Metal Ions Modify In Vitro DNA Damage Yields with High-LET Radiation
Abstract
:1. Introduction
2. Materials and Methods
2.1. Irradiation Conditions
2.2. DNA Solution Preparation and Chemical Treatment
2.3. Electrophoresis and DNA Damage Quantification
2.4. Statistical Analysis
3. Results
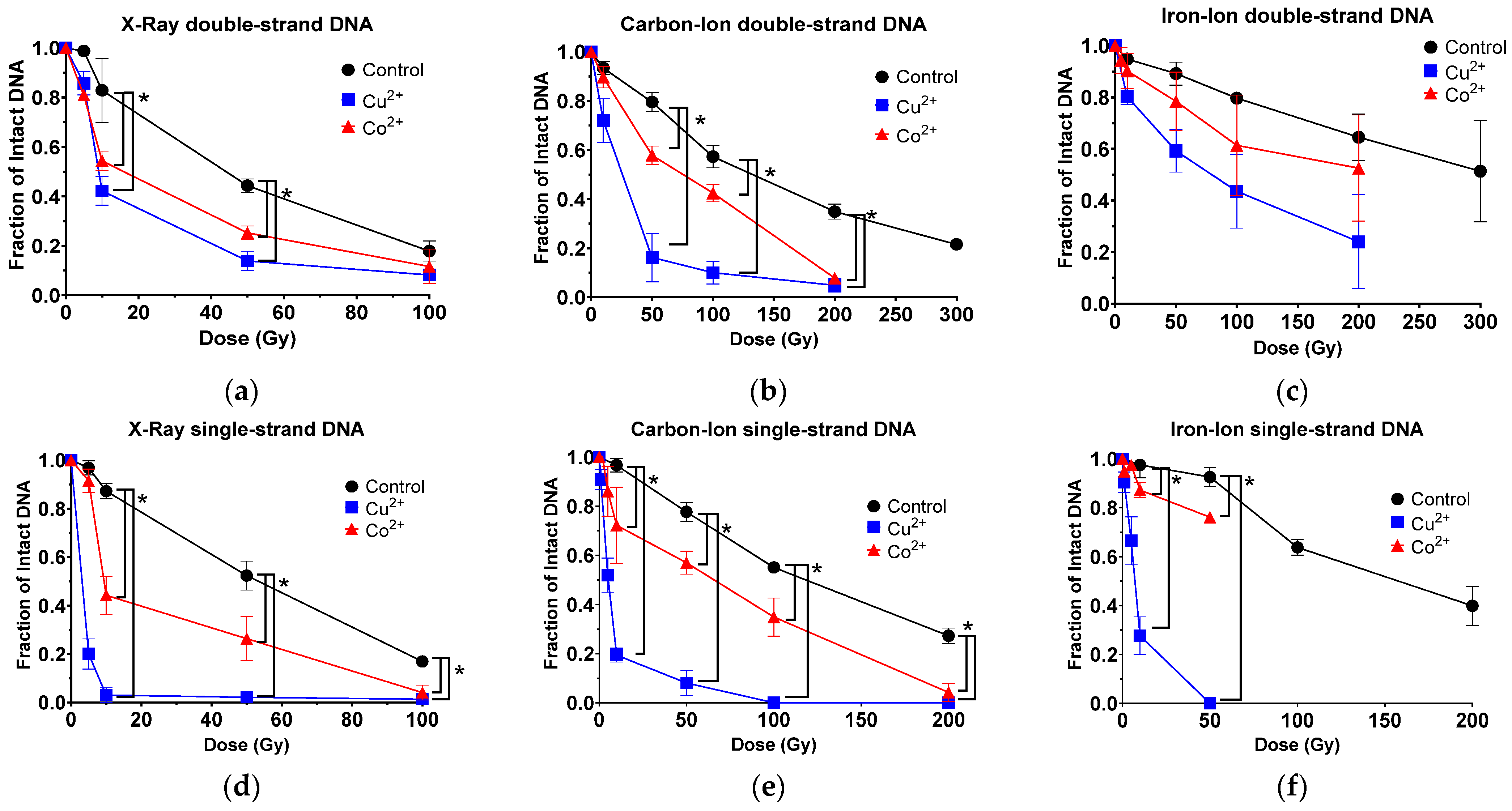
3.1. Copper and Cobalt Ions Increased DNA DSB Following Ionizing Radiation
3.2. Copper and Cobalt Ions Increased DNA SSB Following Ionizing Radiation
3.3. Copper and Cobalt Ions Increased DNA DSB When in Solution with Bleomycin or Hydrogen Peroxide
3.4. Copper and Cobalt Ions Increased DNA SSB When in Solution with Bleomycin or Hydrogen Peroxide
3.5. Metal Enhancement Ratio for DNA Break Formation Was Highest for Hydrogen Peroxide Followed by Ionizing Radiation and Least for Bleomycin
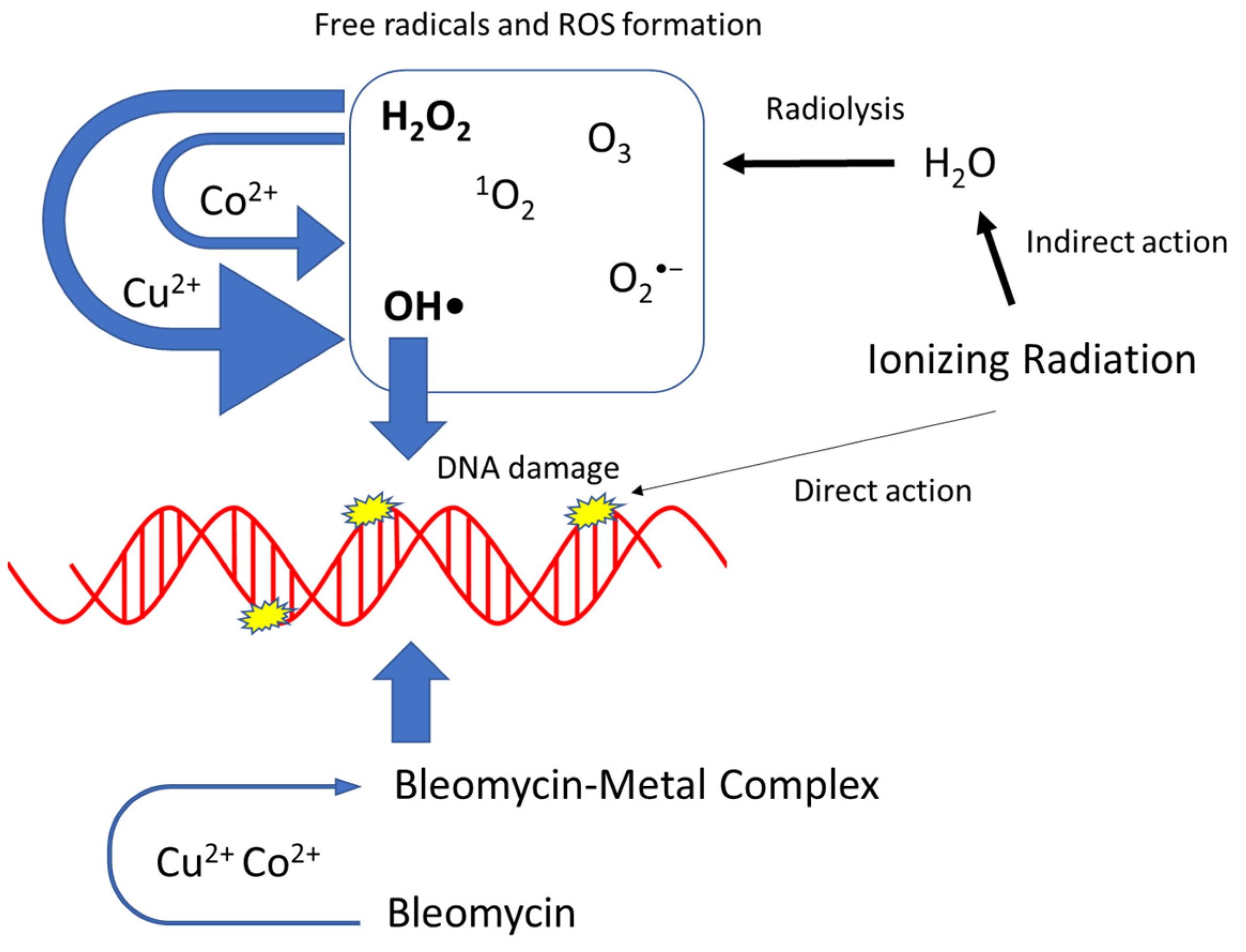
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Nickoloff, J.A.; Sharma, N.; Taylor, L. Clustered DNA Double-Strand Breaks: Biological Effects and Relevance to Cancer Radiotherapy. Genes 2020, 11, 99. [Google Scholar] [CrossRef]
- Pouget, J.P.; Mather, S.J. General aspects of the cellular response to low- and high-LET radiation. Eur. J. Nucl. Med. 2001, 28, 541–561. [Google Scholar] [CrossRef]
- Ward, J.F. Biochemistry of DNA Lesions. Radiat. Res. 1985, 104, S103–S111. [Google Scholar] [CrossRef]
- Holley, W.R.; Chatterjee, A.; Magee, J.L. Production of dna strand breaks by direct effects of heavy charged-particles. Radiat. Res. 1990, 121, 161–168. [Google Scholar] [CrossRef]
- Roots, R.; Chatterjee, A.; Chang, P.; Lommel, L.; Blakely, E.A. Characterization of hydroxyl radical-induced damage after sparsely and densely ionizing irradiation. Int. J. Radiat. Biol. 1985, 47, 157–166. [Google Scholar] [CrossRef]
- Ward, J.F. DNA Damage Produced by Ionizing-Radiation in Mammalian-Cells-Identities, Mechanisms of Formation, and Reparability. Prog. Nucleic Acid Res. Mol. Biol. 1988, 35, 95–125. [Google Scholar] [CrossRef] [PubMed]
