Rapid Measurement of Total Saponins, Mannitol, and Naringenin in Dendrobium officinale by Near-Infrared Spectroscopy and Chemometrics
Abstract
:1. Introduction
2. Materials and Methods
2.1. Materials and Reagents
2.2. NIR Spectral Acquisition
2.3. Reference Assays
2.3.1. Determination of Total Saponins
2.3.2. Determination of Mannitol
2.3.3. Determination of Naringenin
2.4. Chemometrics
2.4.1. Spectral Data Processing
- (1)
- The average spectrum of the calibration sample was calculated.
- (2)
- The linear regression between the spectrum of each calibration sample and the average spectrum was performed.
- (3)
- MSC transforms the spectrum as follows:
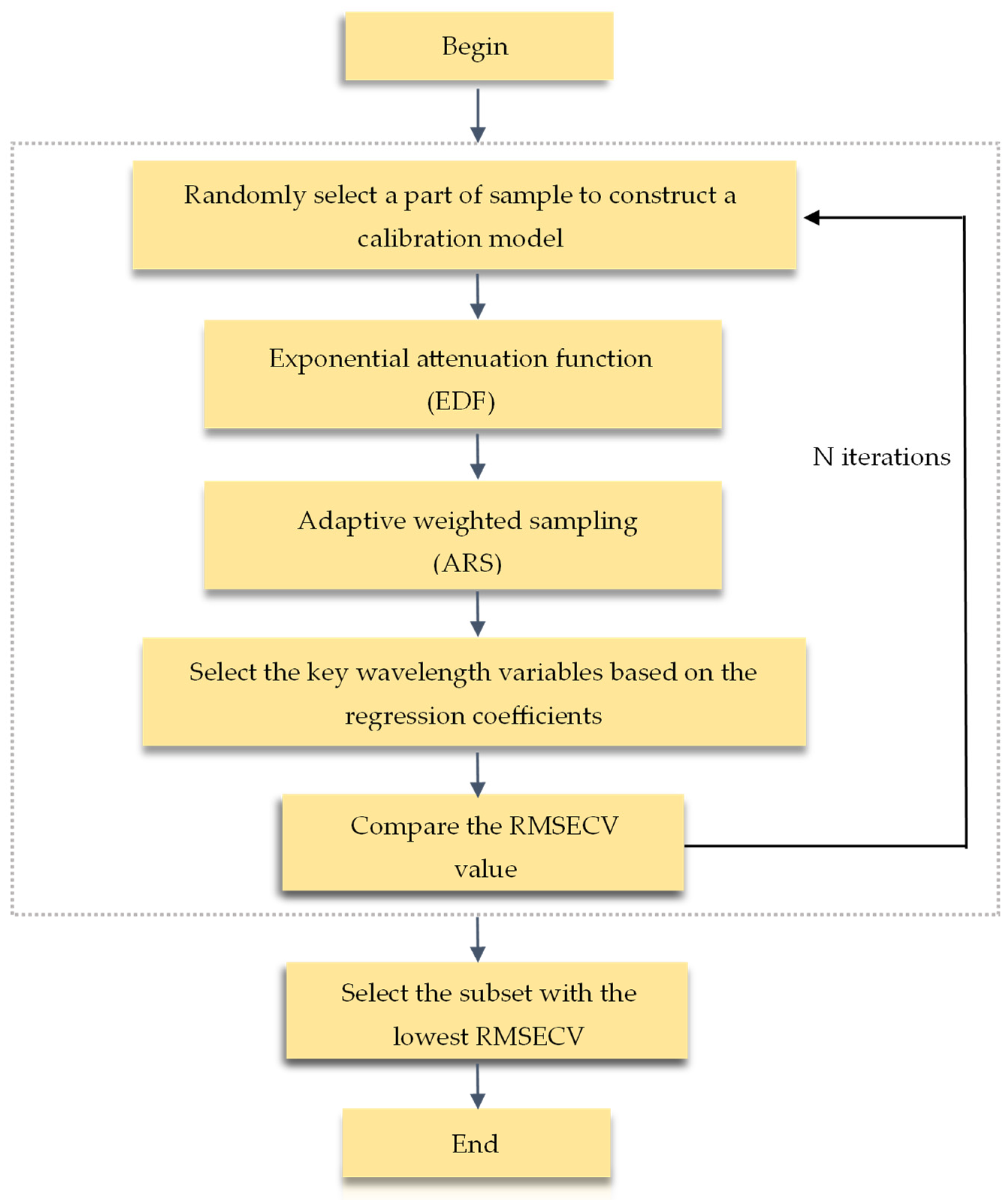
2.4.2. Wavelength Selection Methods
2.5. Model Performance Evaluation
3. Results and Discussion
3.1. Spectral Feature Analysis
3.2. Outlier Detection and Sample Partition
3.3. PLS Models Based on Various Wavelength Selection Methods
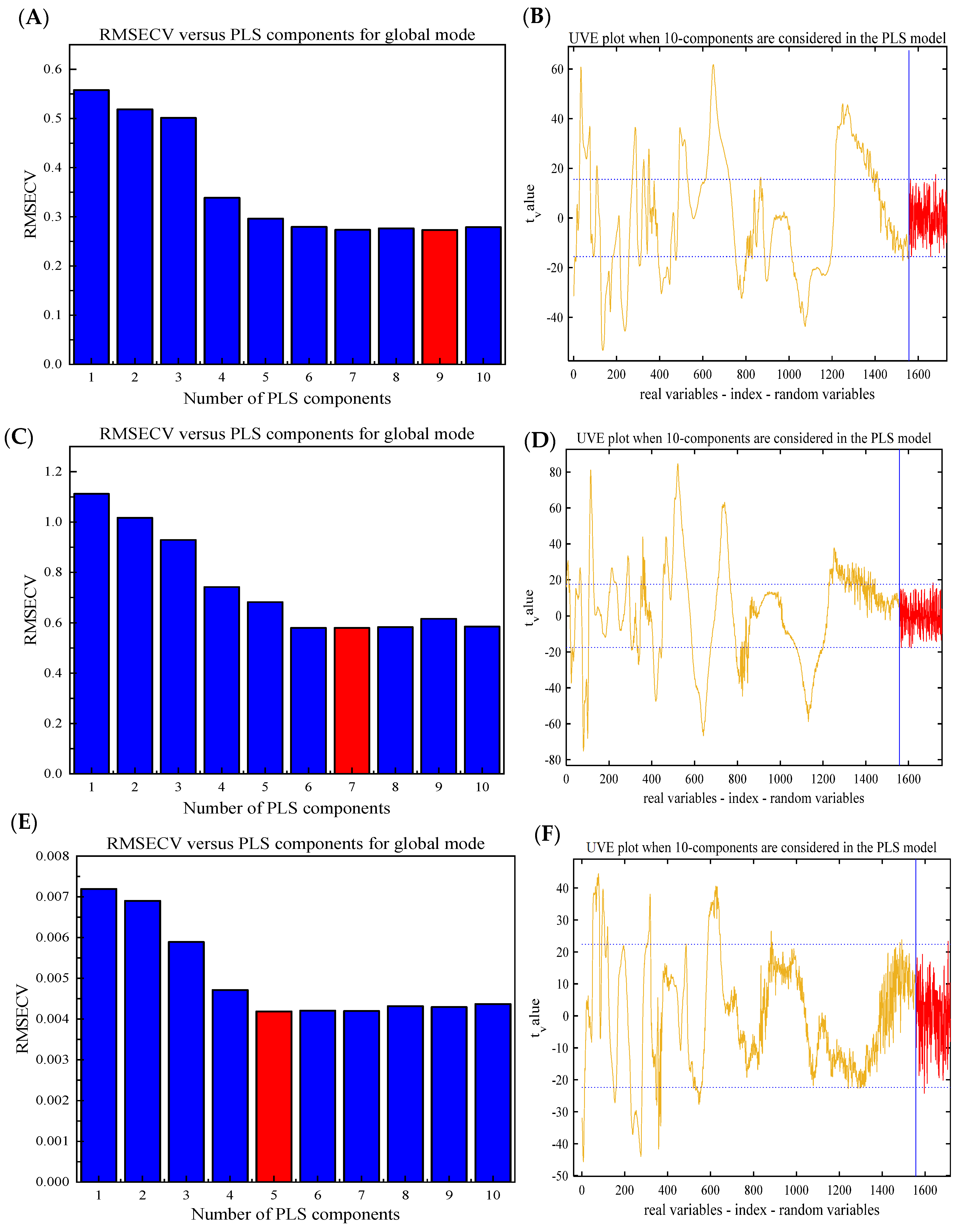
3.3.1. Results of the Full-PLS Model
3.3.2. Results of the UVE-PLS Model
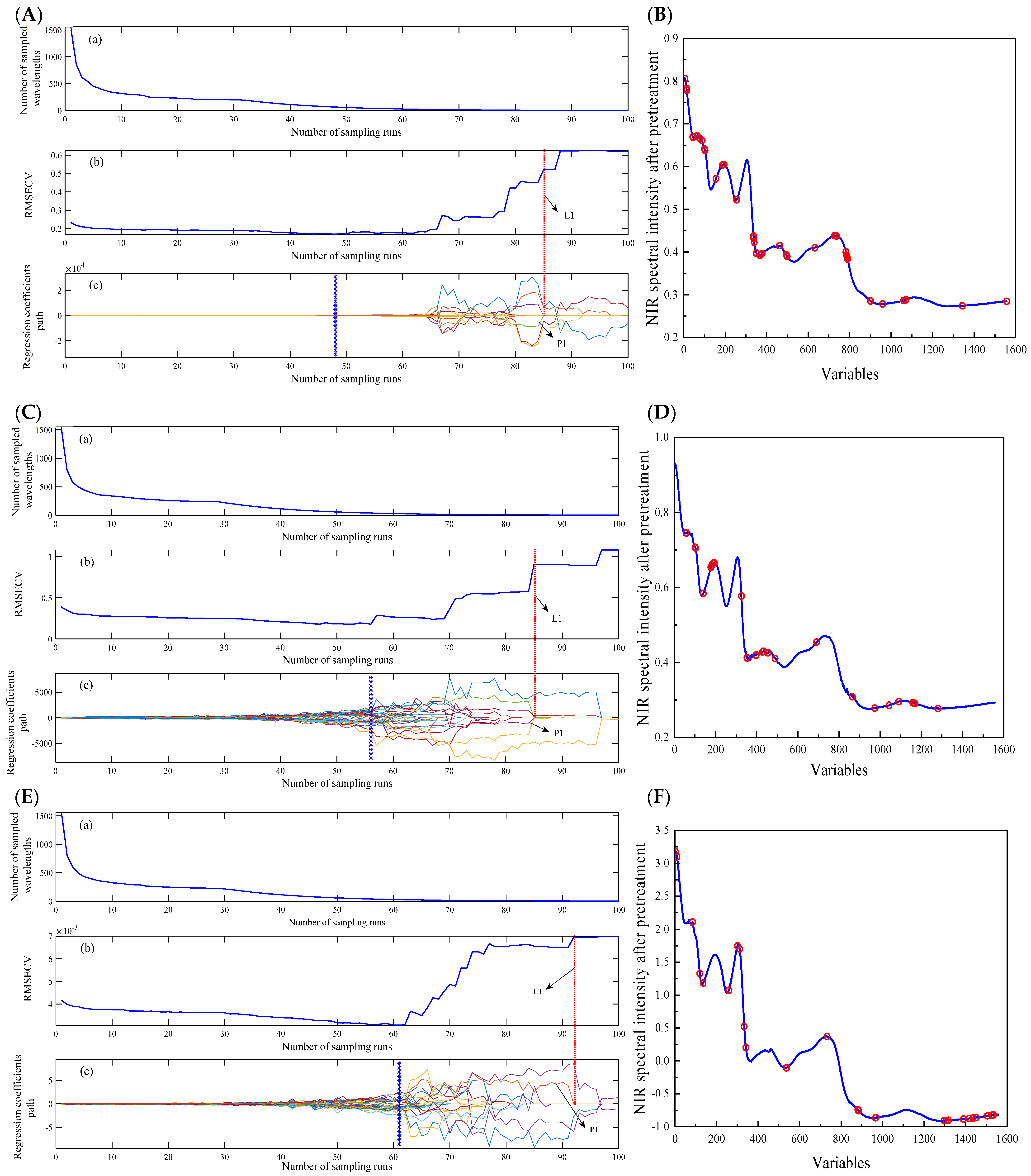
3.3.3. Results of the CARS-PLS Model
3.3.4. Discussion of Results
4. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Yuan, Y.D.; Zuo, J.J.; Zhang, H.Y.; Zu, M.T.; Yu, M.Y.; Liu, S. Transcriptome and metabolome profiling unveil the accumulation of flavonoids in Dendrobium officinale. Genomics 2022, 114, 110324. [Google Scholar] [CrossRef]
- He, Y.; Li, L.; Chang, H.; Cai, B.; Gao, H.J.; Chen, G.Y.; Hou, W.; Jappar, Z.; Yan, Y.Z. Research progress on extraction, purification, structure and biological activity of Dendrobium officinale polysaccharides. Front. Nutr. 2022, 9, 965073. [Google Scholar] [CrossRef] [PubMed]
- Chen, W.Q.; Chen, X.Y.; Xu, J.F.; Cai, J.; Wang, X.F. Identification of Dendrobium officinale Using DNA Barcoding Method Combined with HRM and qPCR Technology. Food Anal. Method 2022, 15, 1310–1320. [Google Scholar] [CrossRef]
