Stability Constant and Potentiometric Sensitivity of Heavy Metal–Organic Fluorescent Compound Complexes: QSPR Models for Prediction and Design of Novel Coumarin-like Ligands
Abstract
1. Introduction
2. Materials and Methods
2.1. Data Pre-Processing
2.2. Construction of QSPR Models Using Statistical and Machine Learning Techniques
2.2.1. Performance Assessment
2.2.2. Internal Validation
2.2.3. External Validation
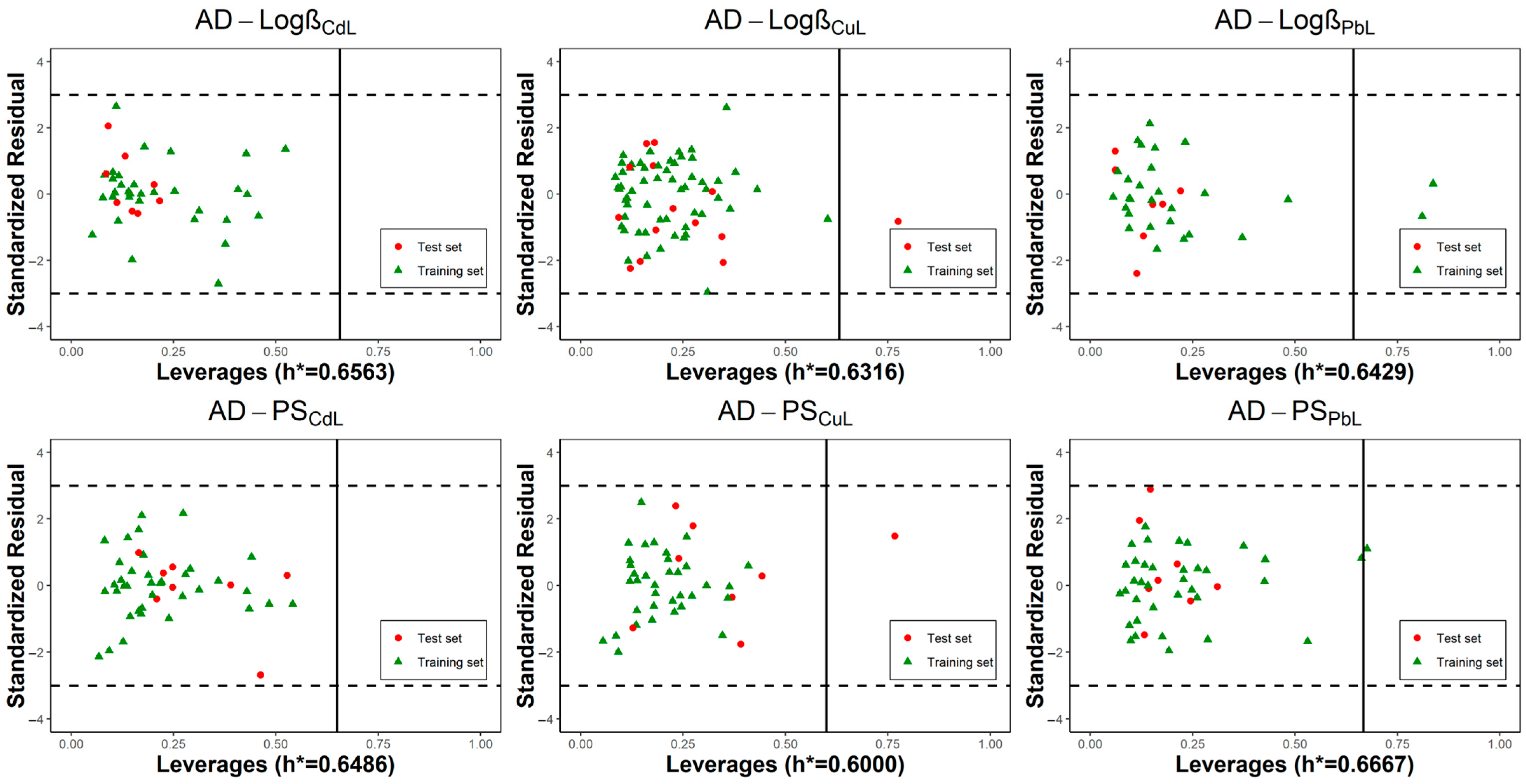
2.3. Applicability Domain
2.4. Design of Novel Coumarin-like Structures
2.5. Quantum Chemical Calculations
2.6. Physicochemical and Toxicological Profiling
2.7. Theoretical Proposal of Synthetic Accessibility for Promising Candidates
3. Results and Discussion
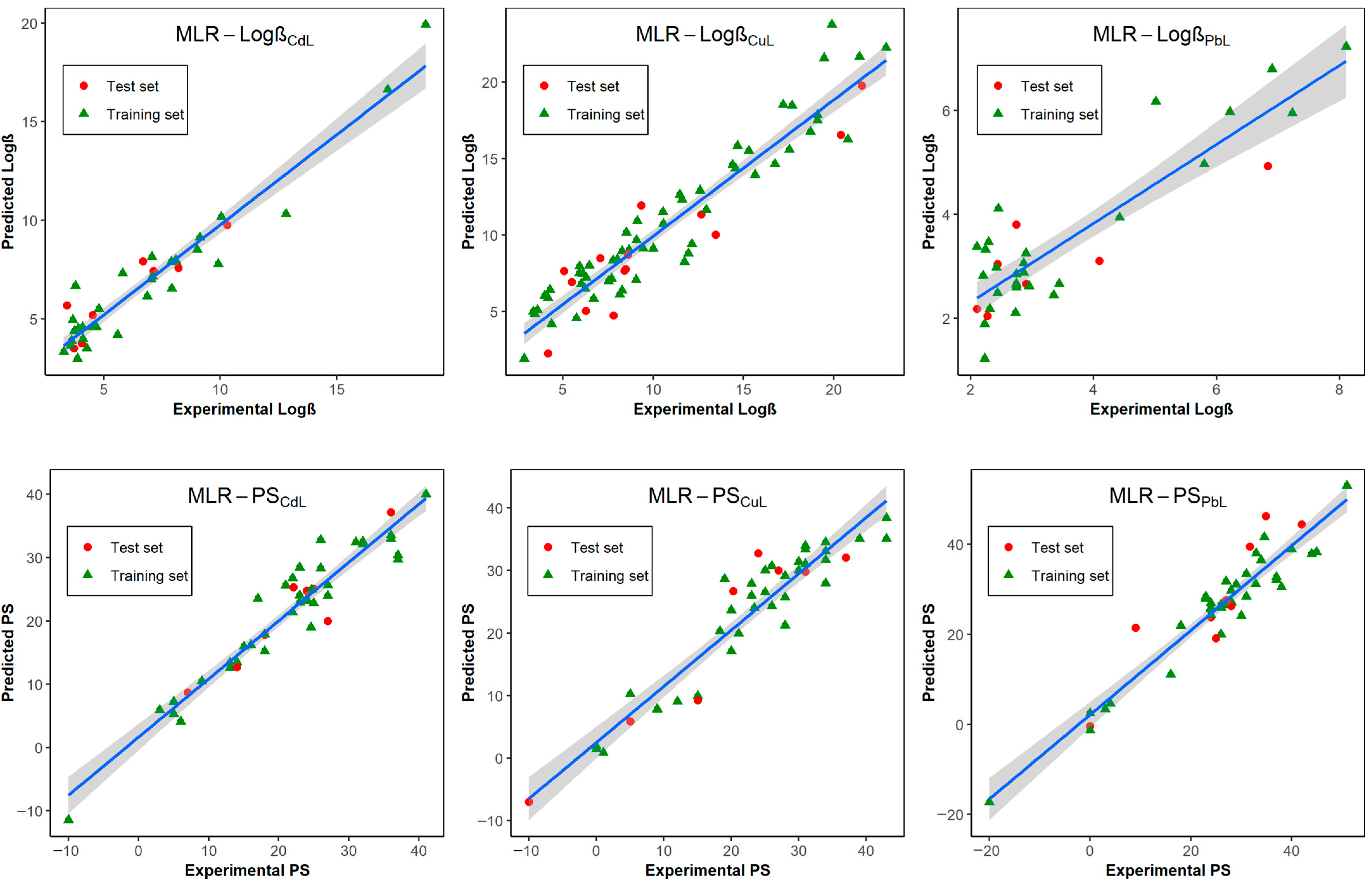
3.1. Development and Selection of GA-MLR QSPR Models
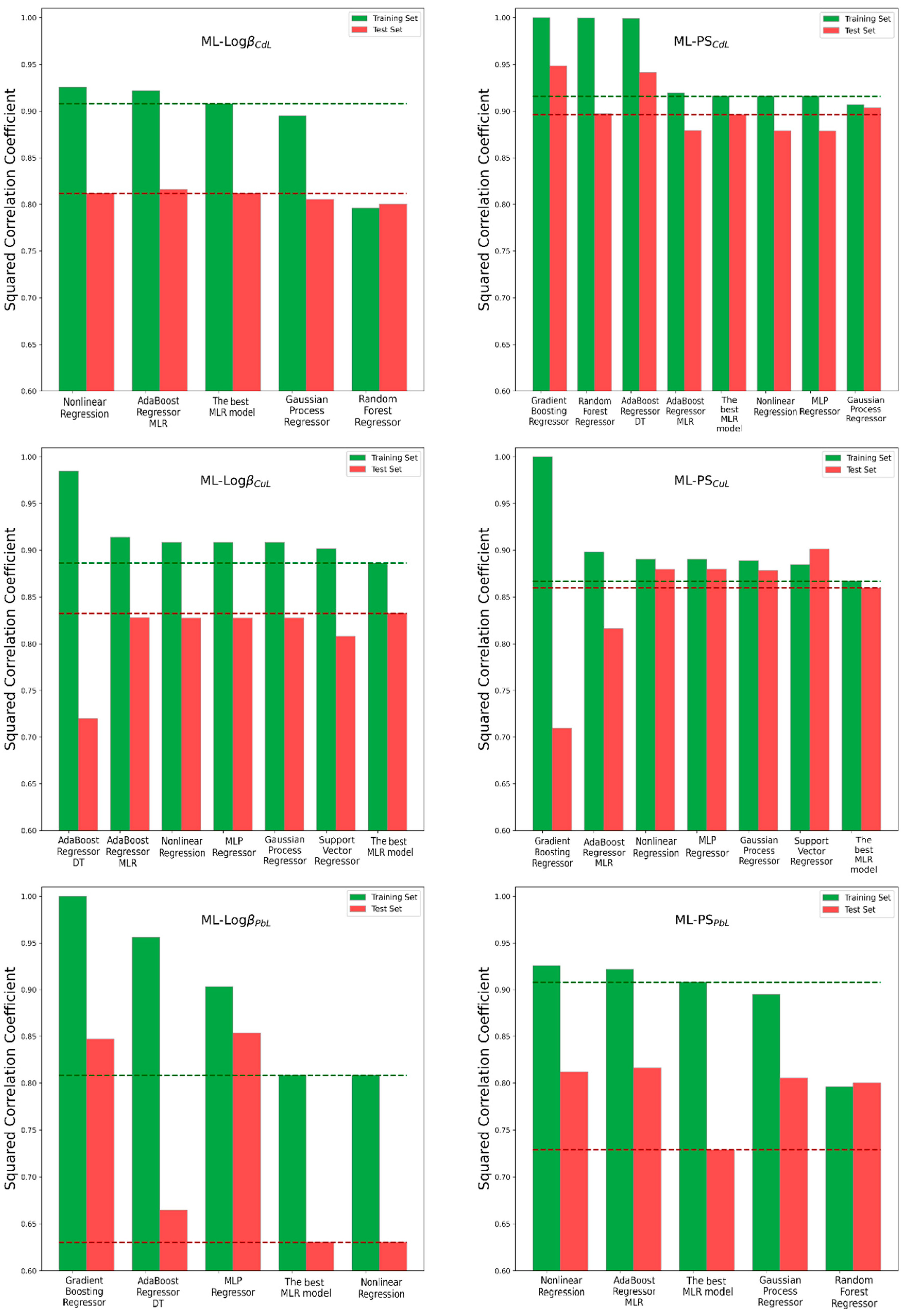
3.2. Machine Learning Algorithms for Improving Model Performance
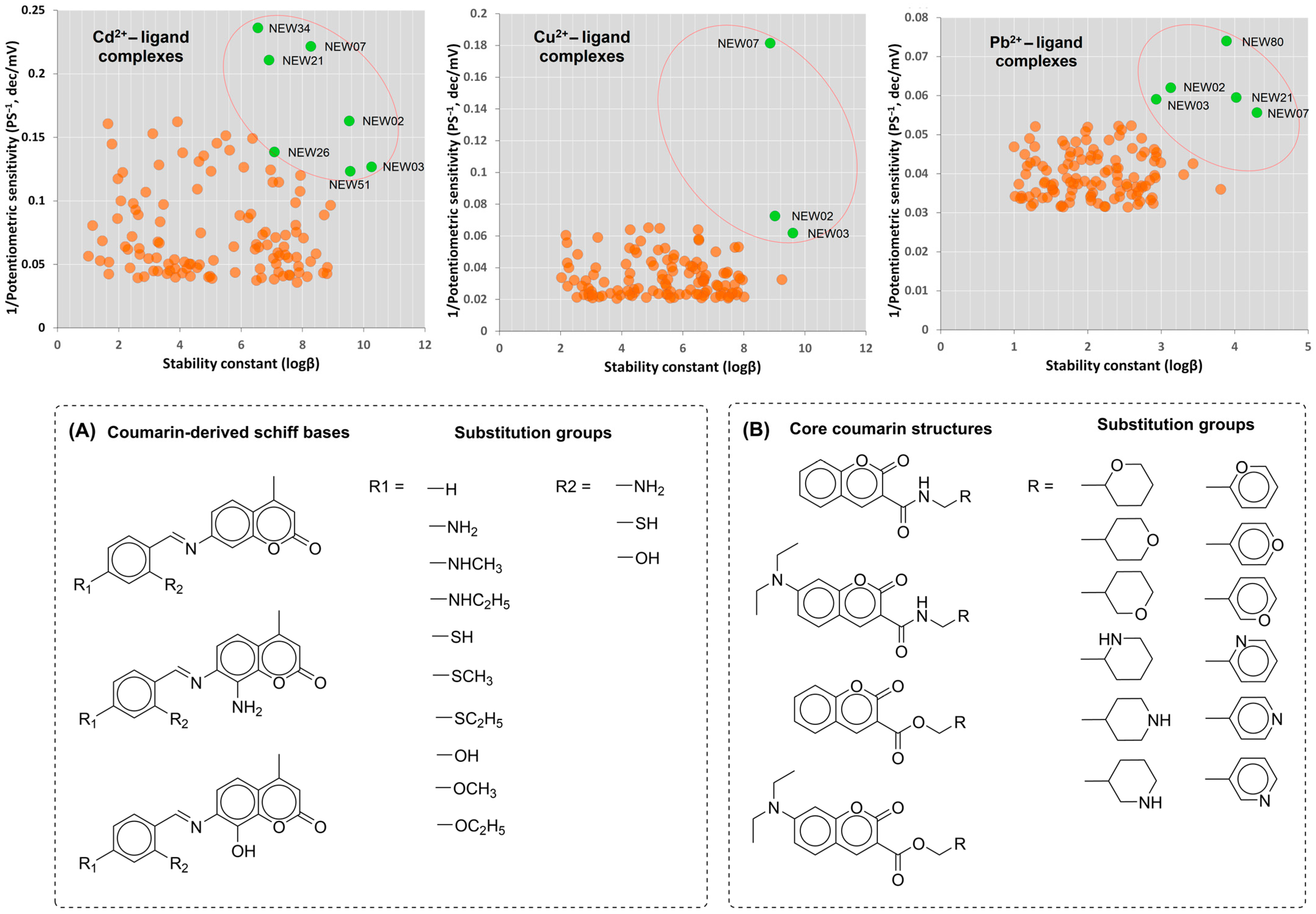
3.3. Design of Coumarin-like Chemical Library for Screening of Novel Chemosensors
3.4. Virtual Screening and Complexation Potential Predictions Using DFT Calculations
3.5. Physicochemical and Toxicological Profiling
3.6. Theoretical Synthetic Routes Proposed for Designed Coumarins
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Romero-Estudillo, I.; Viveros-Ceballos, J.L.; Cazares-Carreño, O.; González-Morales, A.; de Jesús, B.F.; López-Castillo, M.; Razo-Hernández, R.S.; Castañeda-Corral, G.; Ordóñez, M. 000Synthesis of new α-aminophosphonates: Evaluation as anti-inflammatory agents and QSAR studies. Bioorg. Med. Chem. 2019, 27, 2376–2386. [Google Scholar] [CrossRef] [PubMed]
- Lin, L.; Yang, H.; Xu, X. Effects of Water Pollution on Human Health and Disease Heterogeneity: A Review. Front. Environ. Sci. 2022, 10, 880246. [Google Scholar] [CrossRef]
- Noor, R.; Maqsood, A.; Baig, A.; Pande, C.B.; Zahra, S.M.; Saad, A.; Anwar, M.; Singh, S.K. A comprehensive review on water pollution, South Asia Region: Pakistan. Urban Clim. 2023, 48, 101413. [Google Scholar] [CrossRef]
- Md Anawar, H.; Chowdhury, R. Remediation of Polluted River Water by Biological, Chemical, Ecological and Engineering Processes. Sustainability 2020, 12, 7017. [Google Scholar] [CrossRef]
- Vhahangwele, M.; Khathutshelo, L.M. Environmental Contamination by Heavy Metals. In Heavy Metals; Hosam El-Din, M.S., Refaat, F.A., Eds.; IntechOpen: Rijeka, Croatia, 2018; pp. 115–133. [Google Scholar]
- Järup, L. Hazards of heavy metal contamination. Br. Med. Bull. 2003, 68, 167–182. [Google Scholar] [CrossRef]
- Tchounwou, P.B.; Yedjou, C.G.; Patlolla, A.K.; Sutton, D.J. Heavy Metal Toxicity and the Environment. In Molecular, Clinical and Environmental Toxicology: Volume 3: Environmental Toxicology; Luch, A., Ed.; Springer: Basel, Switzerland, 2012; pp. 133–164. [Google Scholar]
- Rafati-Rahimzadeh, M.; Rafati-Rahimzadeh, M.; Kazemi, S.; Moghadamnia, A. Cadmium toxicity and treatment: An update. Caspian J. Intern. Med. 2017, 8, 135–145. [Google Scholar] [CrossRef]
- Ebrahimi, M.; Khalili, N.; Razi, S.; Keshavarz-Fathi, M.; Khalili, N.; Rezaei, N. Effects of lead and cadmium on the immune system and cancer progression. J. Environ. Health Sci. Eng. 2020, 18, 335–343. [Google Scholar] [CrossRef]
- Flemming, C.A.; Trevors, J.T. Copper toxicity and chemistry in the environment: A review. Water Air Soil Pollut. 1989, 44, 143–158. [Google Scholar] [CrossRef]
- Cao, R.; Zhang, Y.; Ju, Y.; Wang, W.; Xi, C.; Liu, W.; Liu, K. Exacerbation of copper pollution toxicity from ocean acidification: A comparative analysis of two bivalve species with distinct sensitivities. Environ. Pollut. 2022, 293, 118525. [Google Scholar] [CrossRef]
- Krämer, J.; Kang, R.; Grimm, L.M.; De Cola, L.; Picchetti, P.; Biedermann, F. Molecular Probes, Chemosensors, and Nanosensors for Optical Detection of Biorelevant Molecules and Ions in Aqueous Media and Biofluids. Chem. Rev. 2022, 122, 3459–3636. [Google Scholar] [CrossRef]
- Punia, P.; Bharti, M.K.; Dhar, R.; Thakur, P.; Thakur, A. Recent Advances in Detection and Removal of Heavy Metals from Contaminated Water. ChemBioEng Rev. 2022, 9, 351–369. [Google Scholar] [CrossRef]
- Wu, D.; Sedgwick, A.C.; Gunnlaugsson, T.; Akkaya, E.U.; Yoon, J.; James, T.D. Fluorescent chemosensors: The past, present and future. Chem. Soc. Rev. 2017, 46, 7105–7123. [Google Scholar] [CrossRef]
