Genetic Diversity of Newcastle Disease Virus Involved in the 2021 Outbreaks in Backyard Poultry Farms in Tanzania
Abstract
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Sampling
2.2. Virus Isolation
2.3. Hemagglutination Assay (HA)
2.4. Viral RNA Extraction and Reverse-Transcription Polymerase Chain Reaction (RT-PCR)
2.5. DNA Sequencing, Genetic Analysis, and Phylogeny Reconstruction
3. Results
3.1. Clinical and Postmortem Findings
3.2. Virus Isolation
3.3. RT-PCR Screening for NDV
3.4. Amino Acid Analyses
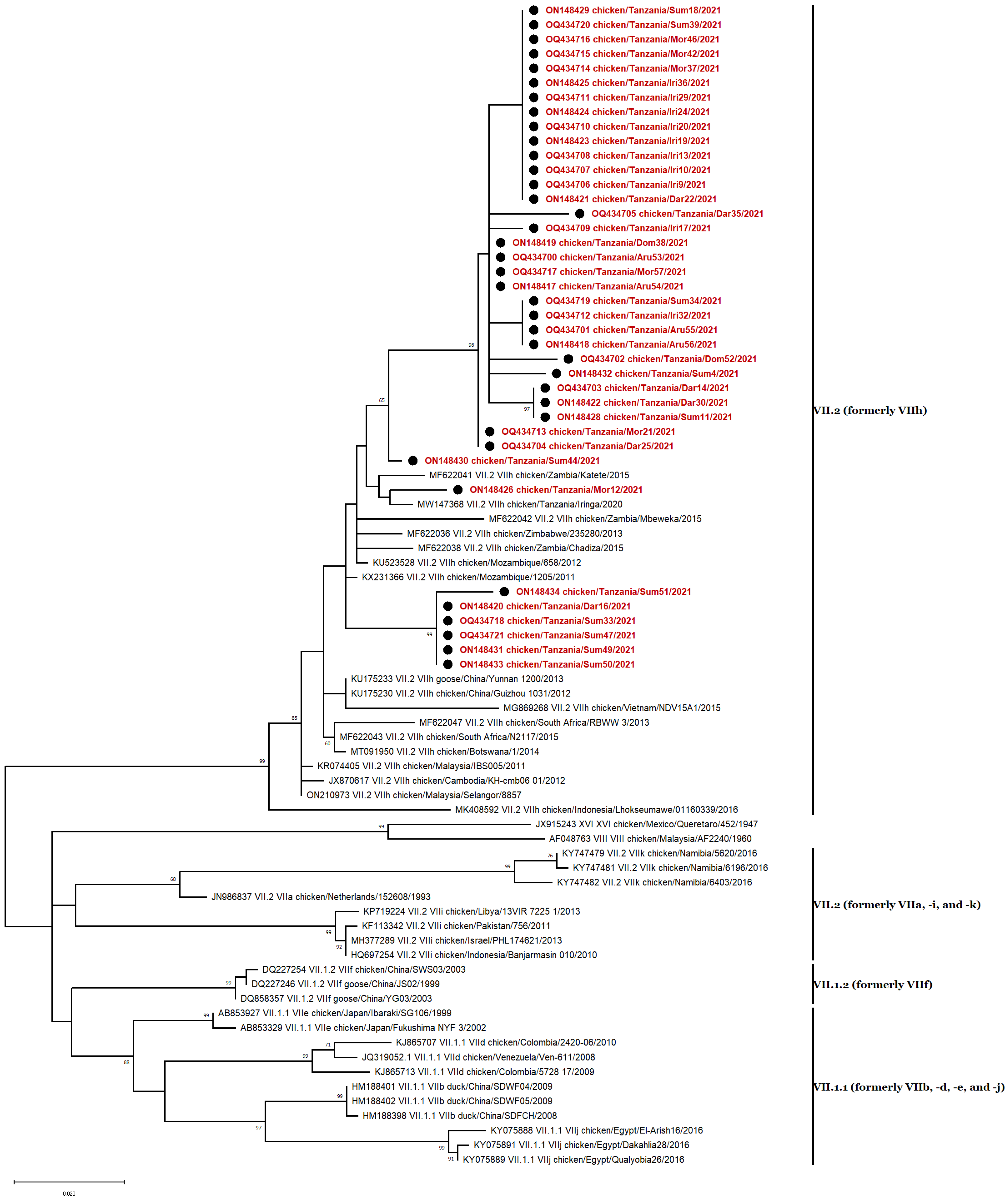
