Multi-Omics Analysis Reveals the Toxicity of Polyvinyl Chloride Microplastics toward BEAS-2B Cells
Abstract
1. Introduction
2. Materials and Methods
2.1. PVC-MPs Characterization
2.2. Cell Culture and Cytotoxicity Testing
2.3. Transcriptomics Analysis
2.4. Untargeted Metabolomics Analysis
2.5. Multi-Omics Analysis
2.6. Statistical Analysis
3. Results
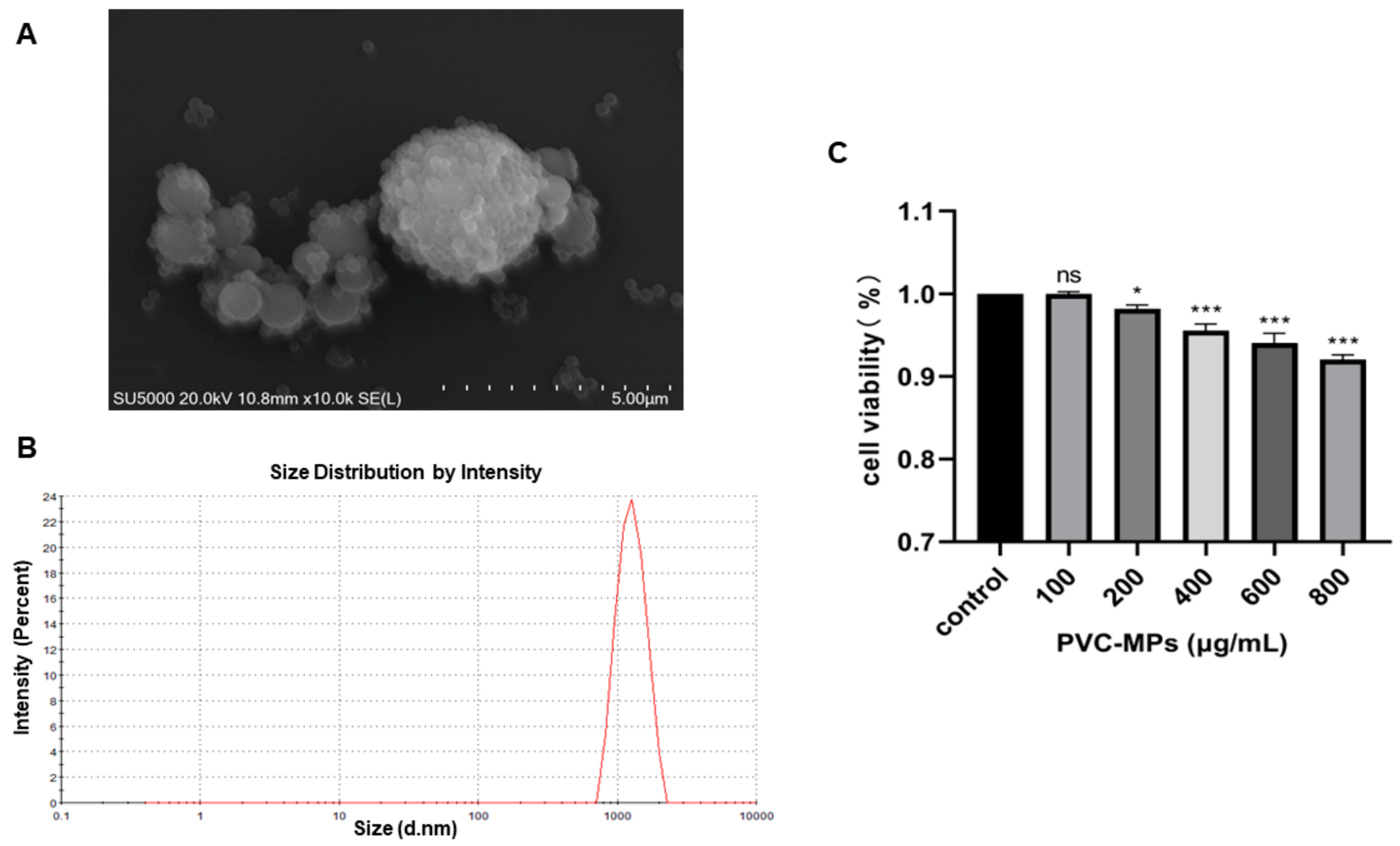
3.1. Characterization of PVC-MPs
3.2. Cytotoxicity Effects of PVC-MPs on BEAS-2B Cells
3.3. Transcriptomics Analysis of BEAS-2B Samples Exposed to PVC-MPs
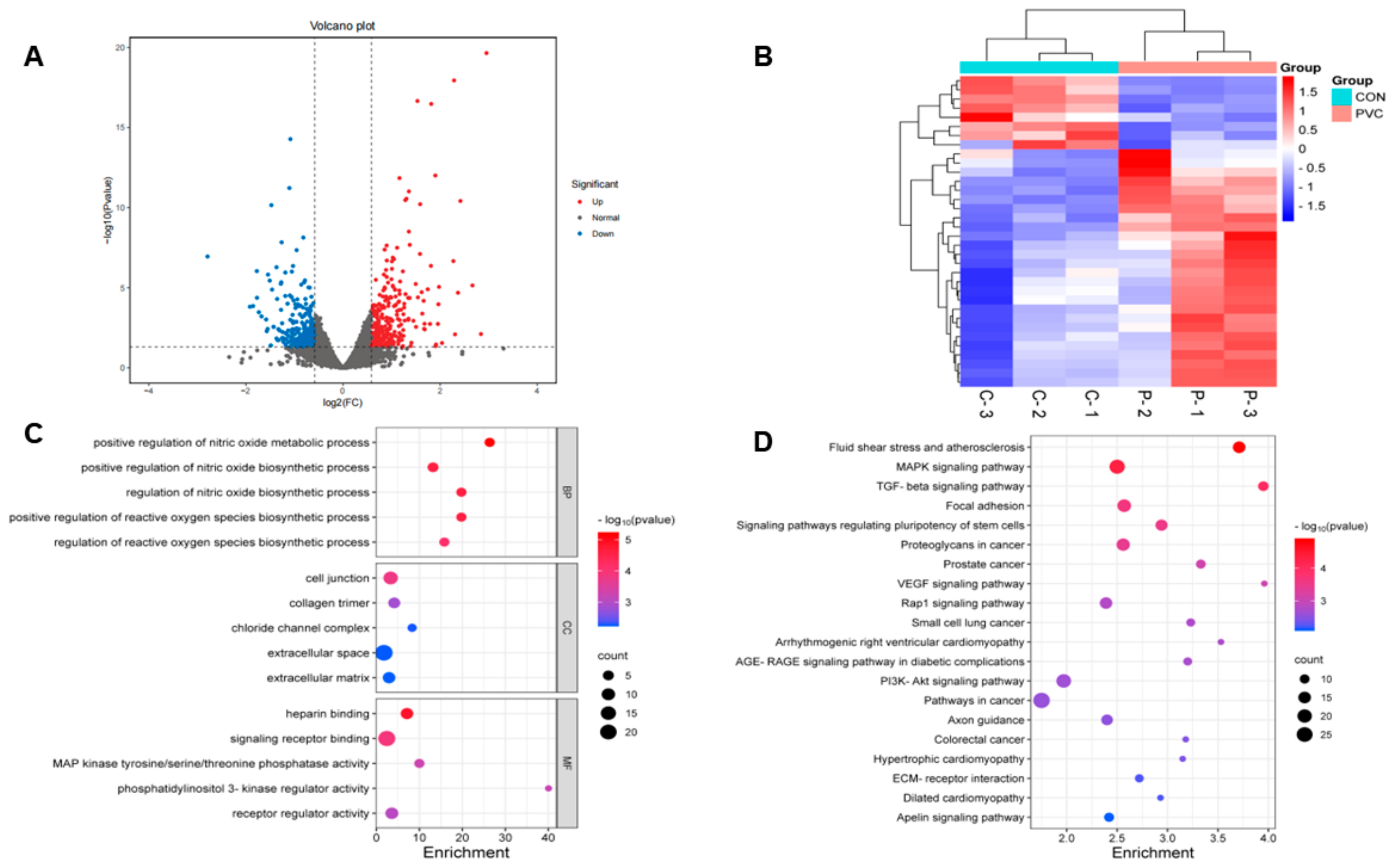
3.3.1. Screening and Analysis of Differentially Expressed Genes
3.3.2. GO and KEGG Analysis
3.4. Metabolomics Analysis of BEAS-2B Samples Exposed to PVC-MPs
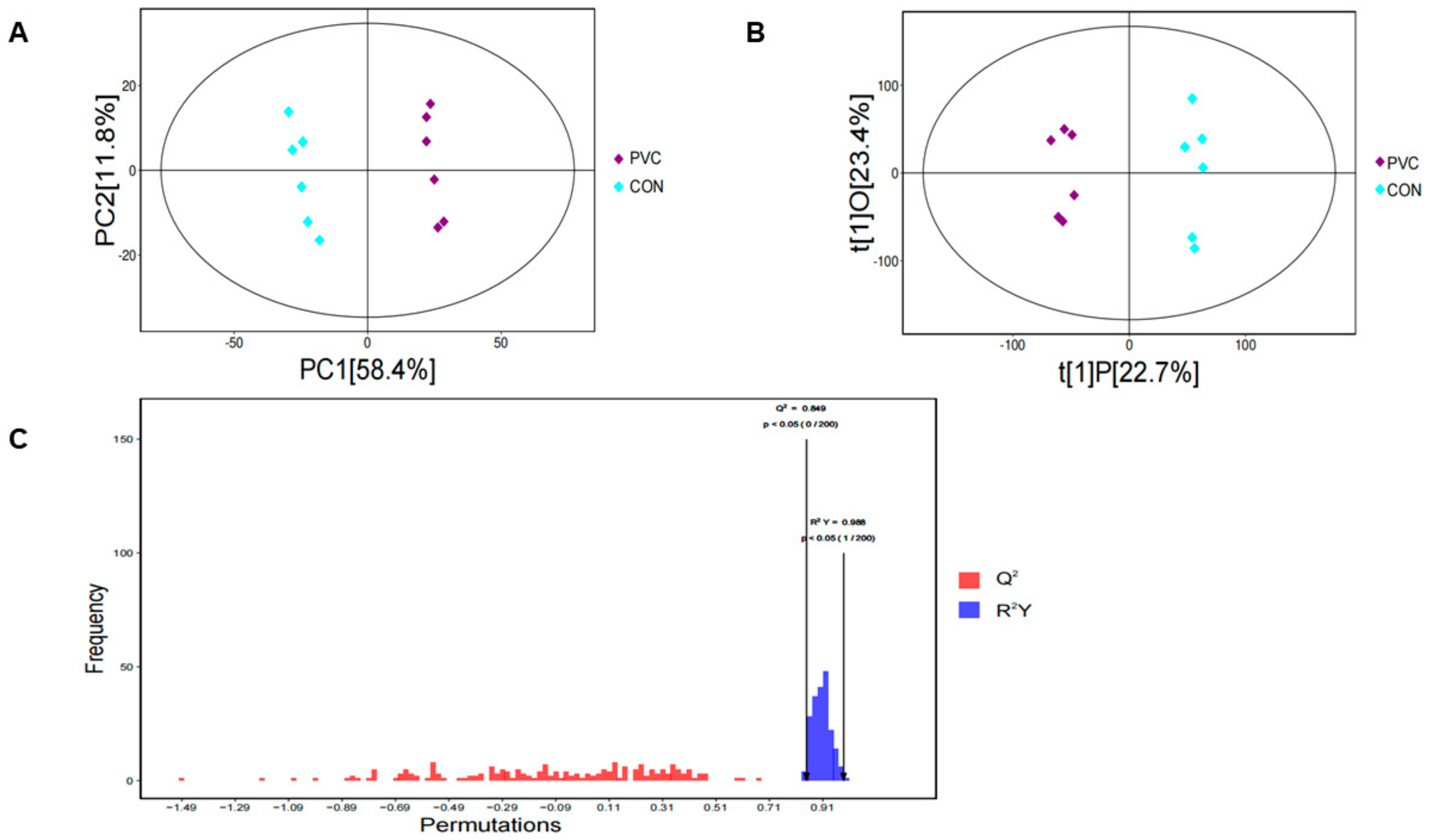
3.4.1. Multivariate Analysis
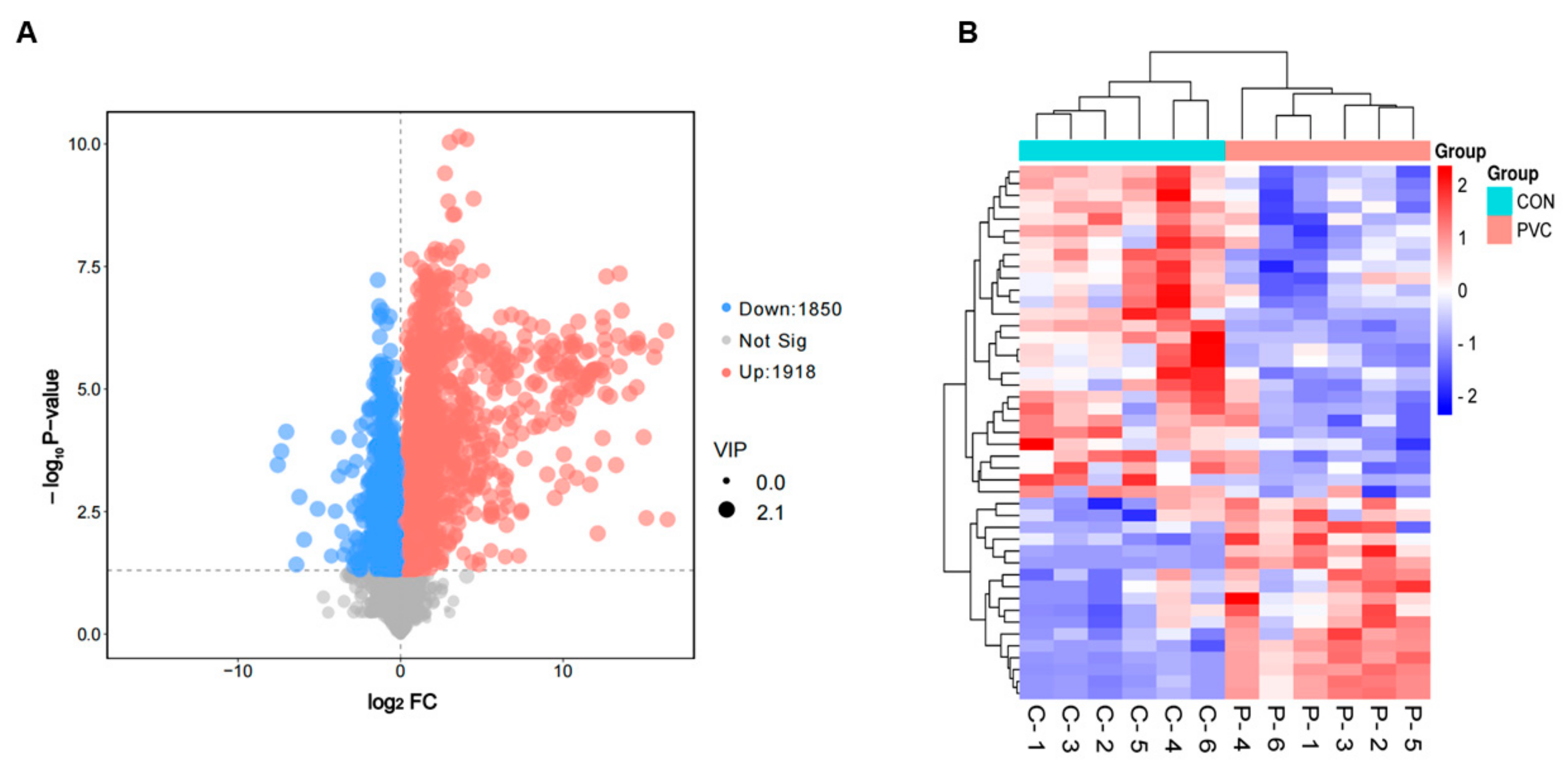
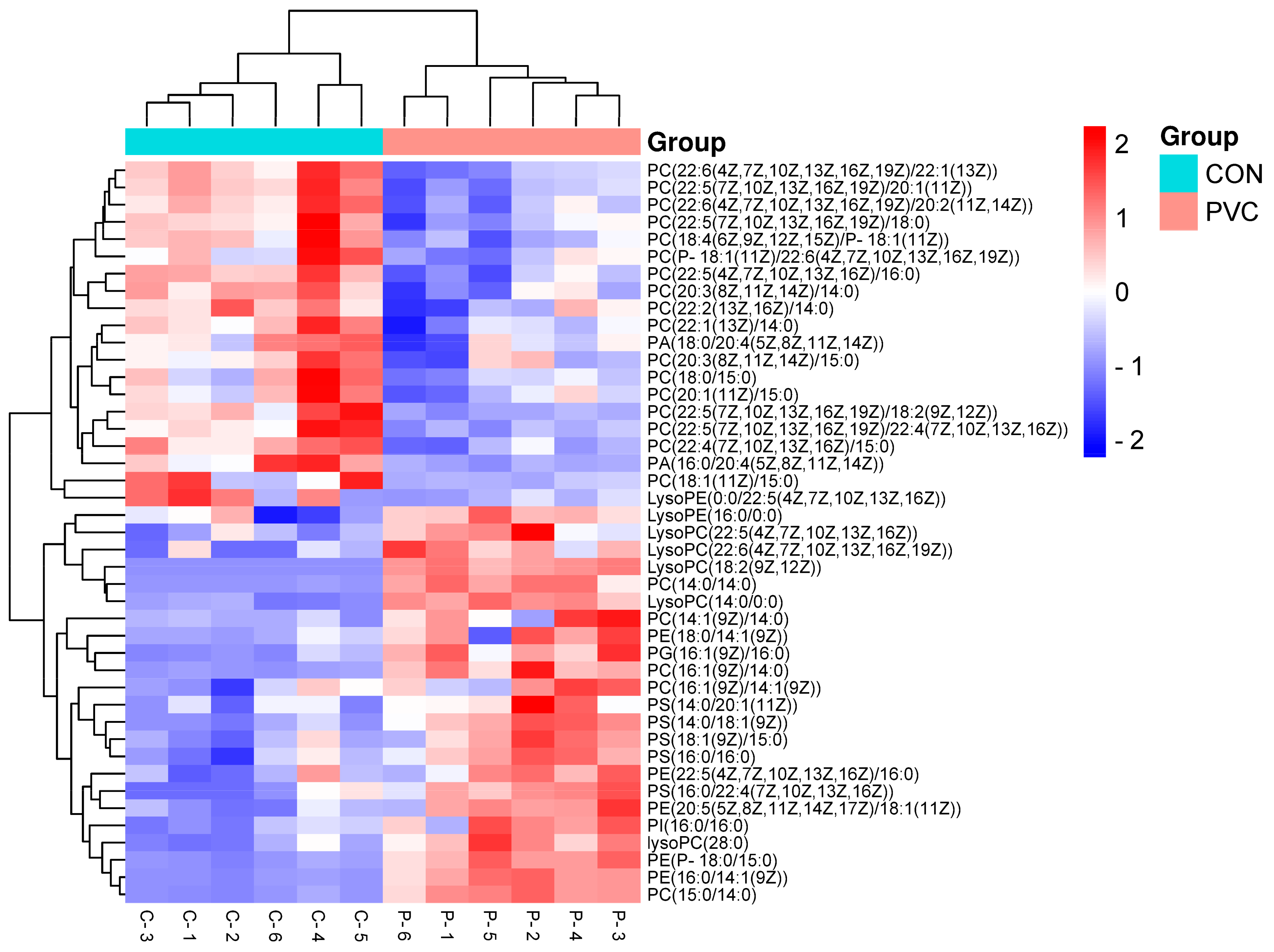
3.4.2. Screening and Analysis of Differentially Expressed Metabolites
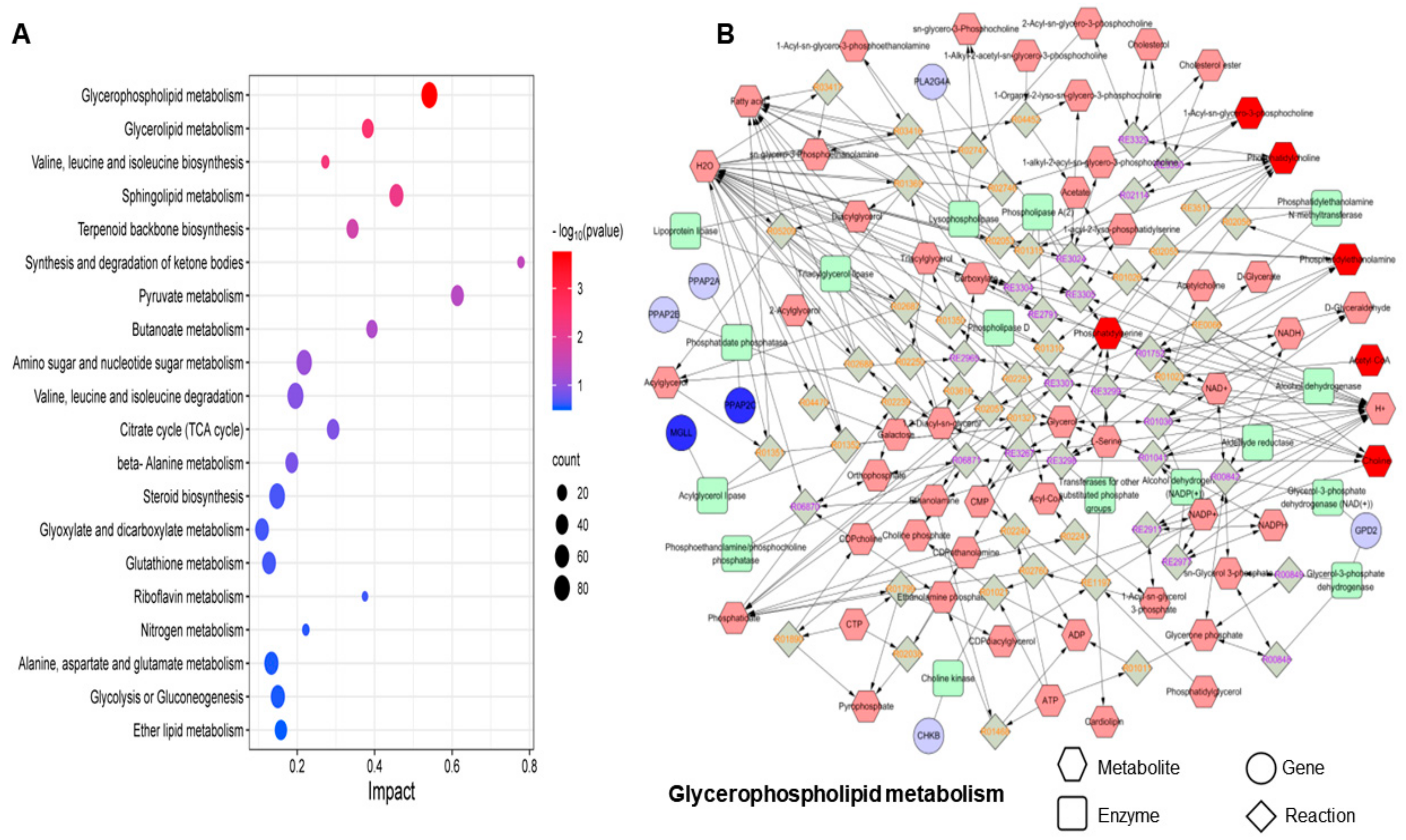
3.4.3. Metabolic Pathway Analysis
3.5. Integrated Analysis of Transcriptomics and Metabolomics
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Joseph, B.; James, J.; Kalarikkal, N.; Thomas, S. Recycling of medical plastics. Adv. Ind. Eng. Polym. Res. 2021, 4, 199–208. [Google Scholar] [CrossRef]
- Romeo, J. Do No Harm: Plastics are playing a major role in giving healthcare professionals the tools and capabilities they need to battle the COVID pandemic. Plast. Eng. 2020, 76, 41. [Google Scholar] [CrossRef]
- Ahmed, N. Utilizing plastic waste in the building and construction industry: A pathway towards the circular economy. Constr. Build. Mater. 2023, 383, 131311. [Google Scholar] [CrossRef]
- Xu, C.; Zhou, G.; Lu, J.; Shen, C.; Dong, Z.; Yin, S.; Li, F. Spatio-vertical distribution of riverine microplastics: Impact of the textile industry. Environ. Res. 2022, 211, 112789. [Google Scholar] [CrossRef] [PubMed]
- Fellner, J.; Brunner, P.H. Plastic waste management: Is circular economy really the best solution? J. Mater. Cycles Waste Manag. 2022, 24, 1–3. [Google Scholar] [CrossRef]
- Walker, T.R.; Fequet, L. Current trends of unsustainable plastic production and micro (nano) plastic pollution. TrAC Trends Anal. Chem. 2023, 160, 116984. [Google Scholar] [CrossRef]
- Chamas, A.; Moon, H.; Zheng, J.; Qiu, Y.; Tabassum, T.; Jang, J.H.; Abu-Omar, M.; Scott, S.L.; Suh, S. Degradation rates of plastics in the environment. ACS Sustain. Chem. Eng. 2020, 8, 3494–3511. [Google Scholar] [CrossRef]
