Microbiological Evaluation of Two Mexican Artisanal Cheeses: Analysis of Foodborne Pathogenic Bacteria in Cotija Cheese and Bola de Ocosingo Cheese by qPCR
Abstract
:1. Introduction
2. Materials and Methods
2.1. Biological Materials
2.2. Cheese Samples
2.3. Culture of Reference Strains and Bacterial Enrichment in Cheese
2.4. DNA Extraction
2.5. qPCR Reaction
2.6. Assessment of Primer–Probe Specificity and Colony Forming Unit (CFU) Limit of Detection
2.7. qPCR of Spiked Microorganisms in Cheese
2.8. Endpoint PCR
2.9. Viable S. aureus Assessment
3. Results and Discussion
3.1. Primer–Probe Set Cross-Reactivity
3.2. Limit of CFU Detection Assessment
3.3. Effect of the Cheese Matrix on Pathogen Detection
3.4. Cheese Analysis: DNA Quality Assessment
3.5. Cheese Analysis: Pathogen Detection
4. Conclusions
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Díaz-Ramírez, M.; García-Garibay, M.; Jiménez-Guzman, J.; Villanueva-Carbajal, A. Food safety management in traditional food products: Balancán’s Poro cheese as a case study. Estud. Soc. 2016, 25, 87–110. Available online: https://www.redalyc.org/journal/417/41744004004/html/ (accessed on 16 July 2024).
- Román, S.; Sánchez-Siles, L.M.; Siegrist, M. The importance of food naturalness for consumers: Results of a systematic review. Trends Food Sci. Tech. 2017, 67, 44–57. [Google Scholar] [CrossRef]
- Cervantes-Escoto, F.; Villegas-de-Gante, A.; Cesín-Vargas, A.; Espinoza-Ortega, A. Chapter 2: El Queso CotijaMC Región de Origen, un caso especial. In Los Quesos Mexicanos Genuinos: Patrimonio Cultural que debe Rescatarse, 2nd ed.; Sandoval, J., Ed.; Editorial del Colegio de Postgraduados, Autonomous University of Chapingo: Texcoco, México, 2013; pp. 145–155. [Google Scholar]
- Yescas, C. Larousse de Quesos Mexicanos; Larousse: Mexico City, México, 2013; pp. 30–33 and 38–41. [Google Scholar]
- Pomeón, T. El Queso Cotija, México. Un Producto con Marca Colectiva Queso “Cotija Región de origen”, en Proceso de Adquisición de una Denominación de Origen. Consultancy Report for FAO and IICA in the Framework of the Joint Study on Origin-Linked Quality Products. Autonomous University of Chapingo. 2007. Available online: https://openknowledge.fao.org/server/api/core/bitstreams/c606f3ad-52ca-489d-a3fc-acfaf1551ac6/content (accessed on 16 July 2024).
- NOM-243-SSA1Mexican Official Standard. Productos y Servicios. Leche, Fórmula Láctea, Producto Lácteo Combinado y Derivados Lácteos. Disposiciones y Especificaciones Sanitarias. Métodos de Prueba. 2010. Available online: https://dof.gob.mx/normasOficiales/4156/salud2a/salud2a.htm (accessed on 16 July 2024).
- NMX-F-735-COFOCALEC. Mexican Standard. Sistema producto Leche-Alimento-Lácteo-Alimento Lácteo Regional-Queso Cotija Artesanal Madurado-Denominación, Especificaciones y métodos de prueba. 2018.
- Escobar-Zepeda, A.; Sanchez-Flores, A.; Quirasco Baruch, M. Metagenomic analysis of a Mexican ripened cheese reveals a unique complex microbiota. Food Microbiol. 2016, 57, 116–127. [Google Scholar] [CrossRef] [PubMed]
- Khattab, A.R.; Guirguis, H.; Tawfik, S.M.; Farag, M.A. Cheese ripening: A review on modern technologies towards flavor enhancement, process acceleration and improved quality assessment. Trends Food Sci. Tech. 2019, 88, 343–360. [Google Scholar] [CrossRef]
- Afshari, R.; Pillidge, C.J.; Dias, D.A.; Osborn, A.M.; Gill, H. Cheesomics: The future pathway to understanding cheese flavour and quality. Crit. Rev. Food Sci. Nut 2018, 6, 33–47. [Google Scholar] [CrossRef] [PubMed]
- Aleksic, B.; Djekic, I.; Miocinovic, J.; Miloradovic, Z.; Savic–Radovanovic, R.; Zdravkovic, N.; Smigic, N. The hygienic assessment of dairy products’ selling places at open markets. Food Control 2023, 148, 109628. [Google Scholar] [CrossRef]
- Olea-Rodríguez, M.A.; Chombo-Morales, P.; Nuño, K.; Vázquez-Paulino, O.; Villagrán-de la Mora, Z.; Garay-Martínez, L.E.; Castro-Rosas, J.; Villarruel-López, A.; Torres-Vitela, M.R. Microbiological Characteristics and Behavior of Staphylococcus aureus, Salmonella spp., Listeria monocytogenes and Staphylococcal Toxin during Making and Maturing Cotija Cheese. Appl. Sci. 2021, 11, 8154. [Google Scholar] [CrossRef]