- Kuang, M.T.; Li, J.Y.; Yang, X.B.; Yang, L.; Xu, J.Y.; Yan, S.; Lv, Y.F.; Ren, F.C.; Hu, J.M.; Zhou, J. Structural characterization and hypoglycemic effect via stimulating glucagon-like peptide-1 secretion of two polysaccharides from Dendrobium officinale. Carbohydr. Polym. 2020, 241, 116326. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.C.; Fan, J.K.; Luo, H.; Liang, Z.W.; Guan, Y.H.; Lei, X.; Bo, N.G.; Zhao, M. Alleviation of Alcoholic Fatty Liver by Dendrobium officinale Flower Extracts due to Regulation of Gut Microbiota and Short-Chain Fatty Acids in Mice Exposed to Chronic Alcohol. Foods 2023, 12, 1428. [Google Scholar] [CrossRef] [PubMed]
- Liang, K.L.; Fang, P.; Shi, Q.Q.; Su, J.; Li, B.; Chen, S.H.; Lv, G.Y. Antihypertensive effect and mechanism of Dendrobium officinale flos on high-blood pressure rats induced by high glucose and high fat compound alcohol. China J. Chin. Mater. Med. 2018, 43, 147–153. [Google Scholar] [CrossRef]
- Meng, Y.J.; Wang, Y.W.; Zhang, L.; Li, J.Y.; Hu, L.; Wu, Z.H.; Yang, L.E.; Wei, G.; Huang, Y.C. Identification of bibenzyls and evaluation of imitative wild planting techniques in Dendrobium officinale by HPLC-ESI-MSn. J. Mass Spectrom. 2023, 58, e4903. [Google Scholar] [CrossRef] [PubMed]
- Yang, J.; Kuang, M.T.; Yang, L.; Huang, W.; Hu, J.M. Modern interpretation of the traditional application of Shihu—A comprehensive review on phytochemistry and pharmacology progress of Dendrobium officinale. J. Ethnopharmacol. 2023, 302, 115912. [Google Scholar] [CrossRef] [PubMed]
- Fang, C.; Xin, G.Z.; Wang, S.L.; Wei, M.M.; Wu, P.; Dong, X.M.; Song, G.Q.; Xie, T.; Zhou, J.L. Discovery and validation of peptide biomarkers for discrimination of Dendrobium species by label-free proteomics and chemometrics. J. Pharm. Biomed. Anal. 2020, 182, 113118. [Google Scholar] [CrossRef]
- Wu, W.J.; Zhao, Z.Q.; Zhao, Z.E.; Zhang, D.D.; Zhang, Q.Y.; Zhang, J.Y.; Fang, Z.Y.; Bai, Y.L.; Guo, X.H. Structure, Health Benefits, Mechanisms, and Gut Microbiota of Dendrobium officinale Polysaccharides: A Review. Nutrients 2023, 15, 4901. [Google Scholar] [CrossRef]
- Miao, X.S.; Cui, Q.Y.; Wang, Z.Y.; Liu, X.N.; Zhao, A.B.; Qiao, Y.J.; Wu, Z.S. Advance in quality assessment of Chinese materia medica using microscopic and morphological methods. Chin. J. Nat. Med. 2017, 15, 653–663. [Google Scholar] [CrossRef] [PubMed]