- Klein, S.; Dimzon, I.K.; Eubeler, J.; Knepper, T.P. Analysis, occurrence, and degradation of microplastics in the aqueous environment. In Freshwater Microplastics: Emerging Environmental Contaminants; Springer: Berlin/Heidelberg, Germany, 2018; pp. 51–67. [Google Scholar]
- Thompson, R.C.; Olsen, Y.; Mitchell, R.P.; Davis, A.; Rowland, S.J.; John, A.W.; McGonigle, D.; Russell, A.E. Lost at sea: Where is all the plastic? Science 2004, 304, 838. [Google Scholar] [CrossRef] [PubMed]
- Arthur, C.; Baker, J.E.; Bamford, H.A. Proceedings of the International Research Workshop on the Occurrence, Effects, and Fate of Microplastic Marine Debris, Tacoma, WA, USA, 9–11 September 2008; University of Washington Tacoma: Tacoma, WA, USA, 2009. [Google Scholar]
- Hernandez, L.M.; Yousefi, N.; Tufenkji, N. Are there nanoplastics in your personal care products? Environ. Sci. Technol. Lett. 2017, 4, 280–285. [Google Scholar] [CrossRef]
- World Health Organization. Dietary and Inhalation Exposure to Nano-and Microplastic Particles and Potential Implications for Human Health; World Health Organization: Geneva, Switzerland, 2022.
- MacLeod, M.; Arp, H.P.H.; Tekman, M.B.; Jahnke, A. The global threat from plastic pollution. Science 2021, 373, 61–65. [Google Scholar] [CrossRef]
- Allen, S.; Allen, D.; Moss, K.; Le Roux, G.; Phoenix, V.R.; Sonke, J.E. Examination of the ocean as a source for atmospheric microplastics. PLoS ONE 2020, 15, e0232746. [Google Scholar] [CrossRef] [PubMed]
- Prata, J.C. Airborne microplastics: Consequences to human health? Environ. Pollut. 2018, 234, 115–126. [Google Scholar] [CrossRef] [PubMed]
- Vianello, A.; Jensen, R.L.; Liu, L.; Vollertsen, J. Simulating human exposure to indoor airborne microplastics using a Breathing Thermal Manikin. Sci. Rep. 2019, 9, 8670. [Google Scholar] [CrossRef] [PubMed]
- Chen, R.; Hu, B.; Liu, Y.; Xu, J.; Yang, G.; Xu, D.; Chen, C. Beyond PM2.5: The role of ultrafine particles on adverse health effects of air pollution. Biochim. Biophys. Acta 2016, 1860, 2844–2855. [Google Scholar] [CrossRef] [PubMed]
- Liu, K.; Wang, X.; Fang, T.; Xu, P.; Zhu, L.; Li, D. Source and potential risk assessment of suspended atmospheric microplastics in Shanghai. Sci. Total Environ. 2019, 675, 462–471. [Google Scholar] [CrossRef] [PubMed]
- Dong, C.D.; Chen, C.W.; Chen, Y.C.; Chen, H.H.; Lee, J.S.; Lin, C.H. Polystyrene microplastic particles: In vitro pulmonary toxicity assessment. J. Hazard. Mater. 2020, 385, 121575. [Google Scholar] [CrossRef]