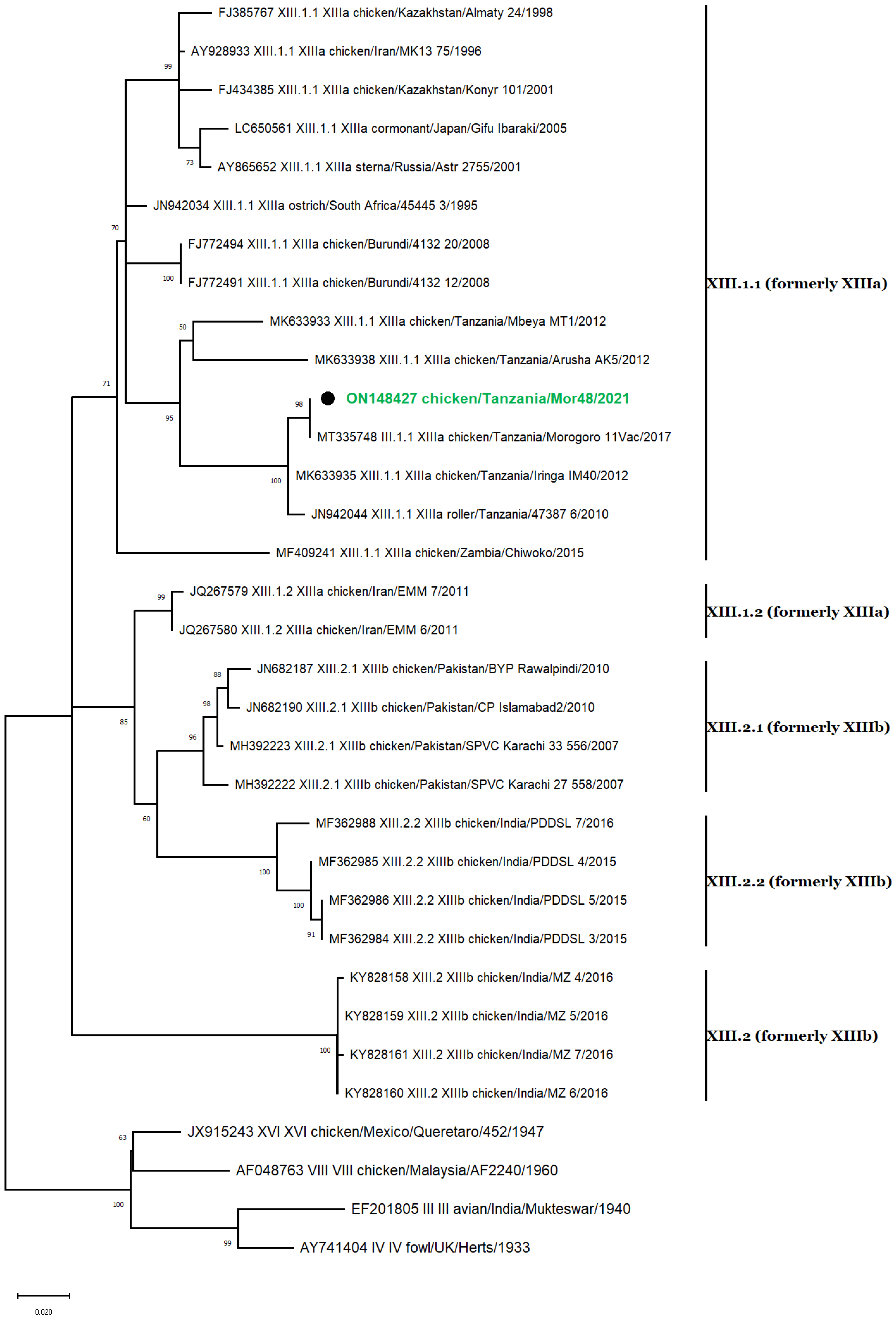
3.5. Phylogenetic Analyses
3.6. Genotype VII Isolates
3.7. Genotype XIII Isolate
3.8. Nucleotide Sequence Similarity
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Dimitrov, K.M.; Afonso, C.L.; Yu, Q.; Miller, P.J. Newcastle disease vaccines—A solved problem or a continuous challenge? Vet. Microbiol. 2017, 206, 126–136. [Google Scholar] [CrossRef] [PubMed]
- ICTV: International Committee on Taxonomy of Viruses. Virus Taxonomy. 2019. Available online: https://talk.ictvonline.org/taxonomy/ (accessed on 11 February 2021).