- Lin, T.; Chen, X.L.; Wu, G.W.; Sha, L.J.; Wang, J.; Hu, Z.J.; Liu, H.C. A simple method for distinguishing Dendrobium devonianum and Dendrobium officinale by ultra performance liquid chromatography-photo diode array detector. Food Sci. Technol. 2023, 43, e110122. [Google Scholar] [CrossRef]
- Liang, X.M.; Jin, Y.; Wang, Y.P.; Jin, G.W.; Fu, Q.; Xiao, Y.S. Qualitative and quantitative analysis in quality control of traditional Chinese medicines. J. Chromatogr. A. 2009, 1216, 2033–2044. [Google Scholar] [CrossRef] [PubMed]
- Long, W.J.; Hu, Z.K.; Wei, L.N.; Chen, H.Y.; Liu, T.K.; Wang, S.Y.; Guan, Y.T.; Yang, X.L.; Yang, J.; Fu, H.Y. Accurate identification of the geographical origins of lily using near-infrared spectroscopy combined with carbon dot-tetramethoxyporphyrin nanocomposite and chemometrics. Spectrochim. Acta A Mol. Biomol. Spectrosc. 2022, 271, 120932. [Google Scholar] [CrossRef] [PubMed]
- del Río Celestino, M.; Font, R. Using Vis-NIR Spectroscopy for Predicting Quality Compounds in Foods. Sensors 2022, 22, 4845. [Google Scholar] [CrossRef] [PubMed]
- Ji, Q.G.; Li, C.F.; Fu, X.S.; Liao, J.Y.; Hong, X.Z.; Yu, X.P.; Ye, Z.H.; Zhang, M.Z.; Qiu, Y.L. Protected Geographical Indication Discrimination of Zhejiang and Non-Zhejiang Ophiopogonis japonicus by Near-Infrared (NIR) Spectroscopy Combined with Chemometrics: The Influence of Different Stoichiometric and Spectrogram Pretreatment Methods. Molecules 2023, 28, 2803. [Google Scholar] [CrossRef] [PubMed]
- Bian, X.H.; Zhang, R.L.; Liu, P.; Xiang, Y.; Wang, S.Y.; Tan, X.Y. Near infrared spectroscopic variable selection by a novel swarm intelligence algorithm for rapid quantification of high order edible blend oil. Spectrochim. Acta A Mol. Biomol. Spectrosc. 2023, 284, 121788. [Google Scholar] [CrossRef] [PubMed]
- Li, H.N.; Li, M.G.; Tang, H.S.; Li, H.; Zhang, T.L.; Yang, X.F. Quantitative analysis of phenanthrene in soil by fluorescence spectroscopy coupled with the CARS-PLS model. RSC Adv. 2023, 13, 9353–9360. [Google Scholar] [CrossRef]
- Sharma, P.; Tyagi, A.; Bhansali, P.; Pareek, S.; Singh, V.; Ilyas, A.; Mishra, R.; Poddar, N.K. Saponins: Extraction, bio-medicinal properties and way forward to anti-viral representatives. Food Chem. Toxicol. 2021, 150, 112075. [Google Scholar] [CrossRef]