- Bajt, O. From plastics to microplastics and organisms. FEBS Open Bio 2021, 11, 954–966. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Kang, S.; Allen, S.; Allen, D.; Gao, T.; Sillanpää, M. Atmospheric microplastics: A review on current status and perspectives. Earth-Sci. Rev. 2020, 203, 103118. [Google Scholar] [CrossRef]
- Chen, X.; Zhuang, J.; Chen, Q.; Xu, L.; Yue, X.; Qiao, D. Chronic exposure to polyvinyl chloride microplastics induces liver injury and gut microbiota dysbiosis based on the integration of liver transcriptome profiles and full-length 16S rRNA sequencing data. Sci. Total Environ. 2022, 839, 155984. [Google Scholar] [CrossRef]
- Zhuang, J.; Chen, Q.; Xu, L.; Chen, X. Combined exposure to polyvinyl chloride and polystyrene microplastics induces liver injury and perturbs gut microbial and serum metabolic homeostasis in mice. Ecotoxicol. Environ. Saf. 2023, 267, 115637. [Google Scholar] [CrossRef] [PubMed]
- Pilechi, A.; Mohammadian, A.; Murphy, E. A numerical framework for modeling fate and transport of microplastics in inland and coastal waters. Mar. Pollut. Bull. 2022, 184, 114119. [Google Scholar] [CrossRef] [PubMed]
- Marcuello, C. Present and future opportunities in the use of atomic force microscopy to address the physico-chemical properties of aquatic ecosystems at the nanoscale level. Int. Aquat. Res. 2022. [Google Scholar]
- Lowe, R.; Shirley, N.; Bleackley, M.; Dolan, S.; Shafee, T. Transcriptomics technologies. PLoS Comput. Biol. 2017, 13, e1005457. [Google Scholar] [CrossRef] [PubMed]
- Galal, A.; Talal, M.; Moustafa, A. Applications of machine learning in metabolomics: Disease modeling and classification. Front. Genet. 2022, 13, 1017340. [Google Scholar] [CrossRef] [PubMed]
- Shah, N.J.; Sureshkumar, S.; Shewade, D.G. Metabolomics: A Tool Ahead for Understanding Molecular Mechanisms of Drugs and Diseases. Indian J. Clin. Biochem. 2015, 30, 247–254. [Google Scholar] [CrossRef] [PubMed]
- Ritchie, M.D.; Holzinger, E.R.; Li, R.; Pendergrass, S.A.; Kim, D. Methods of integrating data to uncover genotype–phenotype interactions. Nat. Rev. Genet. 2015, 16, 85–97. [Google Scholar] [CrossRef] [PubMed]
- Zhang, M.; Shi, J.; Huang, Q.; Xie, Y.; Wu, R.; Zhong, J.; Deng, H. Multi-omics analysis reveals size-dependent toxicity and vascular endothelial cell injury induced by microplastic exposure in vivo and in vitro. Environ. Sci. Nano 2022, 9, 663–683. [Google Scholar] [CrossRef]
- Luo, Y.; Geng, N.; Sun, S.; Cheng, L.; Chen, S.; Zhang, H.; Chen, J. Integration approach of transcriptomics and metabolomics reveals the toxicity of Anthracene and its chlorinated derivatives on human hepatic cells. Sci. Total Environ. 2023, 905, 166886. [Google Scholar] [CrossRef] [PubMed]