- Dongare, P.R.; Gore, A.H. Recent Advances in Colorimetric and Fluorescent Chemosensors for Ionic Species: Design, Principle and Optical Signalling Mechanism. ChemistrySelect 2021, 6, 5657–5669. [Google Scholar] [CrossRef]
- Vladimirova, N.; Polukeev, V.; Ashina, J.; Babain, V.; Legin, A.; Kirsanov, D. Prediction of Carbonate Selectivity of PVC-Plasticized Sensor Membranes with Newly Synthesized Ionophores through QSPR Modeling. Chemosensors 2022, 10, 43. [Google Scholar] [CrossRef]
- Lieberzeit, P.A.; Dickert, F.L. Chemosensors in environmental monitoring: Challenges in ruggedness and selectivity. Anal. Bioanal. Chem. 2009, 393, 467–472. [Google Scholar] [CrossRef]
- Madatov, U.; Rakhimov, S.; Shahidova, D.; Smanova, Z.; Lal, B.; Berdimurodov, E. A new, green, highly effective procedure for manganese determination using alizarin-3-methylamino-N,N-diacetic acid immobilised on a polymer matrix. Int. J. Environ. Anal. Chem. 2022, 1–22. [Google Scholar] [CrossRef]
- Smanova, Z.A.; Usmanova, X.U.; Madusmanova, N.K.; Bobojanov, B.B.; Maxmudova, G.O. Immobilized Oxyazo Compounds as Analytical Reagents for the Sorption-Luminescent Determination of Certain Metals. Turk. Online J. Qual. Inq. 2021, 12, 6113–6119. [Google Scholar]
- Fjodorova, N.; Novic, M.; Gajewicz, A.; Rasulev, B. The way to cover prediction for cytotoxicity for all existing nano-sized metal oxides by using neural network method. Nanotoxicology 2017, 11, 475–483. [Google Scholar] [CrossRef]
- Toropov, A.A.; Toropova, A.P.; Rasulev, B.F.; Benfenati, E.; Gini, G.; Leszczynska, D.; Leszczynski, J. Coral: QSPR modeling of rate constants of reactions between organic aromatic pollutants and hydroxyl radical. J. Comput. Chem. 2012, 33, 1902–1906. [Google Scholar] [CrossRef]
- Sizochenko, N.; Syzochenko, M.; Fjodorova, N.; Rasulev, B.; Leszczynski, J. Evaluating genotoxicity of metal oxide nanoparticles: Application of advanced supervised and unsupervised machine learning techniques. Ecotoxicol. Environ. Saf. 2019, 185, 109733. [Google Scholar] [CrossRef]
- Rasulev, B.; Kušić, H.; Leszczynska, D.; Leszczynski, J.; Koprivanac, N. QSAR modeling of acute toxicity on mammals caused by aromatic compounds: The case study using oral LD50 for rats. J. Environ. Monit. 2010, 12, 1037. [Google Scholar] [CrossRef] [PubMed]
- Turabekova, M.A.; Rasulev, B.F.; Dzhakhangirov, F.N.; Leszczynska, D.; Leszczynski, J. Aconitum and Delphinium alkaloids of curare-like activity. QSAR analysis and molecular docking of alkaloids into AChBP. Eur. J. Med. Chem. 2010, 45, 3885–3894. [Google Scholar] [CrossRef] [PubMed]
- Gooch, A.; Sizochenko, N.; Rasulev, B.; Gorb, L.; Leszczynski, J. In vivo toxicity of nitroaromatics: A comprehensive quantitative structure–activity relationship study. Environ. Toxicol. Chem. 2017, 36, 2227–2233. [Google Scholar] [CrossRef] [PubMed]
- Ahmed, L.; Rasulev, B.; Kar, S.; Krupa, P.; Mozolewska, M.A.; Leszczynski, J. Inhibitors or toxins? Large library target-specific screening of fullerene-based nanoparticles for drug design purpose. Nanoscale 2017, 9, 10263–10276. [Google Scholar] [CrossRef]
- Ukić, Š.; Sigurnjak, M.; Cvetnić, M.; Markić, M.; Stankov, M.N.; Rogošić, M.; Rasulev, B.; Lončarić Božić, A.; Kušić, H.; Bolanča, T. Toxicity of pharmaceuticals in binary mixtures: Assessment by additive and non-additive toxicity models. Ecotoxicol. Environ. Saf. 2019, 185, 109696. [Google Scholar] [CrossRef]
- Turabekova, M.A.; Rasulev, B.F.; Dzhakhangirov, F.N.; Salikhov, S.I. Aconitum and Delphinium alkaloids: “Drug-likeness” descriptors related to toxic mode of action. Environ. Toxicol. Pharmacol. 2008, 25, 310–320. [Google Scholar] [CrossRef]
- Soloviev, V.; Varnek, A.; Babain, V.; Polukeev, V.; Ashina, J.; Legin, E.; Legin, A.; Kirsanov, D. QSPR modeling of potentiometric sensitivity towards heavy metal ions for polymeric membrane sensors. Sens. Actuators B Chem. 2019, 301, 126941. [Google Scholar] [CrossRef]
- Vladimirova, N.; Puchkova, E.; Dar’in, D.; Turanov, A.; Babain, V.; Kirsanov, D. Predicting the Potentiometric Sensitivity of Membrane Sensors Based on Modified Diphenylphosphoryl Acetamide Ionophores with QSPR Modeling. Membranes 2022, 12, 953. [Google Scholar] [CrossRef]
- Kiani-Anbouhi, R.; Ganjali, M.R.; Norouzi, P. Prediction of the complexation stabilities of La3+ ion with ionophores applied in lanthanoid sensors. J. Incl. Phenom. Macrocycl. Chem. 2014, 78, 325–336. [Google Scholar] [CrossRef]