- Dai, Y.W.; Meng, Q.; Mu, W.M.; Zhang, T. Recent advances in the applications and biotechnological production of mannitol. J. Funct. Foods 2017, 36, 404–409. [Google Scholar] [CrossRef]
- Cai, J.; Wen, H.L.; Zhou, H.; Zhang, D.; Lan, D.F.; Liu, S.P.; Li, C.Y.; Dai, X.F.; Song, T.; Wang, X.Y.; et al. Naringenin: A flavanone with anti-inflammatory and anti-infective properties. Biomed. Pharmacother. 2023, 164, 114990. [Google Scholar] [CrossRef]
- Ma, L.H.; Zhang, Z.M.; Zhao, X.B.; Zhang, S.F.; Lu, H.M. The rapid determination of total polyphenols content and antioxidant activity in Dendrobium officinale using near-infrared spectroscopy. Anal. Methods 2016, 8, 4584–4589. [Google Scholar] [CrossRef]
- Yun, Y.H.; Wei, Y.C.; Zhao, X.B.; Wu, W.J.; Liang, Y.Z.; Lu, H.M. A green method for the quantification of polysaccharides in Dendrobium officinale. RSC Adv. 2015, 5, 105057–105065. [Google Scholar] [CrossRef]
- Guo, S.B.; Du, X.M.; Jian, L.Y. Studies on Purification Process of Total Saponins in Radix Astragali with Resin and Structural Identification of Compounds. Asian J. Chem. 2014, 26, 4610–4614. [Google Scholar] [CrossRef]
- Chen, M.; Zhang, W.L.; Wu, H.; Guang, C.; Mu, W.M. Mannitol: Physiological functionalities, determination methods, biotechnological production, and applications. Appl. Microbiol. Biotechnol. 2020, 104, 6941–6951. [Google Scholar] [CrossRef]
- Zhou, G.F.; Chen, S.H.; Lv, G.Y.; Yan, M.Q. Determination of naringenin in Dendrobium officinale by HPLC. China J. Chin. Mater. Med. 2013, 38, 520–523. [Google Scholar]
- Martens, H.; Nielsen, J.P.; Engelsen, S.B. Light scattering and light absorbanceseparated by extended multiplicative signal correction. Application to near-infrared transmission analysis of powder mixtures. Anal. Chem. 2003, 75, 394–404. [Google Scholar] [CrossRef]
- Wang, H.B.; Chen, P.; Dai, J.W.; Liu, D.; Li, J.Y.; Xu, Y.P.; Chu, X.L. Recent advances of chemometric calibration methods in modern spectroscopy: Algorithms, strategy, and related issues. Trends Anal. Chem. 2022, 153, 116648. [Google Scholar] [CrossRef]