- Hewajuli, D.A.; Dharmayanti, N.L.P.I. Patogenitas Virus Newcastle Disease Pada Ayam. Wartazoa 2011, 21, 72–80. [Google Scholar]
- Lamb, R.A.; Parks, G.D. Paramyxoviridae: The Viruses and Their Replication; Lippincott, Williams, and Wilkins: Philadelphia, PA, USA, 2007; pp. 1449–1496. [Google Scholar]
- Connolly, S.A.; Leser, G.P.; Jardetzky, T.S.; Lamb, R.A. Bimolecular Complementation of Paramyxovirus Fusion and Hemag-glutinin-Neuraminidase Proteins Enhances Fusion: Implications for the Mechanism of Fusion Triggering. J. Virol. 2009, 83, 10857–10868. [Google Scholar] [CrossRef] [PubMed]
- Panda, A.; Huang, Z.; Elankumaran, S.; Rockemann, D.D.; Samal, S.K. Role of fusion protein cleavage site in the virulence of Newcastle disease virus. Microb. Pathog. 2003, 36, 1–10. [Google Scholar] [CrossRef]
- Elmardi, N.; Bakheit, M.; Khalafalla, A. Phylogenetic analysis of some Newcastle disease virus isolates from the Sudan. Open Veter. J. 2016, 6, 89–97. [Google Scholar] [CrossRef]
- Dimitrov, K.M.; Abolnik, C.; Afonso, C.L.; Albina, E.; Bahl, J.; Berg, M.; Briand, F.-X.; Brown, I.H.; Choi, K.-S.; Chvala, I.; et al. Updated unified phylogenetic classification system and revised nomenclature for Newcastle disease virus. Infect. Genet. Evol. 2019, 74, 103917. [Google Scholar] [CrossRef]
- Wang, J.; Yu, X.; Zheng, D.; Zhao, Y.; Lv, Y.; Shu, B.; Jiang, W.; Liu, S.; Li, J.; Hou, G.; et al. Continuous surveillance revealing a wide distribution of class I Newcastle disease viruses in China from 2011 to 2020. PLoS ONE 2022, 17, e0264936. [Google Scholar] [CrossRef]
- Hossain, I.; Parvin, R.; Rahman, M.M.; Begum, J.A.; Chowdhury, E.H.; Islam, M.R.; Diel, D.G.; Nooruzzaman, M. Comparative pathogenicity of a genotype XXI.1.2 pigeon Newcastle disease virus isolate in pigeons and chickens. Microb. Pathog. 2023, 178. [Google Scholar] [CrossRef]
- Alexander, D.; Manvell, R.; Banks, J.; Collins, M.; Parsons, G.; Cox, B.; Frost, K.; Speidel, E.; Ashman, S.; Aldous, E. Experimental assessment of the pathogenicity of the Newcastle disease viruses from outbreaks in Great Britain in 1997 for chickens and turkeys, and the protection afforded by vaccination. Avian Pathol. 1999, 28, 501–511. [Google Scholar] [CrossRef] [PubMed]
- Diel, D.G.; Susta, L.; Garcia, S.C.; Killian, M.L.; Brown, C.C.; Miller, P.J.; Afonso, C.L. Complete genome and clinicopathological charac-terization of a virulent newcastle disease virus isolate from South America. J. Clin. Microbiol. 2012, 50, 378–387. [Google Scholar] [CrossRef] [PubMed]
- Amoia, C.F.A.N.; Nnadi, P.A.; Ezema, C.; Couacy-Hymann, E. Epidemiology of Newcastle disease in Africa with emphasis on Côte d’Ivoire: A review. Veter. World 2021, 14, 1727–1740. [Google Scholar] [CrossRef] [PubMed]
- Jin, J.; Zhao, J.; Ren, Y.; Zhong, Q.; Zhang, G. Contribution of HN protein length diversity to Newcastle disease virus virulence, replication and biological activities. Sci. Rep. 2016, 6, 36890. [Google Scholar] [CrossRef] [PubMed]
- Nooruzzaman, M.; Hossain, I.; Begum, J.A.; Moula, M.; Khaled, S.A.; Parvin, R.; Chowdhury, E.H.; Islam, M.R.; Diel, D.G.; Dimitrov, K.M. The First Report of a Virulent Newcastle Disease Virus of Genotype VII.2 Causing Outbreaks in Chickens in Bangladesh. Viruses 2022, 14, 2627. [Google Scholar] [CrossRef]
- Elfatah, K.S.A.; Elabasy, M.A.; El-Khyate, F.; Elmahallawy, E.K.; Mosad, S.M.; El-Gohary, F.A.; Abdo, W.; Al-Brakati, A.; Seadawy, M.G.; Tahoon, A.E.; et al. Molecular Characterization of Velogenic Newcastle Disease Virus (Sub-Genotype VII.1.1) from Wild Birds, with Assessment of Its Pathogenicity in Susceptible Chickens. Animals 2021, 11, 505. [Google Scholar] [CrossRef] [PubMed]