- Lloyd, D.R.; Phillips, D.H. Oxidative DNA damage mediated by copper(II), iron(II) and nickel(II) Fenton reactions: Evidence for site-specific mechanisms in the formation of double-strand breaks, 8-hydroxydeoxyguanosine and putative intrastrand cross-links. Mutat. Res./Fundam. Mol. Mech. Mutagen. 1999, 424, 23–36. [Google Scholar] [CrossRef] [PubMed]
- Feitelson, J.; Hayon, E.; Treinin, A. Photoionization of Phenols in Water-Effects of Light-Intensity, Oxygen, Ph, and Temperature. J. Am. Chem. Soc. 1973, 95, 1025–1029. [Google Scholar] [CrossRef]
- Ward, J.F.; Blakely, W.F.; Joner, E.I. Mammalian-Cells Are Not Killed by DNA Single-Strand Breaks Caused by Hydroxyl Radicals from Hydrogen-Peroxide. Radiat. Res. 1985, 103, 383–392. [Google Scholar] [CrossRef]
- Hoffmann, M.E.; Meneghini, R. Action of hydrogen-peroxide on human fibroblast in culture. Photochem. Photobiol. 1979, 30, 151–155. [Google Scholar] [CrossRef]
- McCormick, M.L.; Buettner, G.R.; Britigan, B.E. Endogenous superoxide dismutase levels regulate iron-dependent hydroxyl radical formation in Escherichia coli exposed to hydrogen peroxide. J. Bacteriol. 1998, 180, 622–625. [Google Scholar] [CrossRef]
- Petering, D.H.; Byrnes, R.W.; Antholine, W.E. The Role of Redox-Active Metals in the Mechanism of Action of Bleomycin. Chem.-Biol. Interact. 1990, 73, 133–182. [Google Scholar] [CrossRef]
- Loeb, K.E.; Zaleski, J.M.; Hess, C.D.; Hecht, S.M.; Solomon, E.I. Spectroscopic investigation of the metal ligation and reactivity of the ferrous active sites of bleomycin and bleomycin derivatives. J. Am. Chem. Soc. 1998, 120, 1249–1259. [Google Scholar] [CrossRef]
- Calafat, A.M.; Won, H.; Marzilli, L.G. A new arrangement for the anticancer antibiotics tallysomycin and bleomycin when bound to zinc: An assessment of metal and ligand chirality by NMR and molecular dynamics. J. Am. Chem. Soc. 1997, 119, 3656–3664. [Google Scholar] [CrossRef]