- Kyung, S.; Zheng, T.; Park, Y.; Lee, J.; Kim, H. Potential toxicity of polystyrene microplastics with different particle size and surface charge in human lung epithelial BEAS-2B cells. Toxicol. Lett. 2022, 368, S135–S136. [Google Scholar] [CrossRef]
- Wu, Y.; Wang, J.; Zhao, T.; Sun, M.; Xu, M.; Che, S.; Pan, Z.; Wu, C.; Shen, L. Polystyrene nanoplastics lead to ferroptosis in the lungs. J. Adv. Res. 2024, 56, 31–41. [Google Scholar] [CrossRef] [PubMed]
- Lou, J.; Chu, G.; Zhou, G.; Jiang, J.; Huang, F.; Xu, J.; Zheng, S.; Jiang, W.; Lu, Y.; Li, X.; et al. Comparison between two kinds of cigarette smoke condensates (CSCs) of the cytogenotoxicity and protein expression in a human B-cell lymphoblastoid cell line using CCK-8 assay, comet assay and protein microarray. Mutat. Res. Toxicol. Environ. Mutagen. 2010, 697, 55–59. [Google Scholar] [CrossRef]
- Zhang, Y.; Li, W.-Y.; Lan, R.; Wang, J.-Y. Quality monitoring of porous zein scaffolds: A novel biomaterial. Engineering 2017, 3, 130–135. [Google Scholar] [CrossRef]
- Cai, L.; Qin, X.; Xu, Z.; Song, Y.; Jiang, H.; Wu, Y.; Ruan, H.; Chen, J. Comparison of cytotoxicity evaluation of anticancer drugs between real-time cell analysis and CCK-8 method. ACS Omega 2019, 4, 12036–12042. [Google Scholar] [CrossRef] [PubMed]
- Gene Ontology Consortium. Gene ontology consortium: Going forward. Nucleic Acids Res. 2015, 43, D1049–D1056. [Google Scholar] [CrossRef] [PubMed]
- Aoki, K.F.; Kanehisa, M. Using the KEGG database resource. Curr. Protoc. Bioinform. 2005, 11, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Amato-Lourenço, L.F.; Carvalho-Oliveira, R.; Júnior, G.R.; dos Santos Galvão, L.; Ando, R.A.; Mauad, T. Presence of airborne microplastics in human lung tissue. J. Hazard. Mater. 2021, 416, 126124. [Google Scholar] [CrossRef] [PubMed]
- Choi, D.; Hwang, J.; Bang, J.; Han, S.; Kim, T.; Oh, Y.; Hwang, Y.; Choi, J.; Hong, J. In vitro toxicity from a physical perspective of polyethylene microplastics based on statistical curvature change analysis. Sci. Total Environ. 2021, 752, 142242. [Google Scholar] [CrossRef] [PubMed]
- Winiarska, E.; Jutel, M.; Zemelka-Wiacek, M. The potential impact of nano-and microplastics on human health: Understanding human health risks. Environ. Res. 2024, 251, 118535. [Google Scholar] [CrossRef] [PubMed]
- Mahadevan, G.; Valiyaveettil, S. Comparison of genotoxicity and cytotoxicity of polyvinyl chloride and poly (methyl methacrylate) nanoparticles on normal human lung cell lines. Chem. Res. Toxicol. 2021, 34, 1468–1480. [Google Scholar] [CrossRef] [PubMed]
