakraborty, S.; Sarangi, B.S.; Dey, G.; et al. Furfural removal from water by bioremediation process by indigenous Pseudomonas putida (OSBH3) and Pseudomonas aeruginosa (OSBH4) using novel suphala media: An optimization for field application. Groundw. Sustain. Dev. 2023, 20, 100895. [Google Scholar] [CrossRef]
- Belay, N.; Boopathy, R.; Voskuilen, G. Anaerobic transformation of furfural by Methanococcus deltae ΔLH. Appl. Environ. Microbiol. 1997, 63, 2092–2094. [Google Scholar] [CrossRef] [PubMed]
- Bischoff, K.M.; Liu, S.; Hughes, S.R.; Rich, J.O. Fermentation of corn fiber hydrolysate to lactic acid by the moderate thermophile Bacillus coagulans. Biotechnol. Lett. 2010, 32, 823–828. [Google Scholar] [CrossRef]
- Crigler, J.; Eiteman, M.A.; Altman, E. Characterization of the Furfural and 5-Hydroxymethylfurfural (HMF) Metabolic Pathway in the Novel Isolate Pseudomonas putida ALS1267. Appl. Biochem. Biotechnol. 2020, 190, 918–930. [Google Scholar] [CrossRef] [PubMed]
- Hunter, W.J.; Manter, D.K. Pre-treatment step with Leuconostoc mesenteroides or L. pseudomesenteroides strains removes furfural from Zymomonas mobilis ethanolic fermentation broth. Bioresour. Technol. 2014, 169, 162–168. [Google Scholar] [CrossRef]
- Lee, S.A.; Wrona, L.J.; Cahoon, A.B.; Crigler, J.; Eiteman, M.A.; Altman, E. Isolation and Characterization of Bacteria That Use Furans as the Sole Carbon Source. Appl. Biochem. Biotechnol. 2016, 178, 76–90. [Google Scholar] [CrossRef]
- Ye, L.; Hudari, M.S.B.; Li, Z.; Wu, J.C. Simultaneous detoxification, saccharification and co-fermentation of oil palm empty fruit bunch hydrolysate for l-lactic acid production by Bacillus coagulans JI12. Biochem. Eng. J. 2014, 83, 16–21. [Google Scholar] [CrossRef]
- Rahmani, A.; Salari, M.; Tari, K.; Shabanloo, A.; Shabanloo, N.; Bajalan, S. Enhanced degradation of furfural by heat-activated persulfate/nZVI-rGO oxidation system: Degradation pathway and improving the biodegradability of oil refinery wastewater. J. Environ. Chem. Eng. 2020, 8, 104468. [Google Scholar] [CrossRef]
- Piculell, M. New Dimensions of Moving Bed Biofilm Carriers. Influence of Biofilm Thickness and Control Possibilities. 2016. Available online: https://portal.research.lu.se/en/publications/new-dimensions-of-moving-bed-biofilm-carriers-influence-of-biofil (accessed on 18 August 2023).
- Guarnieri, M.T.; Ann Franden, M.; Johnson, C.W.; Beckham, G.T. Conversion and assimilation of furfural and 5-(hydroxymethyl)furfural by Pseudomonas putida KT2440. Metab. Eng. Commun. 2017, 4, 22–28. [Google Scholar] [CrossRef] [PubMed]
- Pedrino, M.; Narcizo, J.P.; Aguiar, I.R.; Reginatto, V.; Guazzaroni, M.E. Upgrading Pseudomonas sp. toward Tolerance to a Synthetic Biomass Hydrolysate Enriched with Furfural and 5-Hydroxymethylfurfural. ACS Omega 2025, 10, 5449–5459. [Google Scholar] [CrossRef] [PubMed]
- Zou, L.; Jin, X.; Tao, Y.; Zheng, Z.; Ouyang, J. Unraveling the mechanism of furfural tolerance in engineered Pseudomonas putida by genomics. Front. Microbiol. 2022, 13, 1035263. [Google Scholar] [CrossRef]
- Peng, L.; Wang, L.; Che, C.; Yang, G.; Yu, B.; Ma, Y. Bacillus sp. strain P38: An efficient producer of l-lactate from cellulosic hydrolysate, with high tolerance for 2-furfural. Bioresour. Technol. 2013, 149, 169–176. [Google Scholar] [CrossRef]
- Liu, X.; Zhang, Y.; Shang, M.; Zhu, L.; Yu, M. The Effects of Furoic Acid and Furfuryl Alcohol on Detection of Furfural in Transformer Oil by High—Performance Liquid Chromatography. Chromatographia 2020, 83, 927–932. [Google Scholar] [CrossRef]


Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Farias, A.R.; Panigatti, M.C.; Vullo, D.L. Furfural Biodegradation in a Moving Bed Biofilm Reactor Using Native Bacteria and Agroforestry Waste as Supports. Processes 2025, 13, 1337. https://doi.org/10.3390/pr13051337
Farias AR, Panigatti MC, Vullo DL. Furfural Biodegradation in a Moving Bed Biofilm Reactor Using Native Bacteria and Agroforestry Waste as Supports. Processes. 2025; 13(5):1337. https://doi.org/10.3390/pr13051337
Chicago/Turabian StyleFarias, Alejandro Ruben, Maria Cecilia Panigatti, and Diana Lia Vullo. 2025. "Furfural Biodegradation in a Moving Bed Biofilm Reactor Using Native Bacteria and Agroforestry Waste as Supports" Processes 13, no. 5: 1337. https://doi.org/10.3390/pr13051337
APA StyleFarias, A. R., Panigatti, M. C., & Vullo, D. L. (2025). Furfural Biodegradation in a Moving Bed Biofilm Reactor Using Native Bacteria and Agroforestry Waste as Supports. Processes, 13(5), 1337. https://doi.org/10.3390/pr13051337