- Lusk, T.S.; Strain, E.; Kase, J.A. Comparison of six commercial DNA extraction kits for detection of Brucella neotomae in Mexican and Central American-style cheese and other milk products. Food Microbiol. 2013, 34, 100–105. [Google Scholar] [CrossRef]
- Soto-Varela, Z.E.; Gutiérrez, C.G.; de Moya, Y.; Mattos, R.; Bolívar-Anillo, H.J.; Villarreal, J.L. Detección molecular de Salmonella spp., Listeria spp. y Brucella spp. en queso artesanal fresco comercializado en Barranquilla: Un estudio piloto. Biomedica 2018, 38, 30–36. [Google Scholar] [CrossRef]
- Kabiraz, M.P.; Majumdar, P.R.; Mahmud, M.M.C.; Bhowmik, S.; Ali, A. Conventional and advanced detection techniques of foodborne pathogens: A comprehensive review. Heliyon 2023, 9, e15482. [Google Scholar] [CrossRef]
- Zhou, X.; Zhang, T.; Song, D.; Huang, T.; Peng, Q.; Chen, Y.; Li, A.; Zhang, F.; Wu, Q.; Ye, Y.; et al. Comparison and evaluation of conventional RT-PCR, SYBR green I and TaqMan real-time RT-PCR assays for the detection of porcine epidemic diarrhea virus. Mol. Cell. Probes 2017, 33, 36–41. [Google Scholar] [CrossRef] [PubMed]
- Real-Time PCR Handbook; Thermo Fisher Scientific Inc.: San Jose, CA, USA, 2016; pp. 7 and 12.
- Oon, Y.-L.; Oon, Y.-S.; Ayaz, M.; Deng, M.; Li, L.; Song, K. Waterborne pathogens detection technologies: Advances, challenges, and future perspectives. Front. Microbiol. 2023, 14, 1286923. [Google Scholar] [CrossRef]
- Malorny, B.; Bunge, C.; Helmuth, R. A real-time PCR for the detection of Salmonella Enteritidis in poultry meat and consumption eggs. J. Microbiol. Methods 2007, 70, 245–251. [Google Scholar] [CrossRef]
- Ruiz-Pérez de Pipaón, M.; Torres-Sánchez, M.J.; Arroyo-Pedrero, L.A.; Prados-Blanco, T.; Palomares-Folía, J.C.; Aznar-Martín, J. Detection of methicillin resistance and identification of Staphylococcus spp. in positive blood cultures by amplifying the mecA and nucA genes with the LightCycler system. Enfermedades Infecc. Microbiol. Clin. 2005, 23, 208–212. [Google Scholar] [CrossRef]
- Kim, J.; Demeke, T.; Clear, R.M.; Patrick, S.K. Simultaneous detection by PCR of Escherichia coli, Listeria monocytogenes and Salmonella thyphimurium in artificially inoculated wheat grain. Int. J. Food Microbiol. 2006, 111, 21–35. [Google Scholar] [CrossRef] [PubMed]
- Bounaadja, L.; Albert, D.; Chénais, B.; Hénault, S.; Zygmunt, M.S.; Poliak, S.; Garin-Bastuji, B. Real-time PCR for identification of Brucella spp.: A comparative study of IS711, bcsp31 and per target genes. Vet. Microbiol. 2009, 137, 156–164. [Google Scholar] [CrossRef] [PubMed]
- Nada, R.A.; Shaheen, H.I.; Touni, I.; Fahmy, D.; Armstrong, A.W.; Weiner, M.; Klena, J.D. Design and validation of a multiplex polymerase chain reaction for the identification of enterotoxigenic Escherichia coli and associated colonization factor antigens. Diagn. Micr. Infec. Dis. 2010, 67, 134–142. [Google Scholar] [CrossRef]
- European Food Safety Authority. Technical specifications for the monitoring and reporting of verotoxigenic Escherichia coli (VTEC) on animals and food. EFSA J. 2009, 7, 1366. [Google Scholar] [CrossRef]
- ISO 22174:2005; Microbiology of Food and Animal Feeding Stuffs—Polymerase Chain Reaction (PCR) for the Detection of Food-borne Pathogen—General Requirements and Definitions. International Organization for Standardization: Geneva, Switzerland, 2005.
- World Organization for Animal Health, WOAH. Development and Optimisation of Nucleic Acid Detection Assays. Available online: https://www.woah.org/fileadmin/Home/eng/Health_standards/tahm/2.02.03_NAD_ASSAYS.pdf (accessed on 12 August 2024).
- Nolan, T.; Huggett, J.; Sanchez, E. Good Practice Guide for the Application of Quantitative PCR (qPCR), LGC. 2013. Available online: https://www.gene-quantification.de/national-measurement-system-qpcr-guide.pdf (accessed on 16 July 2024).