- Lotfi, M.; Arab Chamjangali, M.; Mozafari, Z. Ridge regression coupled with a new uninformative variable elimination algorithm as a new descriptor screening method: Application of data reduction in QSAR study of some sulfonated derivatives as c-Met inhibitors. Chemom. Intell. Lab. Syst. 2023, 232, 104714. [Google Scholar] [CrossRef]
- Haruna, S.A.; Li, H.H.; Zareef, M.; Hassan, M.M.; Arslan, M.; Geng, W.H.; Wei, W.Y.; Dandago, M.A.; Yao-Say Solomon Adade, S.; Chen, Q.S. Application of NIR spectroscopy for rapid quantification of acid and peroxide in crude peanut oil coupled multivariate analysis. Spectrochim. Acta A Mol. Biomol. Spectrosc. 2022, 267, 120624. [Google Scholar] [CrossRef]
- Haruna, S.A.; Li, H.H.; Wei, W.Y.; Geng, W.H.; Luo, X.F.; Zareef, M.; Yao-Say Solomon Adade, S.; Ivane, N.M.A.; Isa, A.; Chen, Q.S. Simultaneous quantification of total flavonoids and phenolic content in raw peanut seeds via NIR spectroscopy coupled with integrated algorithms. Spectrochim. Acta A Mol. Biomol. Spectrosc. 2023, 285, 121854. [Google Scholar] [CrossRef]
- Wang, Z.L.; Li, J.B.; Zhang, C.; Fan, S.X. Development of a General Prediction Model of Moisture Content in Maize Seeds Based on LW-NIR Hyperspectral Imaging. Agriculture 2023, 13, 359. [Google Scholar] [CrossRef]
- He, H.J.; Wang, Y.L.; Ou, X.Q.; Ma, H.J.; Liu, H.J.; Yan, J.H. Rapid determination of chemical compositions in chicken flesh by mining hyperspectral data. J. Food Compos. Anal. 2023, 116, 105069. [Google Scholar] [CrossRef]
- Merivaara, A.; Kekkonen, J.; Monola, J.; Koivunotko, E.; Savolainen, M.; Silvast, T.; Svedstrom, K.; Diaz, A.; Holler, M.; Korhonen, O.; et al. Near-infrared analysis of nanofibrillated cellulose aerogel manufacturing. Int. J. Pharm. 2022, 617, 121581. [Google Scholar] [CrossRef]
- Yang, K.; An, C.Q.; Zhu, J.L.; Guo, W.C.; Lu, C.; Zhu, X.H. Comparison of near-infrared and dielectric spectra for quantitative identification of bovine colostrum adulterated with mature milk. J. Dairy Sci. 2022, 105, 8638–8649. [Google Scholar] [CrossRef]