- Solov’ev, V.; Kireeva, N.; Ovchinnikova, S.; Tsivadze, A. The complexation of metal ions with various organic ligands in water: Prediction of stability constants by QSPR ensemble modelling. J. Incl. Phenom. Macrocycl. Chem. 2015, 83, 89–101. [Google Scholar] [CrossRef]
- Martynko, E.; Solov’ev, V.; Varnek, A.; Legin, A.; Kirsanov, D. QSPR Modeling of Potentiometric Mg2+/Ca2+ Selectivity for PVC-plasticized Sensor Membranes. Electroanalysis 2020, 32, 792–798. [Google Scholar] [CrossRef]
- Kiani-Anbouhi, R.; Ganjali, M.R.; Norouzi, P. Application of QSPR for prediction of the complexation stabilities of Sm(III) with ionophores applied in lanthanoid sensors. J. Incl. Phenom. Macrocycl. Chem. 2015, 81, 441–450. [Google Scholar] [CrossRef]
- Kanahashi, K.; Urushihara, M.; Yamaguchi, K. Machine learning-based analysis of overall stability constants of metal–ligand complexes. Sci. Rep. 2022, 12, 11159. [Google Scholar] [CrossRef]
- Solov’ev, V.; Sukhno, I.; Buzko, V.; Polushin, A.; Marcou, G.; Tsivadze, A.; Varnek, A. Stability constants of complexes of Zn2+, Cd2+, and Hg2+ with organic ligands: QSPR consensus modeling and design of new metal binders. J. Incl. Phenom. Macrocycl. Chem. 2012, 72, 309–321. [Google Scholar] [CrossRef]
- Yamasaki, K.; Yasuda, M. Stability of Zinc and Cadmium Complexes with 2,2′-Bipyridine and 1,10-Phenanthroline. J. Am. Chem. Soc. 1956, 78, 1324. [Google Scholar] [CrossRef]
- Golcu, A.; Tumer, M.; Demirelli, H.; Wheatley, R.A. Cd(II) and Cu(II) complexes of polydentate Schiff base ligands: Synthesis, characterization, properties and biological activity. Inorg. Chim. Acta 2005, 358, 1785–1797. [Google Scholar] [CrossRef]
- Sóvágó, I.; Várnagy, K. Cadmium(II) Complexes of Amino Acids and Peptides. In Cadmium: From Toxicity to Essentiality; Sigel, A., Sigel, H., Sigel, R.K.O., Eds.; Springer: Dordrecht, The Netherlands, 2013; pp. 275–302. [Google Scholar]
- Quang, N.M.; Mau, T.X.; Ai Nhung, N.T.; Minh An, T.N.; Van Tat, P. Novel QSPR modeling of stability constants of metal-thiosemicarbazone complexes by hybrid multivariate technique: GA-MLR, GA-SVR and GA-ANN. J. Mol. Struct. 2019, 1195, 95–109. [Google Scholar] [CrossRef]
- Hancock, R.D.; de Sousa, A.S.; Walton, G.B.; Reibenspies, J.H. Metal-Ion Selectivity Produced by C-Alkyl Substituents on the Bridges of Chelating Ligands: The Importance of Short H−H Nonbonded van der Waals Contacts in Controlling Metal-Ion Selectivity. A Thermodynamic, Molecular Mechanics, and Crystallographic Study. Inorg. Chem. 2007, 46, 4749–4757. [Google Scholar] [CrossRef]
- Kotek, J.; Kubíček, V.; Hermann, P.; Lukeš, I. Synthesis and Characterization of Ligands and their Gadolinium(III) Complexes. In The Chemistry of Contrast Agents in Medical Magnetic Resonance Imaging; John Wiley & Sons, Ltd.: West Sussex, UK, 2013; pp. 83–155. [Google Scholar]
- Sigel, H.; Da Costa, C.P.; Song, B.; Carloni, P.; Gregáň, F. Stability and Structure of Metal Ion Complexes Formed in Solution with Acetyl Phosphate and Acetonylphosphonate: Quantification of Isomeric Equilibria. J. Am. Chem. Soc. 1999, 121, 6248–6257. [Google Scholar] [CrossRef]
- Fernández-Botello, A.; Griesser, R.; Holý, A.; Moreno, V.; Sigel, H. Acid−Base and Metal-Ion-Binding Properties of 9-[2-(2-Phosphonoethoxy)ethyl]adenine (PEEA), a Relative of the Antiviral Nucleotide Analogue 9-[2-(Phosphonomethoxy)ethyl]adenine (PMEA). An Exercise on the Quantification of Isomeric Complex Equilibria in Solution. Inorg. Chem. 2005, 44, 5104–5117. [Google Scholar] [CrossRef]
- Hardy, J.G. Metallosupramolecular grid complexes: Towards nanostructured materials with high-tech applications. Chem. Soc. Rev. 2013, 42, 7881–7899. [Google Scholar] [CrossRef] [PubMed]
- Kaczorowska, M.A.; Bożejewicz, D.; Witt, K.; Urbaniak, W. A new application of 2–benzoylpyridine–efficient removal of silver ions from acidic aqueous solutions via adsorption process on polymeric material and classic solvent extraction. Chem. Process Eng. 2022, 43, 369–382. [Google Scholar]
- Jung, H.S.; Kwon, P.S.; Lee, J.W.; Kim, J.I.; Hong, C.S.; Kim, J.W.; Yan, S.; Lee, J.Y.; Lee, J.H.; Joo, T.; et al. Coumarin-Derived Cu2+-Selective Fluorescence Sensor: Synthesis, Mechanisms, and Applications in Living Cells. J. Am. Chem. Soc. 2009, 131, 2008–2012. [Google Scholar] [CrossRef] [PubMed]
- Agarwal, R.P.; Perrin, D.D. Stability constants of complexes of copper(II) ions with some histidine peptides. Dalton Trans. 1975, 3, 268–272. [Google Scholar] [CrossRef]