- Ercolini, D. PCR-DGGE fingerprinting: Novel strategies for detection of microbes in food. J. Microbiol. Methods 2004, 56, 297–314. [Google Scholar] [CrossRef]
- NOM-115-SSA1; Official Mexican Standard. Bienes y Servicios. Método Para la Determinación de Staphylococcus aureus en Alimentos. 1994. Available online: https://www.dof.gob.mx/nota_detalle.php?codigo=4882018&fecha=25/09/1995#gsc.tab=0 (accessed on 27 August 2024).
- González-Córdova, A.F.; Yescas, C.; Ortiz-Estrada, A.M.; De la Rosa-Alcaraz, M.d.l.A.; Hernández-Mendoza, A.; Vallejo-Cordoba, B. Invited review: Artisanal Mexican cheeses. J. Dairy. Sci. 2016, 99, 3250–3262. [Google Scholar] [CrossRef]
- Cuevas-González, P.F.; Heredia-Castro, P.Y.; Méndez-Romero, J.I.; Hernández-Mendoza, A.; Reyes-Díaz, R.; Vallejo-Cordoba, B.; González-Córdova, A.F. Artisanal Sonoran cheese (Cocido cheese): An exploration of its production process, chemical composition and microbiological quality. J. Sci. Food Agric. 2017, 97, 4459–4466. [Google Scholar] [CrossRef]
- Dapkevicius, M.d.L.E.; Sgardioli, B.; Câmara, S.P.A.; Poeta, P.; Malcata, F.X. Current Trends of Enterococci in Dairy Products: A Comprehensive Review of Their Multiple Roles. Foods 2021, 10, 821. [Google Scholar] [CrossRef] [PubMed]
- Altschul, S.F.; Gish, W.; Miller, W.; Myers, E.W.; Lipman, D.J. Basic local alignment search tool. J. Mol. Biol. 1990, 215, 403–410. [Google Scholar] [CrossRef] [PubMed]
- Garafutdinov, R.R.; Galimova, A.A.; Sakhabutdinova, A.R. The influence of quality of primers on the formation of primer dimers in PCR. Nucleos Nucleot Nucl. 2020, 39, 1251–1269. [Google Scholar] [CrossRef]
- Terzić-Vidojević, A.; Veljović, K.; Popović, N.; Tolinački, M.; Golić, N. Enterococci from Raw-Milk Cheeses: Current Knowledge on Safety, Technological, and Probiotic Concerns. Foods 2021, 10, 2753. [Google Scholar] [CrossRef] [PubMed]
- Mendonça, J.F.M.d.; Vieira, F.d.O.; Fonseca, I.; Ribeiro, J.B.; Arcuri, E.F.; Borges, M.d.F.; Borges, C.A.V.; Sá, J.F.O.d.; Martins, M.F. Detection of viable Salmonella Typhimurium and Staphylococcus aureus in Coalho Cheese by Real-Time PCR. Food Sci. Technol. 2019, 39, 690–696. [Google Scholar] [CrossRef]
- Kralik, P.; Ricchi, M. A Basic Guide to Real Time PCR in Microbial Diagnostics: Definitions, Parameters, and Everything. Front. Microbiol. 2017, 8, 108. [Google Scholar] [CrossRef]
- Kadiroğlu, P.; Korel, F.; Ceylan, C. Quantification of Staphylococcus aureus in white cheese by the improved DNA extraction strategy combined with TaqMan and LNA probe-based qPCR. J. Microbiol. Methods 2014, 105, 92–97. [Google Scholar] [CrossRef]
- Heo, E.J.; Kim, H.-Y.; Suh, S.H.; Moon, J.S. Comparison of DNA Extraction Methods for the Quantification of Listeria monocytogenes in Dairy Products by Real-Time Quantitative PCR. J. Food Prot. 2022, 85, 1531–1537. [Google Scholar] [CrossRef]
- Garrido-Maestu, A.; Azinheiro, S.; Carvalho, J.; Espiña, B.; Prado, M. Evaluation and implementation of commercial antibodies for improved nanoparticle-based immunomagnetic separation and real-time PCR for faster detection of Listeria monocytogenes. J. Food Sci. Technol. 2020, 57, 4143–4151. [Google Scholar] [CrossRef]
- NOM-114-SSA1; Mexican Official Standard. Bienes y Servicios. Método Para la Determinación de Salmonella en Alimentos. 1994. Available online: https://www.dof.gob.mx/nota_detalle.php?codigo=4881851&fecha=22/09/1995#gsc.tab=0 (accessed on 27 August 2024).
- NOM-143-SSA1; Mexican Official Standard. Bienes y Servicios. Método de Prueba Microbiológico Para Alimentos. Determinación de Listeria Monocytogenes. 1995. Available online: https://www.dof.gob.mx/nota_detalle.php?codigo=4901269&fecha=19/11/1997#gsc.tab=0 (accessed on 16 July 2024).