- Burger, R.M. Cleavage of nucleic acids by bleomycin. Chem. Rev. 1998, 98, 1153–1169. [Google Scholar] [CrossRef] [PubMed]
- Teicher, B.A.; Jacobs, J.L.; Cathcart, K.N.S.; Abrams, M.J.; Vollano, J.F.; Picker, D.H. Some complexes of cobalt(III) and iron(III) are radiosensitizers of hypoxic EMT6 cells. Radiat. Res. 1987, 109, 36–46. [Google Scholar] [CrossRef]
- Teicher, B.A.; Abrams, M.J.; Rosbe, K.W.; Herman, T.S. Cytotoxicity, radiosensitization, antitumor-activity, and interaction with hyperthermia of a co(III) mustard complex. Cancer Res. 1990, 50, 6971–6975. [Google Scholar]
- Jiang, Y.W.; Gao, G.; Jia, H.R.; Zhang, X.D.; Zhao, J.; Ma, N.N.; Liu, J.B.; Liu, P.D.; Wu, F.G. Copper Oxide Nanoparticles Induce Enhanced Radiosensitizing Effect via Destructive Autophagy. ACS Biomater. Sci. Eng. 2019, 5, 1569–1579. [Google Scholar] [CrossRef] [PubMed]
- Lloyd, D.R.; Phillips, D.H.; Carmichael, P.L. Generation of putative intrastrand cross-links and strand breaks in DNA by transition metal ion-mediated oxygen radical attack. Chem. Res. Toxicol. 1997, 10, 393–400. [Google Scholar] [CrossRef]
- Robison, S.H.; Cantoni, O.; Costa, M. Strand breakage and decreased molecular-weight of dna induced by specific metal-compounds. Carcinogenesis 1982, 3, 657–662. [Google Scholar] [CrossRef]
- Maeda, J.; Fujii, Y.; Fujisawa, H.; Hirakawa, H.; Cartwright, I.M.; Uesaka, M.; Kitamura, H.; Fujimori, A.; Kato, T.A. Hyperthermia-induced radiosensitization in CHO wild-type, NHEJ repair mutant and HR repair mutant following proton and carbon-ion exposure. Oncol. Lett. 2015, 10, 2828–2834. [Google Scholar] [CrossRef] [PubMed]
- Haskins, A.H.; Buglewicz, D.J.; Hirakawa, H.; Fujimori, A.; Aizawa, Y.; Kato, T.A. Palmitoyl ascorbic acid 2-glucoside has the potential to protect mammalian cells from high-LET carbon-ion radiation. Sci. Rep. 2018, 8, 13822. [Google Scholar] [CrossRef]