- Le Fur, M.; Beyler, M.; Le Poul, N.; Lima, L.M.P.; Le Mest, Y.; Delgado, R.; Platas-Iglesias, C.; Patinec, V.; Tripier, R. Improving the stability and inertness of Cu(ii) and Cu(i) complexes with methylthiazolyl ligands by tuning the macrocyclic structure. Dalton Trans. 2016, 45, 7406–7420. [Google Scholar] [CrossRef]
- Nagaj, J.; Stokowa-Sołtys, K.; Kurowska, E.; Frączyk, T.; Jeżowska-Bojczuk, M.; Bal, W. Revised Coordination Model and Stability Constants of Cu(II) Complexes of Tris Buffer. Inorg. Chem. 2013, 52, 13927–13933. [Google Scholar] [CrossRef] [PubMed]
- Royzen, M.; Dai, Z.; Canary, J.W. Ratiometric Displacement Approach to Cu(II) Sensing by Fluorescence. J. Am. Chem. Soc. 2005, 127, 1612–1613. [Google Scholar] [CrossRef] [PubMed]
- Xie, J.; Ménand, M.; Maisonneuve, S.; Métivier, R. Synthesis of Bispyrenyl Sugar-Aza-Crown Ethers as New Fluorescent Molecular Sensors for Cu(II). J. Org. Chem. 2007, 72, 5980–5985. [Google Scholar] [CrossRef]
- Tanaka, M.; Tabata, M. Stability Constants of Metal(II) Complexes with Amines and Aminocarboxylates with Special Reference to Chelation. Bull. Chem. Soc. Jpn. 2009, 82, 1258–1265. [Google Scholar] [CrossRef]
- Türkel, N. Equilibrium Study of the Mixed Complexes of Copper(II) with Adenine and Amino Acids in Aqueous Solution. J. Solut. Chem. 2015, 44, 1267–1280. [Google Scholar] [CrossRef]
- Agrawal, B.R.; Magare, B.K.; Farooqui, M.N.; Janrao, D.M.; Ubale, M.B. Stability Constants of Cu (II), Ni (II) and Mn (II) Metal Complexes with Cetrizine and Benzoic acid. Int. J. Chem. Sci. 2009, 7, 2169–2172. [Google Scholar]
- Pashkina, D.A.; Gusev, V.Y.; Radushev, A.V. Complexation of copper(II) with 2′,2′-dialkyl-para-tert-butylbenzohydrazides. Russ. J. Inorg. Chem. 2014, 59, 394–398. [Google Scholar] [CrossRef]
- Hay, R.W.; Govan, N.; Perotti, A.; Carugo, O. Copper(II), nickel(II) and palladium(II) complexes of the diamide ligand N,N’-bis(2-carbamoylethyl)ethylenediamine (H2L) and the crystal structure of the carbonyl-oxygen-bonded copper(II) complex [Cu(H2L)](ClO4)2. Transit. Met. Chem. 1997, 22, 389–394. [Google Scholar] [CrossRef]
- Polat, F.; Atabey, H.; Sari, H.; Cukurovali, A. Potentiometric study of equilibrium constants of a novel triazine-thione derivative and its stability constants with Hg2+, Cu2+, Ni2+, Pb2+, and Zn2+ metal ions in ethanol and water mixed. Turk. J. Chem. 2013, 37, 439–448. [Google Scholar] [CrossRef]
- Kiani-anboui, R.; Ghasemi, M.H. A QSPR Study for the Prediction of the Selectivity of Pb(II) Sensors by Stability Constants of Ion-Ionophore Complexes. Anal. Bioanal. Electrochem. 2022, 14, 598–609. [Google Scholar]
- Jagvir, S.; Abhay Nanda, S.; Netrapal, S.; Anuradha, S. Stability Constants of Metal Complexes in Solution. In Stability and Applications of Coordination Compounds; Abhay Nanda, S., Ed.; IntechOpen: Rijeka, Croatia, 2019; pp. 1–18. [Google Scholar]
- Todeschini, R.; Consonni, V. Molecular Descriptors for Chemoinformatics; Wiley-VCH Verlag GmbH & Co. KGaA: Weinheim, Germany, 2009; Volume 11, p. 967. [Google Scholar]
- Perez-Castillo, Y.; Sánchez-Rodríguez, A.; Tejera, E.; Cruz-Monteagudo, M.; Borges, F.; Cordeiro, M.N.D.S.; Le-Thi-Thu, H.; Pham-The, H. A desirability-based multi objective approach for the virtual screening discovery of broad-spectrum anti-gastric cancer agents. PLoS ONE 2018, 13, e0192176. [Google Scholar] [CrossRef]
- Pham-The, H.; Casañola-Martin, G.; Diéguez-Santana, K.; Nguyen-Hai, N.; Ngoc, N.T.; Vu-Duc, L.; Le-Thi-Thu, H. Quantitative structure-activity relationship analysis and virtual screening studies for identifying HDAC2 inhibitors from known HDAC bioactive chemical libraries. SAR QSAR Environ. Res. 2017, 28, 199–220. [Google Scholar] [CrossRef]
- Daghighi, A.; Casanola-Martin, G.M.; Timmerman, T.; Milenković, D.; Lučić, B.; Rasulev, B. In Silico Prediction of the Toxicity of Nitroaromatic Compounds: Application of Ensemble Learning QSAR Approach. Toxics 2022, 10, 746. [Google Scholar] [CrossRef]
- Gramatica, P.; Chirico, N.; Papa, E.; Cassani, S.; Kovarich, S. QSARINS: A new software for the development, analysis, and validation of QSAR MLR models. J. Comput. Chem. 2013, 34, 2121–2132. [Google Scholar] [CrossRef]
- Pham-The, H.; Le-Thi-Thu, H. Integrating structure and ligand-based approaches for modelling the Histone deacetylase inhibition activity of hydroxamic acid derivatives. Asian J. Pharm. Clin. Res. 2018, 11, 198–206. [Google Scholar] [CrossRef]