- Miller, P.J.; Haddas, R.; Simanov, L.; Lublin, A.; Rehmani, S.F.; Wajid, A.; Bibi, T.; Khan, T.A.; Yaqub, T.; Setiyaningsih, S.; et al. Identification of new sub-genotypes of virulent Newcastle disease virus with potential panzootic features. Infect. Genet. Evol. 2015, 29, 216–229. [Google Scholar] [CrossRef] [PubMed]
- Nooruzzaman, M.; Mumu, T.T.; Kabiraj, C.K.; Hasnat, A.; Rahman, M.M.; Chowdhury, E.H.; Dimitrov, K.M.; Islam, M.R. Genetic and biological charac-terization of Newcastle disease viruses circulating in Bangladesh during 2010–2017: Further genetic diversification of class II genotype XIII in Southcentral Asia. J. Gen. Virol. 2021, 102, 1554. [Google Scholar] [CrossRef]
- Bello, M.B.; Yusoff, K.M.; Ideris, A.; Hair-Bejo, M.; Peeters, B.P.H.; Jibril, A.H.; Tambuwal, F.M.; Omar, A.R. Genotype Diversity of Newcastle Disease Virus in Nigeria: Disease Control Challenges and Future Outlook. Adv. Virol. 2018, 2018, 1–17. [Google Scholar] [CrossRef]
- Kgotlele, T.; Modise, B.; Nyange, J.F.; Thanda, C.; Cattoli, G.; Dundon, W.G. First molecular characterization of avian paramyx-ovirus-1 (Newcastle disease virus) in Botswana. Virus Genes 2020, 56, 646–650. [Google Scholar] [CrossRef]
- Khan, T.A.; Rue, C.A.; Rehmani, S.F.; Ahmed, A.; Wasilenko, J.L.; Miller, P.J.; Afonso, C.L. Phylogenetic and Biological Characterization of Newcastle Disease Virus Isolates from Pakistan. J. Clin. Microbiol. 2010, 48, 1892–1894. [Google Scholar] [CrossRef]
- Hejazi, Z.; Tabatabaeizadeh, S.; Toroghi, R.; Farzin, H.; Saffarian, P. First detection and characterisation of sub-genotype XIII.2.1 Newcastle disease virus isolated from backyard chickens in Iran. Veter. Med. Sci. 2022, 8, 2521–2531. [Google Scholar] [CrossRef]
- Barman, L.R.; Nooruzzaman, M.; Sarker, R.D.; Rahman, T.; Bin Saife, R.; Giasuddin, M.; Das, B.C.; Das, P.M.; Chowdhury, E.H.; Islam, M.R. Phylogenetic analysis of Newcastle disease viruses from Bangladesh suggests continuing evolution of genotype XIII. Arch. Virol. 2017, 162, 3177–3182. [Google Scholar] [CrossRef]
- Kabiraj, C.K.; Mumu, T.T.; Chowdhury, E.H.; Islam, M.R.; Nooruzzaman, M. Sequential Pathology of a Genotype XIII Newcastle Disease Virus from Bangladesh in Chickens on Experimental Infection. Pathogens 2020, 9, 539. [Google Scholar] [CrossRef] [PubMed]
- Snoeck, C.J.; Owoade, A.A.; Couacy-Hymann, E.; Alkali, B.R.; Okwen, M.P.; Adeyanju, A.T.; Komoyo, G.F.; Nakouné, E.; Le Faou, A.; Muller, C.P. High Genetic Diversity of Newcastle Disease Virus in Poultry in West and Central Africa: Cocirculation of Genotype XIV and Newly Defined Genotypes XVII and XVIII. J. Clin. Microbiol. 2013, 51, 2250–2260. [Google Scholar] [CrossRef]
- Mngumi, E.B.; Bunuma, E. Seroprevalence and risk factors of Newcastle disease virus in local chickens in Njombe and Bahi districts in Tanzania. Trop. Anim. Health Prod. 2022, 54, 53. [Google Scholar] [CrossRef] [PubMed]
- Fisher, H. Newcastle Disease Control in Africa; The Australian Centre for International Agricultural Research (ACIAR): Canberra, ACT, Australia, 2014. [Google Scholar]
- Da Silva, M.; Desta, S.; Stapleton, J. Development of the chicken sector in the Tanzanian livestock master plan. Tanzania. Livest. Master Plan Brief. 2017, 1–4. [Google Scholar]
- Aldous, E.W.; Mynn, J.K.; Banks, J.; Alexander, D.J. A molecular epidemiological study of avian paramyxovirus type 1 (Newcastle disease virus) isolates by phylogenetic analysis of a partial nucleotide sequence of the fusion protein gene. Avian Pathol. 2003, 32, 239–257. [Google Scholar] [CrossRef]