- Lucena-Aguilar, G.; Sánchez-López, A.M.; Barberán-Aceituno, C.; Carrillo-Ávila, J.A.; López-Guerrero, J.A.; Aguilar-Quesada, R. DNA Source Selection for Downstream Applications Based on DNA Quality Indicators Analysis. Biopreserv. Biobank. 2016, 14, 264–270. [Google Scholar] [CrossRef] [PubMed]
- Vargas-Albores, F.; Ortiz-Suárez, L.E.; Villalpando-Canchola, E.; Martínez-Porchas, M. Size-variable zone in V3 region of 16S rRNA. RNA Biol. 2017, 14, 1514–1521. [Google Scholar] [CrossRef] [PubMed]
- Kang, T.S. Basic principles for developing real-time PCR methods used in food analysis: A review. Trends Food Sci. Tech. 2019, 91, 574–585. [Google Scholar] [CrossRef]
- Alessandria, V.; Rantsiou, K.; Dolci, P.; Cocolin, L. Molecular methods to assess Listeria monocytogenes route of contamination in a dairy processing plant. Int. J. Food Microbiol. 2010, 31, S156–S162. [Google Scholar] [CrossRef]
- Barría, C.; Singer, R.S.; Bueno, I.; Estrada, E.; Rivera, D.; Ulloa, S.; Fernández, J.; Mardones, F.O.; Moreno-Switt, A.I. Tracing Listeria monocytogenes contamination in artisanal cheese to the processing environments in cheese producers in southern Chile. Food Microbiol. 2020, 90, 103499. [Google Scholar] [CrossRef]
- Bastam, M.M.; Jalili, M.; Pakzad, I.; Maleki, A.; Ghafourian, S. Pathogenic bacteria in cheese, raw and pasteurised milk. Vet. Med. Sci. 2021, 7, 2445–2449. [Google Scholar] [CrossRef]
- World Health Organization, WHO. Listeriosis. Available online: https://www.who.int/news-room/fact-sheets/detail/listeriosis (accessed on 15 July 2024).
- Pal, M.; Awel, H. Public Health Significance of Listeria monocytogenes in Milk and Milk Products: An Overview. J. Vet. Public Health 2014, 12, 01–05. [Google Scholar]
- Hossain, M.I.; Mizan, M.I.; Hossain, M.F.R.; Roy, P.K.; Nahar, S.; Toushik, S.H.; Ashrafudoulla, M.; Jahid, I.K.; Lee, J.; Ha, S. Listeria monocytogenes biofilm inhibition on food contact surfaces by application of postbiotics from Lactobacillus curvatus B.67 and Lactobacillus plantarum M.2. Food Res. Int. 2021, 148, 110595. [Google Scholar] [CrossRef]
- Herd Verification Program. Servicio Nacional de Sanidad, Inocuidad y Calidad Agroalimentaria (SENASICA). México. 2021. Available online: https://www.gob.mx/senasica/documentos/constatacion-de-hatos-programa-de-hatos-libres (accessed on 16 July 2024).
- NOM-041-ZOO. Mexican Official Standard. National Campaign against Brucellosis in Animal. 1995. Available online: https://www.dof.gob.mx/nota_detalle.php?codigo=4896374&fecha=20/08/1996#gsc.tab=0 (accessed on 13 August 2024).
- Saber Marouf, A.; Hanifian, S.; Shayegh, J. Prevalence of Brucella spp. in raw milk and artisanal cheese tested via real-time qPCR and culture assay. Int. J. Food Microbiol. 2021, 347, 109192. [Google Scholar] [CrossRef]
- Rios, E.A.; Ramos-Pereira, J.; Santos, J.A.; López-Díaz, T.M.; Otero, A.; Rodríguez-Calleja, J.M. Behaviour of Non-O157 STEC and Atypical EPEC during the Manufacturing and Ripening of Raw Milk Cheese. Foods 2020, 9, 1215. [Google Scholar] [CrossRef]
- Food and Drug Administration, FDA. Final Qualitative Assessment of Risk to Public Health from an On-Farm Contamination of Produce. 2015. Available online: https://www.fda.gov/media/116766/download (accessed on 16 July 2024).
- Centers for Disease Control and Prevention, CDC. Data Summary: Persistent Strain of E. coli O157:H7 (REPEXH01) Linked to Multiple Sources. 2024. Available online: https://www.cdc.gov/ecoli/rep-strain/ (accessed on 16 July 2024).
- Currie, A.; Galanis, E.; Chacon, P.A.; Murray, R.; Wilcott, L.; Kirkby, P.; Honish, L.; Franklin, K.; Farber, J.; Parker, R.; et al. Outbreak of Escherichia coli O157:H7 Infections Linked to Aged Raw Milk Gouda Cheese, Canada. J. Food Prot. 2018, 81, 325–331. [Google Scholar] [CrossRef] [PubMed]
- World Health Organization, WHO. E coli. 2018. Available online: https://www.who.int/news-room/fact-sheets/detail/e-coli (accessed on 16 July 2024).
- García-Cano, I.; Serrano-Maldonado, C.E.; Olvera-García, M.; Delgado-Arciniega, E.; Peña-Montes, C.; Mendoza-Hernández, G.; Quirasco, M. Antibacterial activity produced by Enterococcus spp. isolated from an artisanal Mexican dairy product, Cotija cheese. LWT-Food Sci. Technol. 2014, 59, 26–34. [Google Scholar] [CrossRef]
- Bravo-Mendoza, A. Study of Microbial Populations of Biotechnological Interest Isolated from Cotija Cheese. Undergraduate Thesis, School of Chemistry, National Autonomous University of Mexico, Mexico City, México, 2008. Available online: https://tesiunam.dgb.unam.mx/F?current_base=TES01&func=direct&doc_number=000646540 (accessed on 16 July 2024).