- Golbraikh, A.; Tropsha, A. Predictive QSAR modeling based on diversity sampling of experimental datasets for the training and test set selection. Mol. Divers. 2000, 5, 231–243. [Google Scholar] [CrossRef]
- Nimon, K.F.; Oswald, F.L. Understanding the Results of Multiple Linear Regression: Beyond Standardized Regression Coefficients. Organ. Res. Methods 2013, 16, 650–674. [Google Scholar] [CrossRef]
- Jerome, H.F. Multivariate Adaptive Regression Splines. Ann. Statist. 1991, 19, 1–67. [Google Scholar] [CrossRef]
- Bondarchuk, S.V. Friction sensitivity of nitramine energetic materials: A prediction based on genetic function approximation. FirePhysChem 2022, 2, 272–278. [Google Scholar] [CrossRef]
- Bondarchuk, S.V. Prediction of aquatic toxicity of energetic materials using genetic function approximation. FirePhysChem 2023, 3, 23–28. [Google Scholar] [CrossRef]
- Gramatica, P. On the Development and Validation of QSAR Models. In Computational Toxicology: Volume II; Reisfeld, B., Mayeno, A.N., Eds.; Humana Press: Totowa, NJ, USA, 2013; pp. 499–526. [Google Scholar]
- Gramatica, P. Principles of QSAR models validation: Internal and external. QSAR Comb. Sci. 2007, 26, 694–701. [Google Scholar] [CrossRef]
- Netzeva, T.I.; Worth, A.P.; Aldenberg, T.; Benigni, R.; Cronin, M.T.D.; Gramatica, P.; Jaworska, J.S.; Kahn, S.; Klopman, G.; Marchant, C.A.; et al. Current Status of Methods for Defining the Applicability Domain of (Quantitative) Structure-Activity Relationships: The Report and Recommendations of ECVAM Workshop 521,2. Altern. Lab. Anim. 2005, 33, 155–173. [Google Scholar] [CrossRef]
- Cao, D.; Liu, Z.; Verwilst, P.; Koo, S.; Jangjili, P.; Kim, J.S.; Lin, W. Coumarin-Based Small-Molecule Fluorescent Chemosensors. Chem. Rev. 2019, 119, 10403–10519. [Google Scholar] [CrossRef]
- Gill, P.M.W.; Johnson, B.G.; Pople, J.A.; Frisch, M.J. The performance of the Becke—Lee—Yang—Parr (B—LYP) density functional theory with various basis sets. Chem. Phys. Lett. 1992, 197, 499–505. [Google Scholar] [CrossRef]
- Guest, M.F.; Bush, I.J.; Van Dam, H.J.J.; Sherwood, P.; Thomas, J.M.H.; Van Lenthe, J.H.; Havenith, R.W.A.; Kendrick, J. The GAMESS-UK electronic structure package: Algorithms, developments and applications. Mol. Phys. 2005, 103, 719–747. [Google Scholar] [CrossRef]
- Parr, R.G.; Donnelly, R.A.; Levy, M.; Palke, W.E. Electronegativity: The density functional viewpoint. J. Chem. Phys. 1978, 68, 3801–3807. [Google Scholar] [CrossRef]
- Makov, G. Chemical Hardness in Density Functional Theory. J. Phys. Chem. 1995, 99, 9337–9339. [Google Scholar] [CrossRef]
- Parr, R.G.; Szentpály, L.v.; Liu, S. Electrophilicity Index. J. Am. Chem. Soc. 1999, 121, 1922–1924. [Google Scholar] [CrossRef]
- Daina, A.; Michielin, O.; Zoete, V. SwissADME: A free web tool to evaluate pharmacokinetics, drug-likeness and medicinal chemistry friendliness of small molecules. Sci. Rep. 2017, 7, 42717. [Google Scholar] [CrossRef] [PubMed]
- Xiong, G.; Wu, Z.; Yi, J.; Fu, L.; Yang, Z.; Hsieh, C.; Yin, M.; Zeng, X.; Wu, C.; Lu, A.; et al. ADMETlab 2.0: An integrated online platform for accurate and comprehensive predictions of ADMET properties. Nucleic Acids Res. 2021, 49, W5–W14. [Google Scholar] [CrossRef]
- Castillo-Garit, J.A.; Casanola-Martin, G.M.; Le-Thi-Thu, H.; Pham-The, H.; Barigye, S.J. A Simple Method to Predict Blood-Brain Barrier Permeability of Drug- Like Compounds Using Classification Trees. Med. Chem. 2017, 13, 664–669. [Google Scholar] [CrossRef]
- Pham-The, H.; Garrigues, T.; Bermejo, M.; González-Álvarez, I.; Monteagudo, M.C.; Cabrera-Pérez, M. Provisional classification and in silico study of biopharmaceutical system based on caco-2 cell permeability and dose number. Mol. Pharm. 2013, 10, 2445–2461. [Google Scholar] [CrossRef]
- Dieguez-Santana, K.; Pham-The, H.; Villegas-Aguilar, P.J.; Le-Thi-Thu, H.; Castillo-Garit, J.A.; Casañola-Martin, G.M. Prediction of acute toxicity of phenol derivatives using multiple linear regression approach for Tetrahymena pyriformis contaminant identification in a median-size database. Chemosphere 2016, 165, 434–441. [Google Scholar] [CrossRef]
- Pham-The, H.; González-Álvarez, I.; Bermejo, M.; Garrigues, T.; Le-Thi-Thu, H.; Cabrera-Pérez, M.Á. The Use of Rule-Based and QSPR Approaches in ADME Profiling: A Case Study on Caco-2 Permeability. Mol. Inf. 2013, 32, 459–479. [Google Scholar] [CrossRef]
- Yu, J.; Wang, J.; Zhao, H.; Gao, J.; Kang, Y.; Cao, D.; Wang, Z.; Hou, T. Organic Compound Synthetic Accessibility Prediction Based on the Graph Attention Mechanism. J. Chem. Inf. Model. 2022, 62, 2973–2986. [Google Scholar] [CrossRef]