- da Silva, A.P.; Aston, E.J.; Chiwanga, G.H.; Birakos, A.; Muhairwa, A.P.; Kayang, B.B.; Kelly, T.; Zhou, H.; Gallardo, R.A. Molecular Characterization of Newcastle Disease Viruses Isolated from Chickens in Tanzania and Ghana. Viruses 2020, 12, 916. [Google Scholar] [CrossRef]
- Msoffe, P.L.M.; Chiwanga, G.H.; Cardona, C.J.; Miller, P.J.; Suarez, D.L. Isolation and Characterization of Newcastle Disease Virus from Live Bird Markets in Tanzania. Avian Dis. 2019, 63, 634–640. [Google Scholar] [CrossRef] [PubMed]
- Goraichuk, I.V.; Msoffe, P.L.M.; Chiwanga, G.H.; Dimitrov, K.M.; Afonso, C.L.; Suarez, D.L. First Complete Genome Sequence of a Subgenotype Vd Newcastle Disease Virus Isolate. Genome Announc. 2019, 8, e00436-19. [Google Scholar] [CrossRef]
- Kibasa, M.I. Epidemiology of Newcastle Disease in Backyard Chickens Rearing System in Iringa Rural District, Tanzania. Master’s Thesis, Sokoine University of Agriculture, Morogoro, Tanzania, 2020. [Google Scholar]
- Yongolo, M.G.; Christensen, H.; Handberg, K.; Minga, U.; Olsen, J.E. On the origin and diversity of Newcastle disease virus in Tanzania. Onderstepoort J. Veter. Res. 2011, 78, 8. [Google Scholar] [CrossRef] [PubMed]
- OIE. Manual of diagnostic tests and vaccines for terrestrial animals: Mammals, birds and bees. In Biological Standards Commission; World Organization for Animal Health: Paris, France, 2012; pp. 1–19. [Google Scholar]
- Bari, F.D.; Gelaye, E.; Tekola, B.G.; Harder, T.; Beer, M.; Grund, C. Antigenic and Molecular Characterization of Virulent Newcastle Disease Viruses Circulating in Ethiopia Between 1976 and 2008. Veter. Med. Res. Rep. 2021, 12, 129–140. [Google Scholar] [CrossRef] [PubMed]
- Alexander, D.J. Newcastle disease and other avian paramyxoviruses. OIE Rev. Sci. Tech. 2000, 19, 443–462. [Google Scholar] [CrossRef]
- Seal, B.S. Analysis of Matrix Protein Gene Nucleotide Sequence Diversity Among Newcastle Disease Virus Isolates Demonstrates that Recent Disease Outbreaks Are Caused by Viruses of Psittacine Origin. In Molecular Evolution of Viruses—Past and Present; Springer: Boston, MA, USA, 1996; pp. 145–152. [Google Scholar]
- Kumar, S.; Stecher, G.; Li, M.; Knyaz, C.; Tamura, K. MEGA X: Molecular Evolutionary Genetics Analysis across Computing Platforms. Mol. Biol. Evol. 2018, 35, 1547–1549. [Google Scholar] [CrossRef] [PubMed]
- Muhire, B.M.; Varsani, A.; Martin, D.P. SDT: A Virus Classification Tool Based on Pairwise Sequence Alignment and Identity Calculation. PLoS ONE 2014, 9, e108277. [Google Scholar] [CrossRef] [PubMed]
- Jin, Z.; Li, D.; Dai, C.; Cheng, G.; Wang, X.; Zhao, K. Response of live Newcastle disease virus encapsulated in N -2-hydroxypropyl dimethylethyl ammonium chloride chitosan nanoparticles. Carbohydr. Polym. 2017, 171, 267–280. [Google Scholar] [CrossRef]
- Kattenbelt, J.A.; Stevens, M.P.; Gould, A.R. Sequence variation in the Newcastle disease virus genome. Virus Res. 2006, 116, 168–184. [Google Scholar] [CrossRef]
- Aldous, E.W.; Alexander, D.J. Detection and differentiation of Newcastle disease virus (avian paramyxovirus type 1). Avian Pathol. 2001, 30, 117–128. [Google Scholar] [CrossRef]
- Seal, B.S.; King, D.J.; Meinersmann, R.J. Molecular evolution of the Newcastle disease virus matrix protein gene and phylo-genetic relationships among the paramyxoviridae. Virus Res. 2000, 66, 1–11. [Google Scholar] [CrossRef]
- Kimura, M. A simple method for estimating evolutionary rates of base substitutions through comparative studies of nucleotide sequences. J. Mol. Evol. 1980, 16, 111–120. [Google Scholar] [CrossRef]