- Stock, V.; Laurisch, C.; Franke, J.; Dönmez, M.H.; Voss, L.; Böhmert, L.; Braeuning, A.; Sieg, H. Uptake and cellular effects of PE, PP, PET and PVC microplastic particles. Toxicol. Vitr. 2021, 70, 105021. [Google Scholar] [CrossRef] [PubMed]
- Mahadevan, G.; Valiyaveettil, S. Understanding the interactions of poly (methyl methacrylate) and poly (vinyl chloride) nanoparticles with BHK-21 cell line. Sci. Rep. 2021, 11, 2089. [Google Scholar] [CrossRef] [PubMed]
- Salimi, A.; Alavehzadeh, A.; Ramezani, M.; Pourahmad, J. Differences in sensitivity of human lymphocytes and fish lymphocytes to polyvinyl chloride microplastic toxicity. Toxicol. Ind. Health 2022, 38, 100–111. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Liu, Y.; Fu, Y.; Wei, T.; Le Guyader, L.; Gao, G.; Liu, R.-S.; Chang, Y.-Z.; Chen, C. The triggering of apoptosis in macrophages by pristine graphene through the MAPK and TGF-beta signaling pathways. Biomaterials 2012, 33, 402–411. [Google Scholar] [CrossRef]
- Zhang, W.; Liu, H.T. MAPK signal pathways in the regulation of cell proliferation in mammalian cells. Cell Res. 2002, 12, 9–18. [Google Scholar] [CrossRef] [PubMed]
- Junttila, M.R.; Li, S.P.; Westermarck, J. Phosphatase-mediated crosstalk between MAPK signaling pathways in the regulation of cell survival. FASEB J. 2008, 22, 954–965. [Google Scholar] [CrossRef]
- Sun, Q.Y.; Breitbart, H.; Schatten, H. Role of the MAPK cascade in mammalian germ cells. Reprod. Fertil. Dev. 1999, 11, 443–450. [Google Scholar] [CrossRef] [PubMed]
- Moustakas, A.; Pardali, K.; Gaal, A.; Heldin, C.-H. Mechanisms of TGF-β signaling in regulation of cell growth and differentiation. Immunol. Lett. 2002, 82, 85–91. [Google Scholar] [CrossRef] [PubMed]
- Lee, J.-H.; Mellado-Gil, J.M.; Bahn, Y.J.; Pathy, S.M.; Zhang, Y.E.; Rane, S.G. Protection from β-cell apoptosis by inhibition of TGF-β/Smad3 signaling. Cell Death Dis. 2020, 11, 184. [Google Scholar] [CrossRef] [PubMed]
- Heldin, C.-H.; Miyazono, K.; Ten Dijke, P. TGF-β signalling from cell membrane to nucleus through SMAD proteins. Nature 1997, 390, 465–471. [Google Scholar] [CrossRef] [PubMed]
- ten Dijke, P.; Miyazono, K.; Heldin, C.-H. Signaling inputs converge on nuclear effectors in TGF-β signaling. Trends Biochem. Sci. 2000, 25, 64–70. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Jiang, S.; Liu, Y.; Du, X.; Zhang, W.; Zhang, J.; Shen, H. Comprehensive pulmonary metabolome responses to intratracheal instillation of airborne fine particulate matter in rats. Sci. Total Environ. 2017, 592, 41–50. [Google Scholar] [CrossRef]