- Ertl, P.; Schuffenhauer, A. Estimation of synthetic accessibility score of drug-like molecules based on molecular complexity and fragment contributions. J. Cheminform. 2009, 1, 8. [Google Scholar] [CrossRef]
- Austin, P.C.; Steyerberg, E.W. The number of subjects per variable required in linear regression analyses. J. Clin. Epidemiol. 2015, 68, 627–636. [Google Scholar] [CrossRef] [PubMed]
- Pham-The, H.; Nam, N.-H.; Nga, D.-V.; Hai, T.D.; Dieguez-Santana, K.; Marrero-Ponce, Y.; Castillo-Garit, A.J.; Casanola-Martin, M.G.; Le-Thi-Thu, H. Learning from Multiple Classifier Systems: Perspectives for Improving Decision Making of QSAR Models in Medicinal Chemistry. Curr. Top. Med. Chem. 2017, 17, 3269–3288. [Google Scholar] [CrossRef] [PubMed]
- Sun, X.-y.; Liu, T.; Sun, J.; Wang, X.-j. Synthesis and application of coumarin fluorescence probes. RSC Adv. 2020, 10, 10826–10847. [Google Scholar] [CrossRef] [PubMed]
- Sabeel, M.B.; Puthiyavalappil, R.; Vipin, M.; Vishnunarayanan Namboothiri Vadakkedathu, P.; Anandaram, S. Recent Advancements in Schiff Base Fluorescence Chemosensors for the Detection of Heavy Metal Ions. In Schiff Base in Organic, Inorganic and Physical Chemistry; Takashiro, A., Ed.; IntechOpen: Rijeka, Croatia, 2023; pp. 1–22. [Google Scholar]
- Fan, L. Synthesis of Two Coumarin-Derived Schiff Bases and Investigation of theirs Selectivity for Zn2+. J. Fluoresc. 2017, 27, 1331–1337. [Google Scholar] [CrossRef]
- Chang, H.-Q.; Zhao, X.-L.; Wu, W.-N.; Jia, L.; Wang, Y. A highly sensitive on-off fluorescent chemosensor for Cu2+ based on coumarin. J. Lumin. 2017, 182, 268–273. [Google Scholar] [CrossRef]
- Amalia, S.; Lucia, P. Molecular Descriptors and Properties of Organic Molecules. In Symmetry (Group Theory) and Mathematical Treatment in Chemistry; Takashiro, A., Ed.; IntechOpen: Rijeka, Croatia, 2018; pp. 161–176. [Google Scholar]
- Karaca, H.; Kazancı, S. The metal sensing applications of chalcones: The synthesis, characterization and theoretical calculations. J. Mol. Struct. 2022, 1248, 131454. [Google Scholar] [CrossRef]
- Askerov, R.K.; El Bakri, Y.; Osmanov, V.K.; Ahmad, S.; Saravanan, K.; Borisova, G.N.; Nazarov, R.H.o.; Baranov, E.V.; Fukin, G.K.; Fukina, D.G.; et al. Complexes of 1-(2-R(F, CH3, Cl)-phenyl)-1,4-dihydro-5H-tetrazole-5-thiones with cadmium chloride: Synthesis, molecular, crystal structures and computational investigation approach. J. Inorg. Biochem. 2022, 231, 111791. [Google Scholar] [CrossRef]
- Cabrera-Perez, M.A.; Bermejo, M.; Alvarez, I.G.; Alvarez, M.G.; Garrigues, T.M. QSPR in Oral Bioavailability: Specificity or Integrality? Mini-Rev. Med. Chem. 2012, 12, 534–550. [Google Scholar] [CrossRef]
- Tinkov, O.; Polishchuk, P.; Matveieva, M.; Grigorev, V.; Grigoreva, L.; Porozov, Y. The Influence of Structural Patterns on Acute Aquatic Toxicity of Organic Compounds. Mol. Inf. 2021, 40, 2000209. [Google Scholar] [CrossRef]
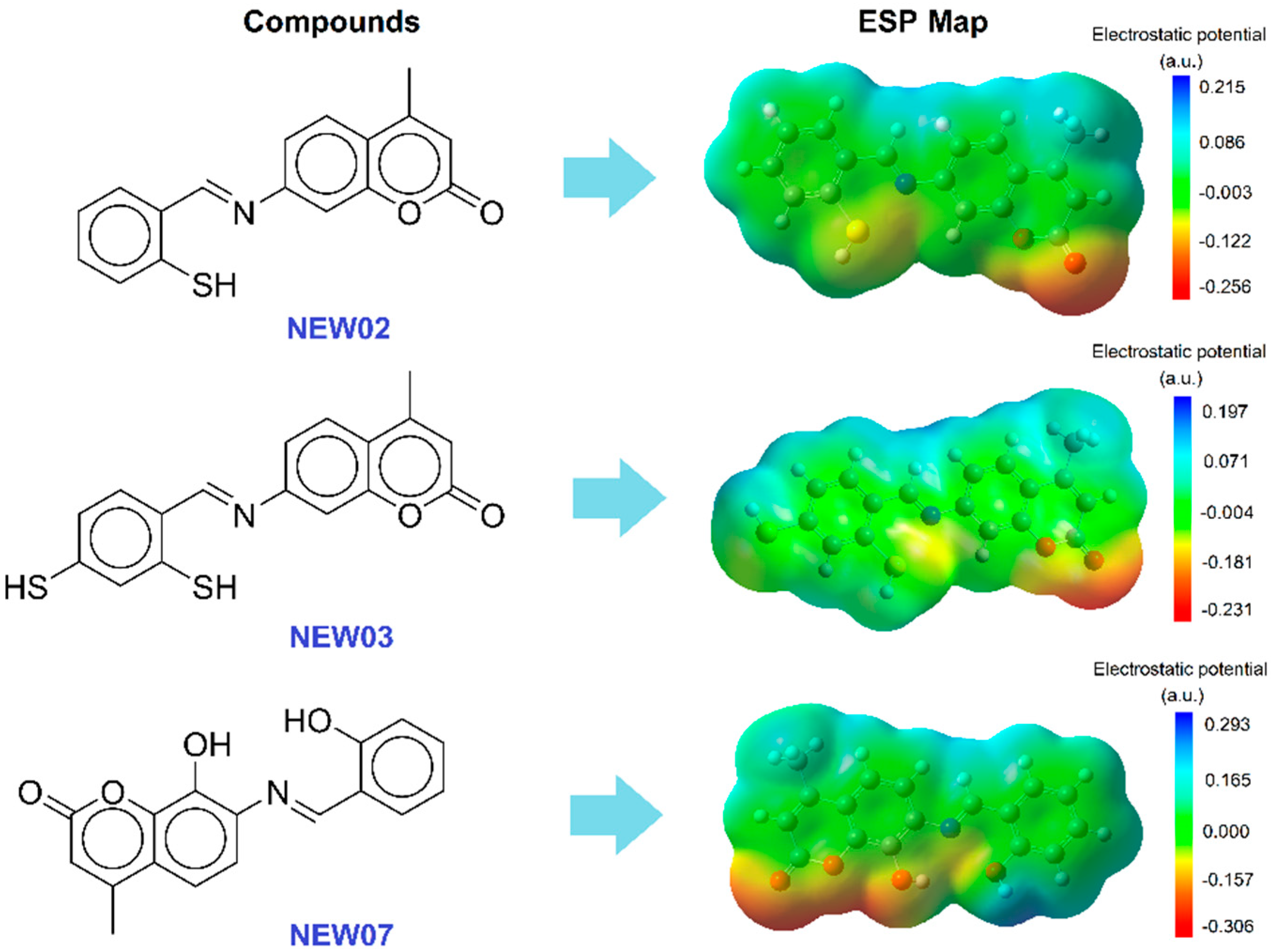
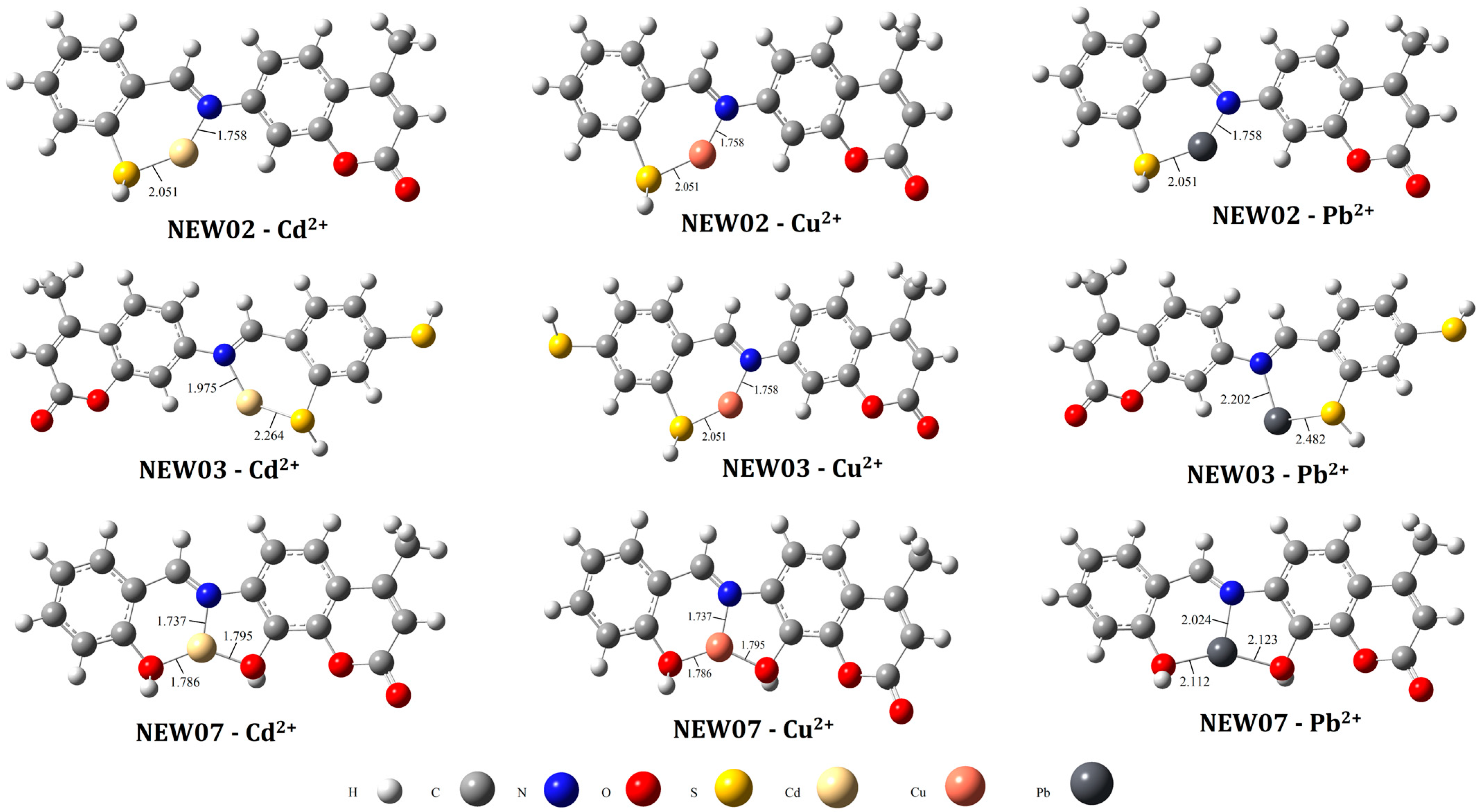

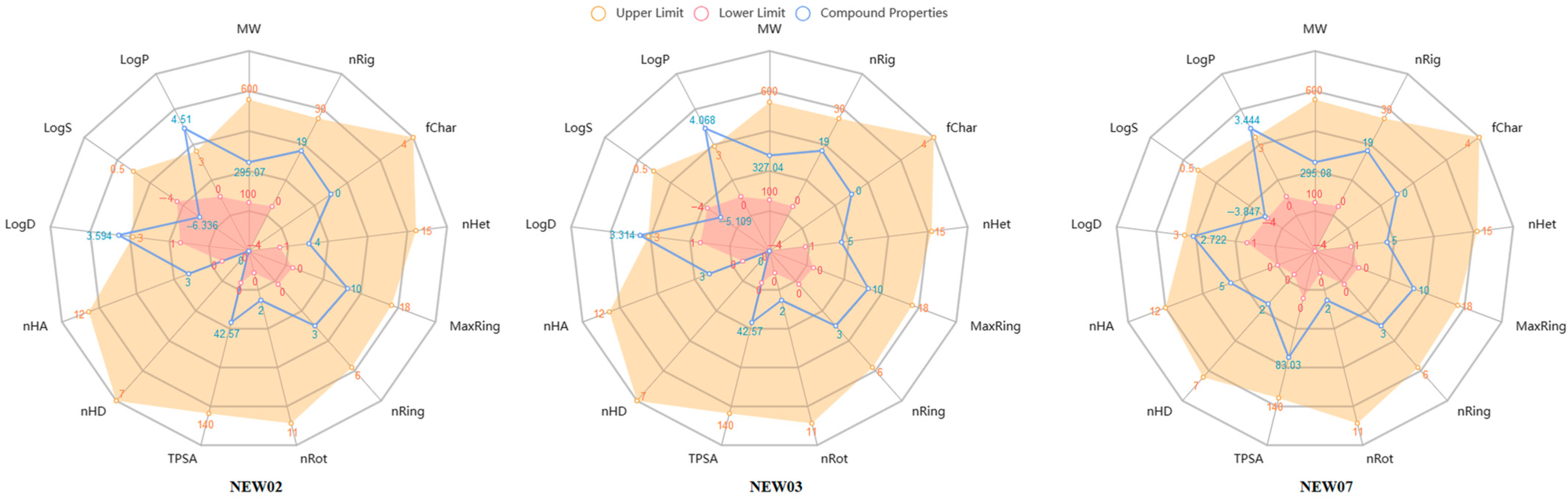

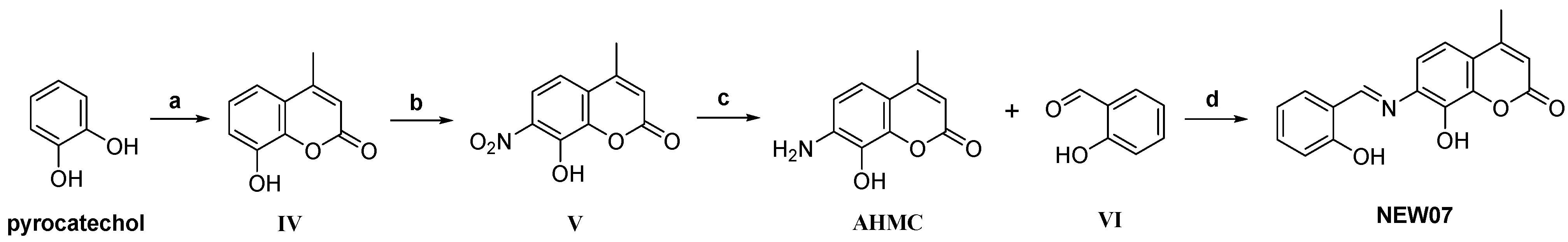
| Year | Chemometric Methods | Metal Ions | Modeling Property | Chemical Structures |
|---|---|---|---|---|
| Kiani-Anbouhi et al. (2014) [31] | Genetic algorithm-multiple linear regression (GA-MLR); ; ; N = 29 ligands; | La3+ | Stability constant (logβL) | Diverse |
| Kiani-Anbouhi et al. (2015) [34] | GA-MLR; ; ; N = 24 ligands; | Sm3+ | Stability constant (logβL) | Diverse |
| Soloviev et al. (2019) [29] | Consensus MLR models; Cu2+: ; RMSE = 8.3; Zn2+: ; RMSE = 4.5; Cd2+: ; RMSE = 4.8; Pb2+: ; RMSE = 5.9; N = 35 ligands; | Cu2+, Zn2+, Cd2+, and Pb2+ | Potentiometric sensitivity (PSML) | Nitrogen-containing ligands (mainly diamides of pyridine and bipyridine acids) |
| Martynko et al. (2020) [33] | MLR models; ; RMSE = 8.8; N = 67 ligands; ; RMSE = 5.3; N = 56 ligands (refined data); | Mg2+/Ca2+ | Selectivity coefficient logK(Mg2+/Ca2+) | Amide ionophores |
| Vladimirova et al. (2022) [30] | Partial least-squares regression (PLS); Cu2+: ; RMSE = 6.9; Cd2+: ; RMSE = 4.2; Pb2+: ; RMSE = 7.5; N = 35 ligands; | Cu2+, Cd2+, and Pb2+ | Potentiometric sensitivity (PSML) | Nitrogen-containing ligands (mainly diamides of pyridine and bipyridine acids) |
| Kanahashi et al. (2022) [35] | Gaussian process regression; Best model (8 cations, 49 ligands, 2 experimental conditions): MAE = 1.31; ; N = 2706 ligands (unpublished data) | 57 cations | Stability constant (logβL) | Diverse |