- Diel, D.G.; da Silva, L.H.; Liu, H.; Wang, Z.; Miller, P.J.; Afonso, C.L. Genetic diversity of avian paramyxovirus type 1: Proposal for a unified nomenclature and classification system of Newcastle disease virus genotypes. Infect. Genet. Evol. 2012, 12, 1770–1779. [Google Scholar] [CrossRef]
- Abolnik, C.; Mubamba, C.; Wandrag, D.B.R.; Horner, R.; Gummow, B.; Dautu, G.; Bisschop, S.P.R. Tracing the origins of genotype VIIh Newcastle disease in southern Africa. Transbound. Emerg. Dis. 2017, 65, e393–e403. [Google Scholar] [CrossRef]
- Mapaco, L.P.; Monjane, I.V.A.; Nhamusso, A.E.; Viljoen, G.J.; Dundon, W.G.; Achá, S.J. Phylogenetic analysis of Newcastle disease viruses isolated from commercial poultry in Mozambique (2011–2016). Virus Genes 2016, 52, 748–753. [Google Scholar] [CrossRef]
- Hakizimana, J.N.; Nyabongo, L.; Ntirandekura, J.B.; Yona, C.; Ntakirutimana, D.; Kamana, O.; Nauwynck, H.; Misinzo, G. Genetic Analysis of African Swine Fever Virus From the 2018 Outbreak in South-Eastern Burundi. Front. Veter. Sci. 2020, 7. [Google Scholar] [CrossRef]
- Abolnik, C.; Mubamba, C.; Dautu, G.; Gummow, B. Complete Genome Sequence of a Newcastle Disease Genotype XIII Virus Isolated from Indigenous Chickens in Zambia. Genome Announc. 2017, 5. [Google Scholar] [CrossRef]
- Cattoli, G.; Fusaro, A.; Monne, I.; Molia, S.; Le Menach, A.; Maregeya, B.; Nchare, A.; Bangana, I.; Maina, A.G.; N’Goran Koffi, J.-N.; et al. Emergence of a new genetic lineage of Newcastle disease virus in West and Central Africa—Implications for diagnosis and control. Veter. Microbiol. 2010, 142, 168–176. [Google Scholar] [CrossRef]
- Das, M.; Kumar, S. Evidence of independent evolution of genotype XIII Newcastle disease viruses in India. Arch. Virol. 2016, 162, 997–1007. [Google Scholar] [CrossRef]
- Khorajiya, J.H.; Pandey, S.; Ghodasara, P.D.; Joshi, B.P.; Prajapati, K.S.; Ghodasara, D.J.; Mathakiya, R.A. Patho-epidemiological study on Genotype-XIII Newcastle disease virus infection in commercial vaccinated layer farms. Veter. World 2015, 8, 372–381. [Google Scholar] [CrossRef] [PubMed]
- Kariithi, H.M.; Ferreira, H.L.; Welch, C.N.; Ateya, L.O.; Apopo, A.A.; Zoller, R.; Volkening, J.D.; Williams-Coplin, D.; Parris, D.J.; Olivier, T.L.; et al. Surveillance and Genetic Characterization of Virulent Newcastle Disease Virus Subgenotype V.3 in Indigenous Chickens from Backyard Poultry Farms and Live Bird Markets in Kenya. Viruses 2021, 13, 103. [Google Scholar] [CrossRef] [PubMed]
- Ogali, I.N.; Wamuyu, L.W.; Lichoti, J.K.; Mungube, E.O.; Agwanda, B.; Ommeh, S.C. Molecular Characterization of Newcastle Disease Virus from Backyard Poultry Farms and Live Bird Markets in Kenya. Int. J. Microbiol. 2018, 2018, 2368597. [Google Scholar] [CrossRef] [PubMed]
- Radin, J.M.; Shaffer, R.A.; Lindsay, S.P.; Araneta, M.R.G.; Raman, R.; Fowler, J.H. International chicken trade and increased risk for introducing or reintroducing highly pathogenic avian influenza A (H5N1) to uninfected countries. Infect. Dis. Model. 2017, 2, 412–418. [Google Scholar] [CrossRef]
- Nath, B.; Kumar, S. Emerging variant of genotype XIII Newcastle disease virus from Northeast India. Acta Trop. 2017, 172, 64–69. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Jiang, Z.; Jin, Z.; Tan, H.; Xu, B. Risk Factors for Infectious Diseases in Backyard Poultry Farms in the Poyang Lake Area, China. PLoS ONE 2013, 8, e67366. [Google Scholar] [CrossRef] [PubMed]
- Marks, F.S.; Rodenbusch, C.R.; Okino, C.H.; Hein, H.E.; Costa, E.F.; Machado, G.; Canal, C.W.; Brentano, L.; Corbellini, L.G. Targeted survey of Newcastle disease virus in backyard poultry flocks located in wintering site for migratory birds from Southern Brazil. Prev. Veter. Med. 2014, 116, 197–202. [Google Scholar] [CrossRef] [PubMed]