- Ling, Z.N.; Jiang, Y.F.; Ru, J.N.; Lu, J.H.; Ding, B.; Wu, J. Amino acid metabolism in health and disease. Signal Transduct. Target. Ther. 2023, 8, 345. [Google Scholar] [CrossRef] [PubMed]
- Holeček, M. Branched-chain amino acids in health and disease: Metabolism, alterations in blood plasma, and as supplements. Nutr. Metab. 2018, 15, 33. [Google Scholar] [CrossRef] [PubMed]
- Ruiz-Canela, M.; Toledo, E.; Clish, C.B.; Hruby, A.; Liang, L.; Salas-Salvado, J.; Razquin, C.; Corella, D.; Estruch, R.; Ros, E. Plasma branched-chain amino acids and incident cardiovascular disease in the PREDIMED trial. Clin. Chem. 2016, 62, 582–592. [Google Scholar] [CrossRef] [PubMed]
- Alvers, A.L.; Fishwick, L.K.; Wood, M.S.; Hu, D.; Chung, H.S.; Dunn, W.A., Jr.; Aris, J.P. Autophagy and amino acid homeostasis are required for chronological longevity in Saccharomyces cerevisiae. Aging Cell 2009, 8, 353–369. [Google Scholar] [CrossRef] [PubMed]
- Liao, X.; Liu, S.; Chen, S.; Shan, X.; He, J.; Li, C. Transcriptomic analysis reveals the role of Glycolysis pathway in Litopenaeus vannamei during DIV1 infection. Fish Shellfish Immunol. 2023, 141, 109036. [Google Scholar] [CrossRef] [PubMed]
- DeBerardinis, R.J.; Thompson, C.B. Cellular metabolism and disease: What do metabolic outliers teach us? Cell 2012, 148, 1132–1144. [Google Scholar] [CrossRef] [PubMed]
- Shi, D.Y.; Xie, F.Z.; Zhai, C.; Stern, J.S.; Liu, Y.; Liu, S.L. The role of cellular oxidative stress in regulating glycolysis energy metabolism in hepatoma cells. Mol. Cancer 2009, 8, 32. [Google Scholar] [CrossRef] [PubMed]
- Gao, L.; Loveless, J.; Shay, C.; Teng, Y. Targeting ROS-mediated crosstalk between autophagy and apoptosis in cancer. In Reviews on New Drug Targets in Age-Related Disorders; Springer: Berlin/Heidelberg, Germany, 2020; pp. 1–12. [Google Scholar]
- Weiner, J., 3rd; Parida, S.K.; Maertzdorf, J.; Black, G.F.; Repsilber, D.; Telaar, A.; Mohney, R.P.; Arndt-Sullivan, C.; Ganoza, C.A.; Fae, K.C.; et al. Biomarkers of inflammation, immunosuppression and stress with active disease are revealed by metabolomic profiling of tuberculosis patients. PLoS ONE 2012, 7, e40221. [Google Scholar] [CrossRef] [PubMed]
- Ding, Z.; Chen, W.; Wu, H.; Li, W.; Mao, X.; Su, W.; Zhang, Y.; Lin, N. Integrative network fusion-based multi-omics study for biomarker identification and patient classification of rheumatoid arthritis. Chin. Med. 2023, 18, 48. [Google Scholar] [CrossRef] [PubMed]