| Statistical Parameters 1 | MLR-logβCdL | MLR-logβCuL | MLR-logβPbL | MLR-PSCdL | MLR-PSCuL | MLR-PSPbL | |
|---|---|---|---|---|---|---|---|
| Fitting criteria | KXX | 0.4121 | 0.2649 | 0.2384 | 0.4845 | 0.3958 | 0.2546 |
| δK | 0.0698 | 0.0577 | 0.0679 | 0.0193 | 0.0679 | 0.0554 | |
| R2 | 0.9257 | 0.9085 | 0.8084 | 0.9160 | 0.8904 | 0.9264 | |
| 0.9079 | 0.8862 | 0.7648 | 0.8957 | 0.8669 | 0.9080 | ||
| LOF | 2.6866 | 7.0495 | 1.3524 | 2.4842 | 3.1127 | 3.7405 | |
| p-value (F-test) | 6.522 × 10−13 | 1.095 × 10−19 | 3.087 × 10−7 | 6.602 × 10−14 | 3.491 × 10−12 | 3.399 × 10−14 | |
| Internal validation criteria | 0.8637 | 0.8434 | 0.6918 | 0.8760 | 0.8423 | 0.8668 | |
| R2 − | 0.0620 | 0.0651 | 0.1165 | 0.0400 | 0.0481 | 0.0596 | |
| 0.8311 | 0.8209 | 0.5872 | 0.8556 | 0.8246 | 0.8394 | ||
| YscrR2 | 0.1935 | 0.1975 | 0.1869 | 0.1938 | 0.1757 | 0.1961 | |
| YscrQ2 | −0.3624 | −0.3143 | −0.5988 | −0.3579 | −0.3038 | −0.4169 | |
| External validation criteria | 0.8324 | 0.8326 | 0.6357 | 0.8791 | 0.8914 | 0.7079 | |
| 0.8120 | 0.8276 | 0.6305 | 0.8789 | 0.8793 | 0.7054 | ||
| 0.9283 | 0.8415 | 0.6961 | 0.9305 | 0.8080 | 0.7734 |
| Data | Selected Model and Method | Optimized Parameter Settings 1 |
|---|---|---|
| logβ (Cd2+ complex) | Model M1/AdaBoost Regressor MLR | Base_estimator=LinearRegression; copy_X=True, fit_intercept=True, n_jobs=None, normalize=‘deprecated’, positive=False); learning_rate=1.0; loss=‘linear’; n_estimators=50; random_state=None; fitting 5 folds for each of 100 candidates; totaling 500 fits {‘n_estimators’: 4, ‘loss’: ‘square’, ‘learning_rate’: 0.01} |
| PSML (Cd2+ complex) | Model M2/Gradient Boosting Regressor | Alpha=0.9; ccp_alpha=0.0; criterion=‘friedman_mse’; init=None; learning_rate=0.1; loss=‘squared_error’; max_depth=3; max_features=None; max_leaf_nodes=None; min_impurity_decrease=0.0; min_samples_leaf=1; min_samples_split=2; min_weight_fraction_leaf=0.0; n_estimators=100; n_iter_no_change=None; random_state=None; subsample=1.0, tol=0.0001; validation_fraction=0.1; verbose=0, warm_start=False {‘n_estimators’: 91, ‘min_samples_split’: 10, ‘min_samples_leaf’: 1, ‘max_features’: ‘sqrt’, ‘max_depth’: 6, ‘learning_rate’: 0.5} |
| logβ (Cu2+ complex) | Model M3/GA-MLR | Equation (11) |
| PSML (Cu2+ complex) | Model M4/Gradient Boosting Regressor | C=1.0, cache_size=200, coef0=0.0, degree=3, epsilon=0.1, gamma=‘scale’, kernel=‘rbf’, max_iter=−1, shrinking=True, tol=0.001, verbose=False {‘kernel’: ‘linear’, ‘gamma’: 10.0, ‘epsilon’: 0.5, ‘C’: 100.0} |
| logβ (Pb2+ complex) | Model M5/Gradient Boosting Regressor | Alpha=0.9; ccp_alpha=0.0; criterion=‘friedman_mse’; init=None, learning_rate=0.1, loss=‘squared_error’; max_depth=3; max_features=None; max_leaf_nodes=None; min_impurity_decrease=0.0; min_samples_leaf=1; min_samples_split=2; min_weight_fraction_leaf=0.0; n_estimators=100; n_iter_no_change=None; random_state=None; subsample=1.0; tol=0.0001; validation_fraction=0.1; verbose=0, warm_start=False {‘n_estimators’: 57, ‘min_samples_split’: 2, ‘min_samples_leaf’: 1, ‘max_features’: ‘sqrt’, ‘max_depth’: 8, ‘learning_rate’: 1} |
| PSML (Pb2+ complex) | Model M6/AdaBoost Regressor MLR | Base_estimator=LinearRegression; copy_X=True; fit_intercept=True; n_jobs=None; normalize=‘deprecated’; positive=False; learning_rate=1.0; loss=‘linear’; n_estimators=50; random_state=None; fitting 5 folds for each of 100 candidates; totaling 500 fits {‘n_estimators’: 42, ‘loss’: ‘linear’, ‘learning_rate’: 0.1} |
| Compound ID | Stability Constant (logβ) | Potentiometric Sensitivity (PSML, mV/dec) | ||||
|---|---|---|---|---|---|---|
| Cd2+ | Cu2+ | Pb2+ | Cd2+ | Cu2+ | Pb2+ | |
| NEW02 | 9.537 | 9.022 | 3.133 | 12.146 | 13.809 | 16.135 |
| NEW03 | 10.263 | 9.610 | 2.935 | 14.901 | 16.194 | 16.947 |
| NEW07 | 8.278 | 8.870 | 4.304 | 4.516 | 5.518 | 17.973 |
| NEW21 | 6.912 | 5.506 | 4.024 | 4.746 | 22.307 | 16.812 |
| NEW26 | 7.087 | 7.701 | 2.539 | 7.230 | 34.360 | 27.286 |
| NEW34 | 6.550 | 6.607 | 3.435 | 4.236 | 28.975 | 23.507 |
| NEW51 | 9.566 | 9.252 | 2.608 | 8.119 | 30.902 | 21.520 |
| Global Reactivity Index | DFT Energy (eV) | ||||||||
|---|---|---|---|---|---|---|---|---|---|
| Cd2+ Complexes | Cu2+ Complexes | Pb2+ Complexes | |||||||
| NEW02 | NEW03 | NEW07 | NEW02 | NEW03 | NEW07 | NEW02 | NEW03 | NEW07 | |
| HOMO–LUMO Gap, ΔE | 4.004 | 3.839 | 4.328 | 4.586 | 4.419 | 4.439 | 3.983 | 3.839 | 4.300 |