- Campbell, Z.A.; Otieno, L.; Shirima, G.M.; Marsh, T.L.; Palmer, G.H. Drivers of vaccination preferences to protect a low-value livestock resource: Willingness to pay for Newcastle disease vaccines by smallholder households. Vaccine 2018, 37, 11–18. [Google Scholar] [CrossRef]
- Osman, N.; Goovaerts, D.; Sultan, S.; Salt, J.; Grund, C. Vaccine quality is a key factor to determine thermal stability of com-mercial newcastle disease (ND) vaccines. Vaccines 2021, 9, 363. [Google Scholar] [CrossRef] [PubMed]




| Zone | Regions | Number of Samples | Test | |
|---|---|---|---|---|
| HA | RT-PCR | |||
| Positive | Positive | |||
| Northern | Arusha | 7 | 6 | 6 |
| Coastal | Dar es Salaam | 9 | 8 | 7 |
| Coastal | Morogoro | 8 | 7 | 7 |
| Central | Dodoma | 16 | 7 | 2 |
| Southern Highlands | Iringa | 12 | 12 | 12 |
| Southern Highlands | Rukwa | 27 | 17 | 16 |
| Total | 79 | 57/79 | 50/79 | |
| GenBank Accession | Isolate ID | Subgenotype | Location | Collection Date | Sample Type | Vaccination Status | Vaccine | Cleavage Site Motif b | Pathotype |
|---|---|---|---|---|---|---|---|---|---|
| OQ434700 | Aru53 | VII.2 | Arusha | 15 October 2021 | Tissue | Vaccinated | LaSota | RRRKRF | Virulent (velogenic) |
| ON148417 | Aru54 | VII.2 | Arusha | 15 October 2021 | Swab | Unvaccinated | N/A a | RRRKRF | Virulent (velogenic) |
| OQ434701 | Aru55 | VII.2 | Arusha | 15 October 2021 | Tissue | Vaccinated | I-2 | RRRKRF | Virulent (velogenic) |
| ON148418 | Aru56 | VII.2 | Arusha | 15 October 2021 | Swab | Unvaccinated | N/A | RRRKRF | Virulent (velogenic) |
| ON148419 | Dom38 | VII.2 | Dodoma | 18 November 2021 | Swab | Unvaccinated | N/A | RRRKRF | Virulent (velogenic) |
| OQ434702 | Dom52 | VII.2 | Dodoma | 19 August 2021 | Tissue | Vaccinated | LaSota | RRRKRF | Virulent (velogenic) |
| OQ434703 | Dar14 | VII.2 | Dar es Salaam | 19 August 2021 | Swab | Unvaccinated | N/A | RRRKRF | Virulent (velogenic) |
| ON148420 | Dar16 | VII.2 | Dar es Salaam | 19 August 2021 | Swab | Unvaccinated | N/A | RRRKRF | Virulent (velogenic) |
| ON148421 | Dar22 | VII.2 | Dar es Salaam | 29 August 2021 | Swab | Unvaccinated | N/A | RRRKRF | Virulent (velogenic) |
| OQ434704 | Dar25 | VII.2 | Dar es Salaam | 29 August 2021 | Swab | Vaccinated | LaSota | RRRKRF | Virulent (velogenic) |
| ON148422 | Dar30 | VII.2 | Dar es Salaam | 4 September 2021 | Swab | Vaccinated | I-2 | RRRKRF | Virulent (velogenic) |
| OQ434705 | Dar35 | VII.2 | Dar es Salaam | 4 September 2021 | Tissue | Vaccinated | I-2 | RRRKRF | Virulent (velogenic) |
| OQ434706 | Iri9 | VII.2 | Iringa | 14 July 2021 | Tissue | Vaccinated | I-2 | RRRKRF | Virulent (velogenic) |
| OQ434707 | Iri10 | VII.2 | Iringa | 14 July 2021 | Swab | Unvaccinated | N/A | RRRKRF | Virulent (velogenic) |
| OQ434708 | Iri13 | VII.2 | Iringa | 14 July 2021 | Swab | Unvaccinated | N/A | RRRKRF | Virulent (velogenic) |
| OQ434709 | Iri17 | VII.2 | Iringa | 14 July 2021 | Tissue | Vaccinated | I-2 | RRRKRF | Virulent (velogenic) |
| ON148423 | Iri19 | VII.2 | Iringa | 23 August 2021 | Swab | Vaccinated | LaSota | RRRKRF | Virulent (velogenic) |
| OQ434710 | Iri20 | VII.2 | Iringa | 23 August 2021 | Swab | Vaccinated | LaSota | RRRKRF | Virulent (velogenic) |
| ON148424 | Iri24 | VII.2 | Iringa | 23 August 2021 | Swab | Vaccinated | LaSota | RRRKRF | Virulent (velogenic) |
| OQ434711 | Iri29 | VII.2 | Iringa | 23 August 2021 | Swab | Vaccinated | LaSota | RRRKRF | Virulent (velogenic) |
| OQ434712 | Iri32 | VII.2 | Iringa | 23 August 2021 | Swab | Vaccinated | LaSota | RRRKRF | Virulent (velogenic) |