- Yan, J.; Risacher, S.L.; Shen, L.; Saykin, A.J. Network approaches to systems biology analysis of complex disease: Integrative methods for multi-omics data. Briefings Bioinform. 2018, 19, 1370–1381. [Google Scholar] [CrossRef]
- Huang, C.; Freter, C. Lipid metabolism, apoptosis and cancer therapy. Int. J. Mol. Sci. 2015, 16, 924–949. [Google Scholar] [CrossRef] [PubMed]
- Santos, C.R.; Schulze, A. Lipid metabolism in cancer. FEBS J. 2012, 279, 2610–2623. [Google Scholar] [CrossRef]
- Dolce, V.; Rita Cappello, A.; Lappano, R.; Maggiolini, M. Glycerophospholipid synthesis as a novel drug target against cancer. Curr. Mol. Pharmacol. 2011, 4, 167–175. [Google Scholar] [CrossRef] [PubMed]
- Vance, D.E. Glycerolipid biosynthesis in eukaryotes. In New Comprehensive Biochemistry; Elsevier: Amsterdam, The Netherlands, 1996; Volume 31, pp. 153–181. [Google Scholar]
- Wang, X.; Xu, Y.; Song, X.; Jia, Q.; Zhang, X.; Qian, Y.; Qiu, J. Analysis of glycerophospholipid metabolism after exposure to PCB153 in PC12 cells through targeted lipidomics by UHPLC-MS/MS. Ecotoxicol. Environ. Saf. 2019, 169, 120–127. [Google Scholar] [CrossRef] [PubMed]
- Feng, J.; Cavallero, S.; Hsiai, T.; Li, R. Impact of air pollution on intestinal redox lipidome and microbiome. Free Radic. Biol. Med. 2020, 151, 99–110. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Li, M.; Pu, Z.; Chi, X.; Yang, J. Multi-omics data reveals the disturbance of glycerophospholipid metabolism and linoleic acid metabolism caused by disordered gut microbiota in PM2.5 gastrointestinal exposed rats. Ecotoxicol. Environ. Saf. 2023, 262, 115182. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Zeng, Y.; Song, J.; Zhu, M.; Zhu, G.; Cai, H.; Chen, C.; Jin, M.; Song, Y. Perturbation of arachidonic acid and glycerolipid metabolism promoted particulate matter-induced inflammatory responses in human bronchial epithelial cells. Ecotoxicol. Environ. Saf. 2023, 256, 114839. [Google Scholar] [CrossRef] [PubMed]
- Novgorodov, S.A.; Gudz, T.I. Ceramide and mitochondria in ischemic brain injury. Int. J. Biochem. Mol. Biol. 2011, 2, 347–361. [Google Scholar] [PubMed]
- Nagahara, Y.; Shinomiya, T.; Kuroda, S.; Kaneko, N.; Nishio, R.; Ikekita, M. Phytosphingosine induced mitochondria-involved apoptosis. Cancer Sci. 2005, 96, 83–92. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Wen, J.; Sun, C.; Zhou, Y.; Xu, J.; MacIsaac, H.J.; Chang, X.; Cui, Q. Phytosphingosine-induced cell apoptosis via a mitochondrially mediated pathway. Toxicology 2022, 482, 153370. [Google Scholar] [CrossRef] [PubMed]