- García-Saturnino, V. Isolation of Microorganisms with a Higher Lipolytic Activity from Cotija Cheese. Undergraduate Thesis, School of Chemistry, National Autonomous University of Mexico, Mexico City, México, 2006. Available online: https://tesiunam.dgb.unam.mx/F/K8PFECHQLBQP66NVY7F5IG88HU3M1YH6B7RIY7CQL5CYA92137-05282?func=full-set-set&set_number=216357&set_entry=000068&format=999 (accessed on 26 August 2024).
- Aldrete-Tapia, A.; Escobar-Ramírez, C.M.; Tamplin, M.L.; Hernández-Iturriaga, M. Characterization of Bacterial Communities in Mexican Artisanal Raw Milk “Bola de Ocosingo” Cheese by High-Throughput Sequencing. Front. Microbiol. 2018, 9, 417280. [Google Scholar] [CrossRef]
- European Food Safety Authority, EFSA. Control of Salmonella. Available online: https://food.ec.europa.eu/safety/biological-safety/food-borne-diseases-zoonoses/control-salmonella_en (accessed on 16 July 2024).
- Hennekinne, J.A.; De Buyser, M.L.; Dragacci, S. Staphylococcus aureus and its food poisoning toxins: Characterization and outbreak investigation. FEMS Microbiol. Rev. 2012, 36, 815–836. [Google Scholar] [CrossRef]
- Radoslava, S.; Nemanja, Z.; Branko, V. Occurrence and Characterization of Enterotoxigenic Staphylococci Isolated from Soft Cheeses in Serbia. Acta Vet. 2020, 70, 238–254. [Google Scholar] [CrossRef]

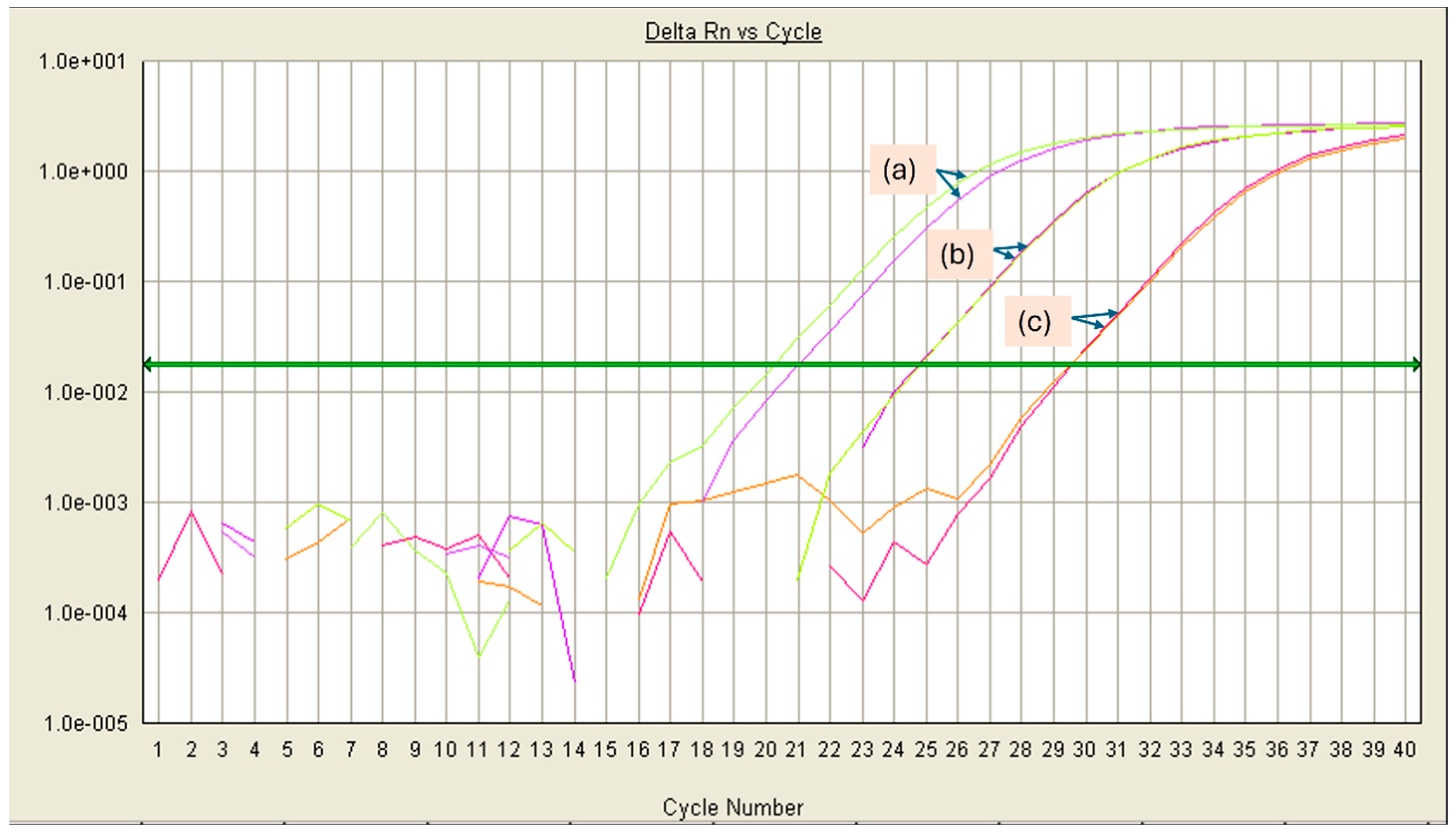
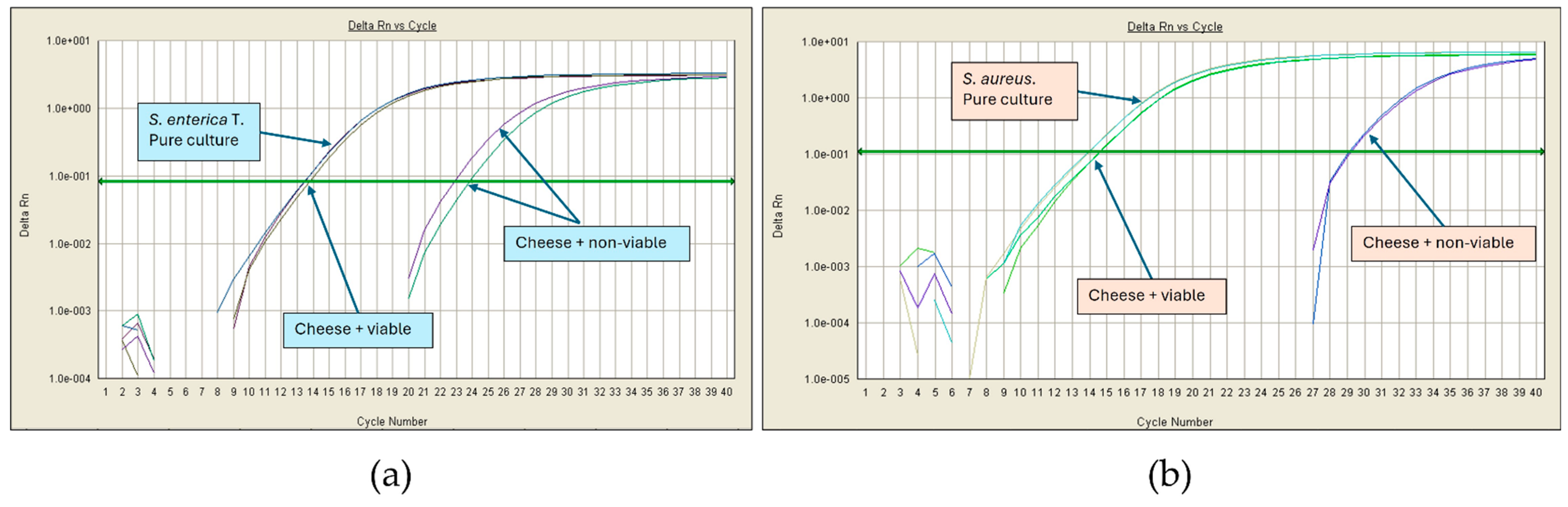

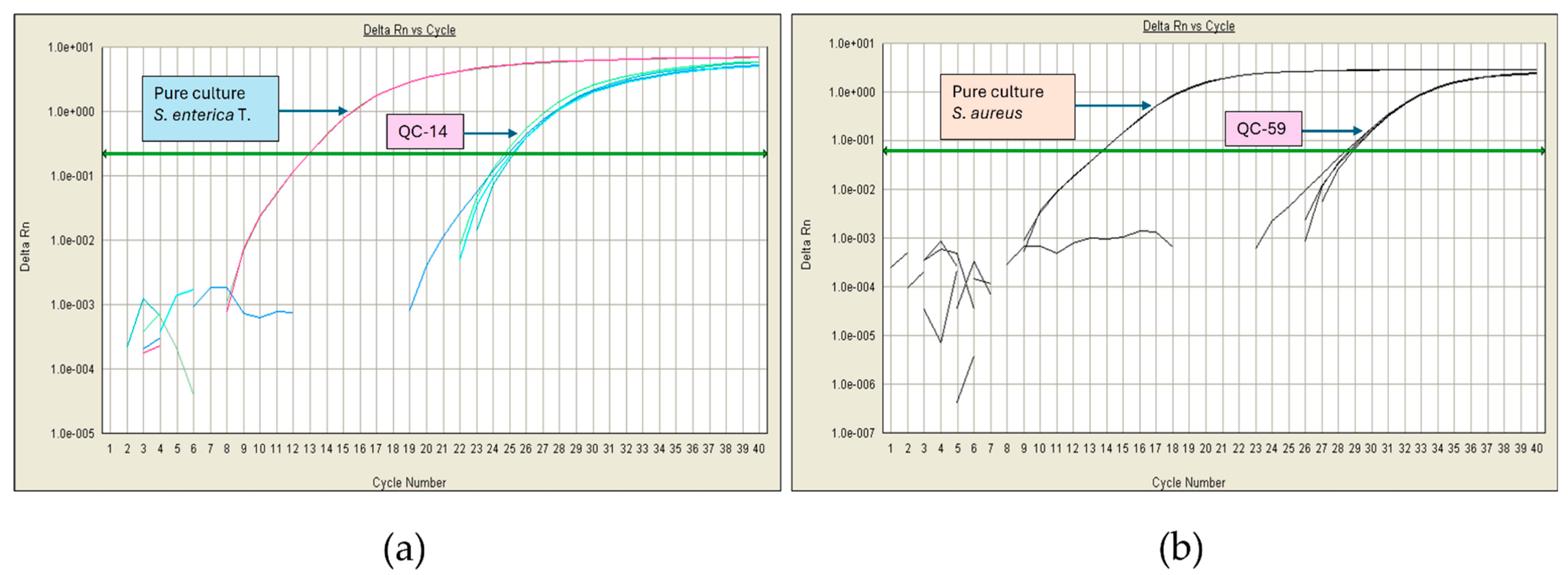

| Microorganism | Enrichment Broth | Culture Conditions |
|---|---|---|
| Salmonella spp. | Buffered peptone water | 37 °C, 24 h, 250 rpm |
| S. aureus | Nutrient broth (OXOID, Basingstoke, UK) | 37 °C, 48 h, 250 rpm |
| L. monocytogenes | Fraser broth (DIFCO, Detroit, MI, USA) | 37 °C, 48 h, 250 rpm |
| Brucella spp. | Brucella broth (DIFCO, Detroit, MI, USA) + amphotericin B (1 mg/mL) (Sigma-Aldrich, St. Louis, MO, USA), and vancomycin (20 mg/mL) (Sigma-Aldrich, St. Louis, MO, USA) | 37 °C, 7 days, static |