- Ma, H.; Xiao, L.L.; Xu, D.C.; Geng, Y.R.; Liu, X.S.; Chen, Y.; Wu, Y.J. Non-Invasive Detection of Anti-Inflammatory Bioactivity and Key Chemical Indicators of the Commercial Lanqin Oral Solution by Near Infrared Spectroscopy. Molecules 2022, 27, 2955. [Google Scholar] [CrossRef]
- Yang, Y.; Yang, L.C.; He, S.Y.; Cao, X.Q.; Huang, J.M.; Ji, X.L.; Tong, H.B.; Zhang, X.; Wu, M.J. Use of near-infrared spectroscopy and chemometrics for fast discrimination of Sargassum fusiforme. J. Food Compos. Anal. 2022, 110, 104537. [Google Scholar] [CrossRef]
- Kennard, R.W.; Stone, L.A. Computer Aided Design of Experiments. Technometrics 1969, 11, 137–148. [Google Scholar] [CrossRef]
- Yang, Y.; Mao, R.; Yang, L.C.; Liu, J.; Wu, S.Y.; Wu, M.J.; Zhang, X.; Tong, H.B.; Ji, X.L. Rapid and comprehensive quality assessment of Bupleuri Radix through near-infrared spectroscopy combined with chemometrics. Infrared Phys. Technol. 2022, 121, 104051. [Google Scholar] [CrossRef]
- Lu, Y.Z.; Du, C.W.; Yu, C.B.; Zhou, J.M. Determination of the contents of magnesium and potassium in rapeseeds using FTIR-PAS combined with least squares support vector machines and uninformative variable elimination. Anal. Methods 2014, 6, 2586–2591. [Google Scholar] [CrossRef]
- Yang, G.; Li, S.L.; Ji, S.X.; Wang, Y.J.; Wang, J.M.; Ji, L.Q.; Li, C.Q. Determination of Total Saccharide Content in Auricularia auricula Based on Near-Infrared Spectroscopy. J. Food Qual. 2022, 2022, 8858235. [Google Scholar] [CrossRef]
- Li, Y.M.; Yang, X.W. Quantitative analysis of near infrared spectroscopic data based on dual-band transformation and competitive adaptive reweighted sampling. Spectrochim. Acta A Mol. Biomol. Spectrosc. 2023, 285, 121924. [Google Scholar] [CrossRef]
- Chen, Q.M.; Xie, Y.F.; Yu, H.; Guo, Y.H.; Yao, W.R. Non-destructive prediction of colour and water-related properties of frozen/thawed beef meat by Raman spectroscopy coupled multivariate calibration. Food Chem. 2023, 413, 135513. [Google Scholar] [CrossRef]
- Cao, X.Q.; Ding, H.C.; Yang, L.C.; Huang, J.M.; Zeng, L.; Tong, H.B.; Su, L.J.; Ji, X.L.; Wu, M.J.; Yang, Y. Near-infrared spectroscopy as a tool to assist Sargassum fusiforme quality grading: Harvest time discrimination and polyphenol prediction. Postharvest Biol. Technol. 2022, 192, 112030. [Google Scholar] [CrossRef]