- Yu, H.; Haskins, J.S.; Su, C.; Allum, A.; Haskins, A.H.; Salinas, V.A.; Sunada, S.; Inoue, T.; Aizawa, Y.; Uesaka, M.; et al. In vitro screening of radioprotective properties in the novel glucosylated flavonoids. Int. J. Mol. Med. 2016, 38, 1525–1530. [Google Scholar] [CrossRef]
- Elmegerhi, S.; Su, C.; Buglewicz, D.J.; Aizawa, Y.; Kato, T.A. Effect of hydroxyl group position in flavonoids on inducing singlestranded DNA damage mediated by cupric ions. Int. J. Mol. Med. 2018, 42, 658–664. [Google Scholar] [CrossRef]
- Jenner, T.J.; Delara, C.M.; Oneill, P.; Stevens, D.L. Induction and rejoining of dna double-strand breaks in V79-4 mammalian-cells following gamma-irradiation and alpha-irradiation. Int. J. Radiat. Biol. 1993, 64, 265–273. [Google Scholar] [CrossRef] [PubMed]
- Heilmann, J.; TaucherScholz, G.; Haberer, T.; Scholz, M.; Kraft, G. Measurement of intracellular DNA double-strand break induction and rejoining along the track of carbon and neon particle beams in water. Int. J. Radiat. Oncol. Biol. Phys. 1996, 34, 599–608. [Google Scholar] [CrossRef]
- Takata, H.; Hanafusa, T.; Mori, T.; Shimura, M.; Iida, Y.; Ishikawa, K.; Yoshikawa, K.; Yoshikawa, Y.; Maeshima, K. Chromatin Compaction Protects Genomic DNA from Radiation Damage. PLoS ONE 2013, 8, e75622. [Google Scholar] [CrossRef]
- Penninckx, S.; Pariset, E.; Cekanaviciute, E.; Costes, S.V. Quantification of radiation-induced DNA double strand break repair foci to evaluate and predict biological responses to ionizing radiation. NAR Cancer 2021, 3, zcab046. [Google Scholar] [CrossRef] [PubMed]
- Sabella, S.; Carney, R.P.; Brunetti, V.; Malvindi, M.A.; Al-Juffali, N.; Vecchio, G.; Janes, S.M.; Bakr, O.M.; Cingolani, R.; Stellacci, F.; et al. A general mechanism for intracellular toxicity of metal-containing nanoparticles. Nanoscale 2014, 6, 7052–7061. [Google Scholar] [CrossRef] [PubMed]