| E. coli ETEC and E. coli O157:H7 | BHI (DIFCO, Detroit, MI, USA) + casamino acids (2%) (OXOID, Basingstoke, UK) | 37 °C, 24 h, static |
| Microorganism | Target Gene | Nucleotide Sequences 5′ → 3′ | Observations/Reference |
|---|---|---|---|
| Salmonella enterica | invA (invasion protein) | Fw-ACCGTGGTCCAGTTTATCGTTATT Rv-GGGCATACCATCCAGAGAAAATCG FAM-TCCGCGACACGTTCTG | This work. Target gene reported by Malorny et al. [19] |
| Staphylococcus aureus | nucA (staphylococcal thermonuclease) | Fw-CCTGAAGCAAGTGCATTTACGAAAA Rv-CGCTAAGCCACGTCCATATTTATCA FAM-CTCGACTTCAATTTTC | This work. Target gene reported by Ruiz-Pérez et al. [20] |
| Listeria monocytogenes | hly (listeriolysin) | Fw-AAGGTGCTACTTTTAACCGGGAAA Rv-CATTGTCTTTTAAGAAGTTTGTTGTATAGGCA FAM-CACCAGGAGTTCCC | This work. Target gene reported by Kim et al. [21] |
| Brucella spp. | per (perosamine synthetase) | Fw-GTTTAGTTTCTTTGGGAACAAGACAA Rv-GAGGATTGCGCGCTAGCA FAM-TACGACCGGTGAAGGCGGGATG | Individual synthesis of primers and probe as reported by Bounaadja et al. [22] |
| Escherichia coli ETEC | eltBI (thermolabile toxin subunit B) | Fw-GAGTACTTCGATAGAGGAACTCAAATGAAT Rv-TCATCATATCTGACAAAGCCGGTTT FAM-CCTCTCGCGTGATCAT | This work. Target gene reported by Nada et al. [23] |
| Escherichia coli O157:H7 | eae (intimin) | Fw-CATTGATCAGGATTTTTCTGGTGATA Rv-CTCATGCGGAAATAGCCGTTA VIC-ATAGTCTCGCCAGTATTCGCCACCAATACC | Individual synthesis of primers and probe as reported by the European Food Safety Authority [24] |
| Bacterium (Target Gene) | S. enterica Typhimurium | S. aureus | L. monocytogenes | B. abortus | E. coli ETEC | E. coli O157:H7 | E. faecalis | E. faecium | L. fermentum | E. coli DH5α |
|---|---|---|---|---|---|---|---|---|---|---|