| ON148425 | Iri36 | VII.2 | Iringa | 23 August 2021 | Swab | Unvaccinated | N/A | RRRKRF | Virulent (velogenic) |
| ON148426 | Mor12 | VII.2 | Morogoro | 2 October 2021 | Tissue | Vaccinated | LaSota | RRQKRF | Virulent (velogenic) |
| OQ434713 | Mor21 | VII.2 | Morogoro | 2 October 2021 | Swab | Vaccinated | LaSota | RRRKRF | Virulent (velogenic) |
| OQ434714 | Mor37 | VII.2 | Morogoro | 2 October 2021 | Swab | Vaccinated | I-2 | RRRKRF | Virulent (velogenic) |
| OQ434715 | Mor42 | VII.2 | Morogoro | 2 October 2021 | Swab | Unvaccinated | N/A | RRRKRF | Virulent (velogenic) |
| OQ434716 | Mor46 | VII.2 | Morogoro | 2 October 2021 | Swab | Unvaccinated | N/A | RRRKRF | Virulent (velogenic) |
| ON148427 | Mor48 | XIII.1.1 | Morogoro | 2 October 2021 | Swab | Vaccinated | LaSota | RRQKRF | Virulent (velogenic) |
| OQ434717 | Mor57 | VII.2 | Morogoro | 9 October 2021 | Tissue | Vaccinated | LaSota | RRRKRF | Virulent (velogenic) |
| ON148432 | Sum4 | VII.2 | Sumbawanga | 25 July 2021 | Swab | Unvaccinated | N/A | RRRKRF | Virulent (velogenic) |
| ON148428 | Sum11 | VII.2 | Sumbawanga | 21 June 2021 | Tissue | Vaccinated | LaSota | RRRKRF | Virulent (velogenic) |
| ON148429 | Sum18 | VII.2 | Sumbawanga | 21 June 2021 | Tissue | Vaccinated | LaSota | RRRKRF | Virulent (velogenic) |
| OQ434718 | Sum33 | VII.2 | Sumbawanga | 22 June 2021 | Swab | Unvaccinated | N/A | RRRKRF | Virulent (velogenic) |
| OQ434719 | Sum34 | VII.2 | Sumbawanga | 22 June 2021 | Swab | Vaccinated | I-2 | RRRKRF | Virulent (velogenic) |
| OQ434720 | Sum39 | VII.2 | Sumbawanga | 22 June 2021 | Swab | Vaccinated | LaSota | RRRKRF | Virulent (velogenic) |
| ON148430 | Sum44 | VII.2 | Sumbawanga | 22 June 2021 | Swab | Unvaccinated | N/A | RRRKRF | Virulent (velogenic) |
| OQ434721 | Sum47 | VII.2 | Sumbawanga | 22 June 2021 | Swab | Unvaccinated | N/A | RRRKRF | Virulent (velogenic) |
| ON148431 | Sum49 | VII.2 | Sumbawanga | 22 June 2021 | Tissue | Vaccinated | LaSota | RRRKRF | Virulent (velogenic) |
| ON148433 | Sum50 | VII.2 | Sumbawanga | 23 June 2021 | Tissue | Vaccinated | LaSota | RRRKRF | Virulent (velogenic) |
| ON148434 | Sum51 | VII.2 | Sumbawanga | 27 June 2021 | Tissue | Vaccinated | LaSota | RRRKRF | Virulent (velogenic) |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Amoia, C.F.; Hakizimana, J.N.; Duggal, N.K.; Chengula, A.A.; Rohaim, M.A.; Munir, M.; Weger-Lucarelli, J.; Misinzo, G. Genetic Diversity of Newcastle Disease Virus Involved in the 2021 Outbreaks in Backyard Poultry Farms in Tanzania. Vet. Sci. 2023, 10, 477. https://doi.org/10.3390/vetsci10070477
Amoia CF, Hakizimana JN, Duggal NK, Chengula AA, Rohaim MA, Munir M, Weger-Lucarelli J, Misinzo G. Genetic Diversity of Newcastle Disease Virus Involved in the 2021 Outbreaks in Backyard Poultry Farms in Tanzania. Veterinary Sciences. 2023; 10(7):477. https://doi.org/10.3390/vetsci10070477
Chicago/Turabian StyleAmoia, Charlie F., Jean N. Hakizimana, Nisha K. Duggal, Augustino A. Chengula, Mohammed A. Rohaim, Muhammad Munir, James Weger-Lucarelli, and Gerald Misinzo. 2023. "Genetic Diversity of Newcastle Disease Virus Involved in the 2021 Outbreaks in Backyard Poultry Farms in Tanzania" Veterinary Sciences 10, no. 7: 477. https://doi.org/10.3390/vetsci10070477
APA StyleAmoia, C. F., Hakizimana, J. N., Duggal, N. K., Chengula, A. A., Rohaim, M. A., Munir, M., Weger-Lucarelli, J., & Misinzo, G. (2023). Genetic Diversity of Newcastle Disease Virus Involved in the 2021 Outbreaks in Backyard Poultry Farms in Tanzania. Veterinary Sciences, 10(7), 477. https://doi.org/10.3390/vetsci10070477