- Zhao, J.; Tian, X.C.; Zhang, J.Q.; Li, T.T.; Qiao, S.; Jiang, S.L. Tribulus terrestris L. induces cell apoptosis of breast cancer by regulating sphingolipid metabolism signaling pathways. Phytomedicine 2023, 120, 155014. [Google Scholar] [CrossRef] [PubMed]
- D’Angelo, G.; Moorthi, S.; Luberto, C. Role and Function of Sphingomyelin Biosynthesis in the Development of Cancer. Adv. Cancer Res. 2018, 140, 61–96. [Google Scholar] [CrossRef] [PubMed]
- Guo, Y.; Chang, L.; Zhang, G.; Gao, Z.; Lin, H.; Zhang, Y.; Hu, L.; Chen, S.; Fan, B.; Zhang, S.; et al. The role of Sphingomyelin synthase 2 (SMS2) in platelet activation and its clinical significance. Thromb. J. 2021, 19, 27. [Google Scholar] [CrossRef] [PubMed]
- Bian, X.; Liu, R.; Meng, Y.; Xing, D.; Xu, D.; Lu, Z. Lipid metabolism and cancer. J. Exp. Med. 2021, 218. [Google Scholar] [CrossRef] [PubMed]
- Choi, D.; Kim, C.; Kim, T.; Park, K.; Im, J.; Hong, J. Potential threat of microplastics to humans: Toxicity prediction modeling by small data analysis. Environ. Sci. Nano 2023, 10, 1096–1108. [Google Scholar] [CrossRef]

| Concentration (μg/mL) | Zeta Potential (mV) |
|---|---|
| 100 | −27.53 |
| 200 | −25.17 |
| 400 | −25.10 |
| 600 | −25.52 |
| 800 | −31.83 |
| KEGG ID | Pathway | p-Value |
|---|---|---|
| ko00564 | Glycerophospholipid metabolism | 0.0001756 |
| hsa00561 | Glycerolipid metabolism | 0.0033649 |
| ko00290 | Valine, leucine, and isoleucine biosynthesis | 0.0046556 |
| map00600 | Sphingolipid metabolism | 0.0068775 |
| map00900 | Terpenoid backbone biosynthesis | 0.021066 |
| ko00072 | Synthesis and degradation of ketone bodies | 0.034016 |
| ko00620 | Pyruvate metabolism | 0.043594 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Liu, C.; Chen, S.; Chu, J.; Yang, Y.; Yuan, B.; Zhang, H. Multi-Omics Analysis Reveals the Toxicity of Polyvinyl Chloride Microplastics toward BEAS-2B Cells. Toxics 2024, 12, 399. https://doi.org/10.3390/toxics12060399
Liu C, Chen S, Chu J, Yang Y, Yuan B, Zhang H. Multi-Omics Analysis Reveals the Toxicity of Polyvinyl Chloride Microplastics toward BEAS-2B Cells. Toxics. 2024; 12(6):399. https://doi.org/10.3390/toxics12060399
Chicago/Turabian StyleLiu, Chengzhi, Shuang Chen, Jiangliang Chu, Yifan Yang, Beilei Yuan, and Huazhong Zhang. 2024. "Multi-Omics Analysis Reveals the Toxicity of Polyvinyl Chloride Microplastics toward BEAS-2B Cells" Toxics 12, no. 6: 399. https://doi.org/10.3390/toxics12060399
APA StyleLiu, C., Chen, S., Chu, J., Yang, Y., Yuan, B., & Zhang, H. (2024). Multi-Omics Analysis Reveals the Toxicity of Polyvinyl Chloride Microplastics toward BEAS-2B Cells. Toxics, 12(6), 399. https://doi.org/10.3390/toxics12060399