| Salmonella spp. (invA) | 13.59 ± 0.23 | >39 | >39 | >36 | >36 | >35 | >33 | >33 | >33 | >32 |
| S. aureus (nucA) | >32 | 13.95 ± 0.53 | >33 | >33 | >31 | >31 | >33 | >37 | >37 | >33 |
| L. monocytogenes (hly) | >34 | >30 | 13.83 ± 0.70 | >35 | NA | NA | NA | >39 | >37 | >38 |
| Brucella spp. (per) | NA | NA | NA | 12.20 ± 0.09 | NA | NA | NA | NA | NA | NA |
| E. coli ETEC (eltBI) | >34 | >30 | >30 | >36 | 11.57 ± 0.29 | >30 | >33 | >35 | >37 | NA |
| E. coli O157:H7 (eae) | >34 | >31 | >30 | >36 | >32 | 11.72 ± 0.69 | >31 | >34 | >37 | NA |
| Microorganism | Ct (DNA from Pure Bacterium Culture) | Ct (DNA from the Bacterium Spiked in Cheese) |
|---|---|---|
| S. enterica | 13.59 ± 0.23 | 13.55 ± 0.24 |
| S. aureus | 13.95 ± 0.53 | 14.53 ± 0.02 |
| L. monocytogenes | 13.83 ± 0.70 | 13.99 ± 0.84 |
| B. abortus | 12.20 ± 0.09 | 13.17 ± 0.71 |
| E. coli ETEC | 11.57 ± 0.29 | 11.35 ± 0.65 |
| E. coli O157:H7 | 11.72 ± 0.69 | 13.08 ± 1.15 |
| Microorganism | Number of Positive Samples/Total (%) | |
|---|---|---|
| Cotija Cheese | Bola de Ocosingo Cheese | |
| S. enterica | 10/95 (10.5%) | 0/16 |
| S. aureus | 13/95 (13.7%) | 0/16 |
| L. monocytogenes | 0/95 | 0/16 |
| Brucella spp. | 0/95 | 0/16 |
| E. coli ETEC | 0/95 | 0/16 |
| E. coli O157:H7 | 0/95 | 0/16 |
| Sample Identifier | Salmonella spp. Ct Value | S. aureus Ct Value |
|---|---|---|
| QC-10 | >30 | 28.92 ± 0.77 |
| QC-11 | 24.76 ± 0.85 | >30 |
| QC-12 | 23.54 ± 0.14 | >30 |
| QC-13 | 25.23 ± 0.90 | >30 |
| QC-14 | 23.85 ± 0.21 | >30 |
| QC-15 | 26.03 ± 0.52 | 28.15 ± 0.21 |
| QC-16 | 24.75 ± 0.37 | >30 |
| QC-17 | 23.78 ± 0.15 | >30 |
| QC-18 | 24.17 ± 0.49 | >30 |
| QC-19 | 27.93 ± 0.63 | >30 |
| QC-20 | 26.17 ± 0.63 | >30 |
| QC-53 | >30 | 29.31 ± 2.39 |
| QC-56 | >30 | 26.77 ± 0.21 |
| QC-59 | >30 | 28.61 ± 0.20 |
| QC-61 | >30 | 29.34 ± 1.83 |
| QC-70 | >30 | 29.45 ± 0.16 |
| QC-72 | >30 | 27.46 ± 1.98 |