- Guo, Z.M.; Wang, M.M.; Agyekum, A.A.; Wu, J.Z.; Chen, Q.S.; Zuo, M.; El-Seedi, H.R.; Tao, F.F.; Shi, J.; Ouyang, Q.; et al. Quantitative detection of apple watercore and soluble solids content by near infrared transmittance spectroscopy. J. Food Eng. 2020, 279, 109955. [Google Scholar] [CrossRef]
- Guo, Z.M.; Guo, C.; Chen, Q.S.; Ouyang, Q.; Shi, J.Y.; El-Seedi, H.R.; Zou, X.B. Classification for Penicillium expansum Spoilage and Defect in Apples by Electronic Nose Combined with Chemometrics. Sensors 2020, 20, 2130. [Google Scholar] [CrossRef]
- Jiang, H.; Wang, J.N.; Chen, Q.S. Comparison of wavelength selected methods for improving of prediction performance of PLS model to determine aflatoxin B1 (AFB1) in wheat samples during storage. Microchem. J. 2021, 170, 106642. [Google Scholar] [CrossRef]
- Sokač Cvetnić, T.; Krog, K.; Valinger, D.; Gajdoš Kljusurić, J.; Benković, M.; Jurina, T.; Jakovljević, T.; Radojčić Redovniković, I.; Jurinjak Tušek, A. Application of Multivariate Regression and Artificial Neural Network Modelling for Prediction of Physicochemical Properties of Grape-Skin Compost. Bioengineering 2024, 11, 285. [Google Scholar] [CrossRef]


| Parameter | Calibration (g kg−1) | Prediction (g kg−1) | ||||
|---|---|---|---|---|---|---|
| Minimum | Maximum | Average | Minimum | Maximum | Average | |
| Total saponins (117) | 0.79 | 5.79 | 1.81 | 1.13 | 3.07 | 1.79 |
| Naringenin (120) | 0.0038 | 0.0366 | 0.0130 | 0.0041 | 0.0369 | 0.0129 |
| Mannitol (120) | 1.65 | 5.91 | 3.24 | 1.57 | 5.69 | 3.09 |
| Parameter | Raw | Smooth | 1D + SG | 2D + SG | MSC | SNV | |
|---|---|---|---|---|---|---|---|
| Total saponins | RC2 | 0.9480 | 0.9494 | 0.9246 | 0.9082 | 0.9513 | 0.9401 |
| RP2 | 0.8421 | 0.8506 | 0.2626 | 0.0704 | 0.7343 | 0.7205 | |
| RMSEC (g kg−1) | 0.1441 | 0.1421 | 0.1739 | 0.1978 | 0.1414 | 0.1567 | |
| RMSEP (g kg−1) | 0.1464 | 0.1439 | 0.2272 | 0.2056 | 0.1441 | 0.1446 | |
| Mannitol | RC2 | 0.8960 | 0.9444 | 0.9282 | 0.9579 | 0.9716 | 0.9517 |
| RP2 | 0.8682 | 0.9233 | 0.7984 | 0.5476 | 0.9385 | 0.9303 | |
| RMSEC (g kg−1) | 0.3509 | 0.2565 | 0.3073 | 0.2402 | 0.1914 | 0.2495 | |