- Penninckx, S.; Heuskin, A.-C.; Michiels, C.; Lucas, S. The role of thioredoxin reductase in gold nanoparticle radiosensitization effects. Nanomedicine 2018, 13, 2917–2937. [Google Scholar] [CrossRef]
- Penninckx, S.; Heuskin, A.-C.; Michiels, C.; Lucas, S. Gold Nanoparticles as a Potent Radiosensitizer: A Transdisciplinary Approach from Physics to Patient. Cancers 2020, 12, 2021. [Google Scholar] [CrossRef]
- Lomax, M.E.; Folkes, L.K.; O’Neill, P. Biological Consequences of Radiation-induced DNA Damage: Relevance to Radiotherapy. Clin. Oncol. (R. Coll. Radiol.) 2013, 25, 578–585. [Google Scholar] [CrossRef]
- Regulus, P.; Duroux, B.; Baylet, P.A.; Favier, A.; Cadet, J.; Ravanat, J.L. Oxidation of the sugar radiation or bleomycin of a cluster DNA lesion moiety of DNA by ionizing could induce the formation. Proc. Natl. Acad. Sci. USA 2007, 104, 14032–14037. [Google Scholar] [CrossRef]
- Stubbe, J.; Kozarich, J.W. Mechanisms of bleomycin-induced DNA-degradation. Chem. Rev. 1987, 87, 1107–1136. [Google Scholar] [CrossRef]
- Suzuki, T.; Kuwahara, J.; Sugiura, Y. Copper-bleomycin has no significant DNA cleavage activity. Biochemistry 1985, 24, 4719–4721. [Google Scholar] [CrossRef] [PubMed]
- Abraham, A.T.; Zhou, X.; Hecht, S.M. Metallobleomycin-mediated cleavage of DNA not involving a threading-intercalation mechanism. J. Am. Chem. Soc. 2001, 123, 5167–5175. [Google Scholar] [CrossRef]
- Ehrenfeld, G.M.; Shipley, J.B.; Heimbrook, D.C.; Sugiyama, H.; Long, E.C.; Vanboom, J.H.; Vandermarel, G.A.; Oppenheimer, N.J.; Hecht, S.M. Copper-dependent cleavage of DNA by bleomycin. Biochemistry 1987, 26, 931–942. [Google Scholar] [CrossRef] [PubMed]
- Hecht, S.M. Bleomycin: New perspectives on the mechanism of action. J. Nat. Prod. 2000, 63, 158–168. [Google Scholar] [CrossRef]