| QC-76 | >30 | 28.94 ± 0.80 |
| QC-77 | >30 | 29.17 ± 0.71 |
| QC-82 | >36 | 29.43 ± 0.48 |
| QC-85 | >36 | 28.62 ± 1.13 |
| QC-93 | >30 | 29.20 ± 0.80 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Estrada-Hernández, C.A.; Becerra-Cedillo, M.B.; Hernández Velázquez, I.A.; Mejía-Buenfil, H.E.; Olivera-Martínez, T.; Salto-González, I.B.; Torres-López, F.; Quirasco, M. Microbiological Evaluation of Two Mexican Artisanal Cheeses: Analysis of Foodborne Pathogenic Bacteria in Cotija Cheese and Bola de Ocosingo Cheese by qPCR. Foods 2024, 13, 2824. https://doi.org/10.3390/foods13172824
Estrada-Hernández CA, Becerra-Cedillo MB, Hernández Velázquez IA, Mejía-Buenfil HE, Olivera-Martínez T, Salto-González IB, Torres-López F, Quirasco M. Microbiological Evaluation of Two Mexican Artisanal Cheeses: Analysis of Foodborne Pathogenic Bacteria in Cotija Cheese and Bola de Ocosingo Cheese by qPCR. Foods. 2024; 13(17):2824. https://doi.org/10.3390/foods13172824
Chicago/Turabian StyleEstrada-Hernández, Cindy Adriana, María Belén Becerra-Cedillo, Irma Angélica Hernández Velázquez, Hermann E. Mejía-Buenfil, Tania Olivera-Martínez, I. Berenice Salto-González, Frida Torres-López, and Maricarmen Quirasco. 2024. "Microbiological Evaluation of Two Mexican Artisanal Cheeses: Analysis of Foodborne Pathogenic Bacteria in Cotija Cheese and Bola de Ocosingo Cheese by qPCR" Foods 13, no. 17: 2824. https://doi.org/10.3390/foods13172824
APA StyleEstrada-Hernández, C. A., Becerra-Cedillo, M. B., Hernández Velázquez, I. A., Mejía-Buenfil, H. E., Olivera-Martínez, T., Salto-González, I. B., Torres-López, F., & Quirasco, M. (2024). Microbiological Evaluation of Two Mexican Artisanal Cheeses: Analysis of Foodborne Pathogenic Bacteria in Cotija Cheese and Bola de Ocosingo Cheese by qPCR. Foods, 13(17), 2824. https://doi.org/10.3390/foods13172824