| RMSEP (g kg−1) | 0.4822 | 0.3704 | 0.5303 | 0.7035 | 0.2876 | 0.3073 | |
| Naringenin | RC2 | 0.8053 | 0.8012 | 0.9236 | 0.8828 | 0.8581 | 0.8589 |
| RP2 | 0.8298 | 0.8313 | 0.2245 | 0.1377 | 0.8415 | 0.8432 | |
| RMSEC (g kg−1) | 0.003117 | 0.003150 | 0.002149 | 0.002679 | 0.002723 | 0.002715 | |
| RMSEP (g kg−1) | 0.003529 | 0.003515 | 0.003839 | 0.003938 | 0.003202 | 0.003195 |
| Parameter | Model | PCs | Variables | Calibration | Prediction | ||||
|---|---|---|---|---|---|---|---|---|---|
| Rc2 | RMSECV (g kg−1) | RMSEC (g kg−1) | Rp2 | RMSEP (g kg−1) | RPD | ||||
| Total saponins | Full-PLS | 18 | 1577 | 0.9494 | 0.2340 | 0.1421 | 0.8506 | 0.1439 | 2.54 |
| CARS-PLS | 16 | 66 | 0.9626 | 0.1691 | 0.1221 | 0.8949 | 0.1250 | 2.92 | |
| UVE-PLS | 17 | 872 | 0.9404 | 0.2622 | 0.1541 | 0.8210 | 0.1623 | 2.25 | |
| Mannitol | Full-PLS | 18 | 1557 | 0.9716 | 0.3901 | 0.1914 | 0.9385 | 0.2876 | 4.01 |
| CARS-PLS | 17 | 39 | 0.9868 | 0.1799 | 0.1307 | 0.9664 | 0.2192 | 5.26 | |
| UVE-PLS | 18 | 767 | 0.9722 | 0.3907 | 0.1894 | 0.9437 | 0.2795 | 4.13 | |
| Naringenin | Full-PLS | 16 | 1557 | 0.8589 | 0.0041 | 0.002715 | 0.8432 | 0.003195 | 2.48 |
| CARS-PLS | 17 | 28 | 0.8888 | 0.0031 | 0.002411 | 0.8570 | 0.003159 | 2.51 | |
| UVE-PLS | 18 | 256 | 0.8623 | 0.0040 | 0.002682 | 0.8326 | 0.003390 | 2.34 | |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
She, X.; Huang, J.; Cao, X.; Wu, M.; Yang, Y. Rapid Measurement of Total Saponins, Mannitol, and Naringenin in Dendrobium officinale by Near-Infrared Spectroscopy and Chemometrics. Foods 2024, 13, 1199. https://doi.org/10.3390/foods13081199
She X, Huang J, Cao X, Wu M, Yang Y. Rapid Measurement of Total Saponins, Mannitol, and Naringenin in Dendrobium officinale by Near-Infrared Spectroscopy and Chemometrics. Foods. 2024; 13(8):1199. https://doi.org/10.3390/foods13081199
Chicago/Turabian StyleShe, Xiangting, Jing Huang, Xiaoqing Cao, Mingjiang Wu, and Yue Yang. 2024. "Rapid Measurement of Total Saponins, Mannitol, and Naringenin in Dendrobium officinale by Near-Infrared Spectroscopy and Chemometrics" Foods 13, no. 8: 1199. https://doi.org/10.3390/foods13081199