- Nightingale, K.P.; Fox, K.R. Light-activated cleavage of DNA by cobalt-bleomycin. Eur. J. Biochem. 1994, 220, 173–181. [Google Scholar] [CrossRef] [PubMed]


| Control | Cu2+ | Co2+ | |
|---|---|---|---|
| DSB | Initial dsDNA amount: 30 ng Lambda phage dsDNA in 10 μL of reaction solution | ||
| X-ray | 44 Gy | 8.5 Gy | 15.5 Gy |
| Carbon-ion | 142.9 Gy | 28.1 Gy | 80.9 Gy |
| Iron-ions | 299.1 Gy | 80 Gy | 187 Gy |
| Bleomycin | 46,625 nM | 29,882.4 nM | 39,560.6 nM |
| H2O2 | 20,073 nM | 22 nM | 177 nM |
| SSB | Initial ssDNA amount: 83 ng M13 bacteriophage ssDNA in 10 μL of reaction solution | ||
| X-ray | 57.5 Gy | 3.1 Gy | 9.6 Gy |
| Carbon-ion | 129.8 Gy | 2.9 Gy | 65 Gy |
| Iron-ions | 160 Gy | 7.0 Gy | 102.0 Gy |
| Bleomycin | 8689.2 nM | 4591.9 nM | 26,138.2 nM |
| H2O2 | 17,237 nM | 0.156 nM | 1138 nM |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Buglewicz, D.J.; Su, C.; Banks, A.B.; Stenger-Smith, J.; Elmegerhi, S.; Hirakawa, H.; Fujimori, A.; Kato, T.A. Metal Ions Modify In Vitro DNA Damage Yields with High-LET Radiation. Toxics 2023, 11, 773. https://doi.org/10.3390/toxics11090773
Buglewicz DJ, Su C, Banks AB, Stenger-Smith J, Elmegerhi S, Hirakawa H, Fujimori A, Kato TA. Metal Ions Modify In Vitro DNA Damage Yields with High-LET Radiation. Toxics. 2023; 11(9):773. https://doi.org/10.3390/toxics11090773
Chicago/Turabian StyleBuglewicz, Dylan J., Cathy Su, Austin B. Banks, Jazmine Stenger-Smith, Suad Elmegerhi, Hirokazu Hirakawa, Akira Fujimori, and Takamitsu A. Kato. 2023. "Metal Ions Modify In Vitro DNA Damage Yields with High-LET Radiation" Toxics 11, no. 9: 773. https://doi.org/10.3390/toxics11090773
APA StyleBuglewicz, D. J., Su, C., Banks, A. B., Stenger-Smith, J., Elmegerhi, S., Hirakawa, H., Fujimori, A., & Kato, T. A. (2023). Metal Ions Modify In Vitro DNA Damage Yields with High-LET Radiation. Toxics, 11(9), 773. https://doi.org/10.3390/toxics11090773