| EHOMO | −6.074 | −5.947 | −6.305 | −6.950 | −6.774 | −6.803 | −6.225 | −6.044 | −6.410 |
| ELUMO | −2.070 | −2.108 | −1.977 | −2.363 | −2.354 | −2.364 | −2.242 | −2.205 | −2.109 |
| Ionization Energy, IE | 6.074 | 5.947 | 6.305 | 6.950 | 6.774 | 6.803 | 6.225 | 6.044 | 6.410 |
| Electron Affinity, EA | 2.070 | 2.108 | 1.977 | 2.363 | 2.355 | 2.364 | 2.242 | 2.205 | 2.109 |
| Global Hardness, η | 2.002 | 1.919 | 2.164 | 2.293 | 2.210 | 2.219 | 1.992 | 1.920 | 2.150 |
| Global Softness, σ | 0.500 | 0.521 | 0.462 | 0.436 | 0.453 | 0.451 | 0.502 | 0.521 | 0.465 |
| Electronegativity, χ | 4.072 | 4.027 | 4.141 | 4.656 | 4.564 | 4.584 | 4.234 | 4.125 | 4.259 |
| Electrophilicity, ω | 4.142 | 4.225 | 3.962 | 4.727 | 4.715 | 4.734 | 4.500 | 4.431 | 4.219 |
| Parameters | NEW02 | NEW03 | NEW07 | Description |
|---|---|---|---|---|
| Oral bioavailability (F20%) | 0.982 | 0.999 | 0.968 | The probability of oral bioavailability of 20% (F20%+). Category 1: F20%+ (bioavailability < 20%); Category 0: F20%− (bioavailability ≥ 20%). |
| Skin permeability | −5.47 | −5.56 | −6.39 | logKp (cm/s). |
| Blood–brain barrier (BBB) penetration | 0.039 | 0.107 | 0.023 | The output value is the probability of being permeable (BBB+). Category 1: BBB+; Category 0: BBB−. |
| Acute oral toxicity | 0.049 | 0.097 | 0.381 | The output value is the probability of being highly toxic (Category 0: low–toxicity; Category 1: high–toxicity). |
| Toxicophore predictions | ||||
| Acute toxicity rule | 0 alert | 0 alert | 0 alert | Predicted as toxic towards water sources based on 99 substructures |
| Non-biodegradable rule | 0 alert | 0 alert | 0 alert | Predicted as non-biodegradable substances based on 19 substructures. |
| Skin sensitization rules | 1 alert | 1 alert | 0 alert | Predicted as skin irritants based on 155 substructures. |
| Eye irritation | 0.991 | 0.978 | 0.917 | The output value is the probability of being irritants (Category 1: irritants; Category 0: non-irritants). |
| Environmental toxicity | ||||
| IGC50 | −5.367 | −5.796 | −4.465 | Tetrahymena pyriformis 50% growth inhibition concentration. The unit is log10[(mg/L)/(MW × 103)]. |
| LC50FM | −5.981 | −6.466 | −4.975 | 96 h fathead minnow 50% lethal concentration. The unit is log10[(mg/L)/(MW × 103)]. |
| LC50DM | −6.191 | −6.547 | −5.153 | 48 h daphnia magna 50% lethal concentration. The unit is log10[(mg/L)/(MW × 103)]. |
| Cpd. ID | Synthetic Probability 1 | SwissADME SA Score 2 | ADMETlab 2.0 SA Score 3 | Consensus Classification |
|---|---|---|---|---|
| NEW02 | 0.879 | 3.14 | 2.62 | ES (easy-to-synthesize) |
| NEW03 | 0.900 | 3.17 | 3.01 | ES (easy-to-synthesize) |
| NEW07 | 0.899 | 3.36 | 2.63 | ES (easy-to-synthesize) |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Diem-Tran, P.T.; Ho, T.-T.; Tuan, N.-V.; Bao, L.-Q.; Phuong, H.T.; Chau, T.T.G.; Minh, H.T.B.; Nguyen, C.-T.; Smanova, Z.; Casanola-Martin, G.M.; et al. Stability Constant and Potentiometric Sensitivity of Heavy Metal–Organic Fluorescent Compound Complexes: QSPR Models for Prediction and Design of Novel Coumarin-like Ligands. Toxics 2023, 11, 595. https://doi.org/10.3390/toxics11070595
Diem-Tran PT, Ho T-T, Tuan N-V, Bao L-Q, Phuong HT, Chau TTG, Minh HTB, Nguyen C-T, Smanova Z, Casanola-Martin GM, et al. Stability Constant and Potentiometric Sensitivity of Heavy Metal–Organic Fluorescent Compound Complexes: QSPR Models for Prediction and Design of Novel Coumarin-like Ligands. Toxics. 2023; 11(7):595. https://doi.org/10.3390/toxics11070595
Chicago/Turabian StyleDiem-Tran, Phan Thi, Tue-Tam Ho, Nguyen-Van Tuan, Le-Quang Bao, Ha Tran Phuong, Trinh Thi Giao Chau, Hoang Thi Binh Minh, Cong-Truong Nguyen, Zulayho Smanova, Gerardo M. Casanola-Martin, and et al. 2023. "Stability Constant and Potentiometric Sensitivity of Heavy Metal–Organic Fluorescent Compound Complexes: QSPR Models for Prediction and Design of Novel Coumarin-like Ligands" Toxics 11, no. 7: 595. https://doi.org/10.3390/toxics11070595
APA StyleDiem-Tran, P. T., Ho, T.-T., Tuan, N.-V., Bao, L.-Q., Phuong, H. T., Chau, T. T. G., Minh, H. T. B., Nguyen, C.-T., Smanova, Z., Casanola-Martin, G. M., Rasulev, B., Pham-The, H., & Cuong, L. C. V. (2023). Stability Constant and Potentiometric Sensitivity of Heavy Metal–Organic Fluorescent Compound Complexes: QSPR Models for Prediction and Design of Novel Coumarin-like Ligands. Toxics, 11(7), 595. https://doi.org/10.3390/toxics11070595










