Analysis of Fecal Microbial Changes in Young Calves Following Bovine Rotavirus Infection
Abstract
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Ethics Statement
2.2. Experimental Design, Animals, and Diet
2.3. Real-Time RT-PCR Detection of Bovine Rotavirus
2.4. Metagenome Analysis
2.5. Statistical Analysis
3. Results
3.1. Clinical Findings of Rotavirus in Calves
3.2. Quality Evaluation and Sample Statistics
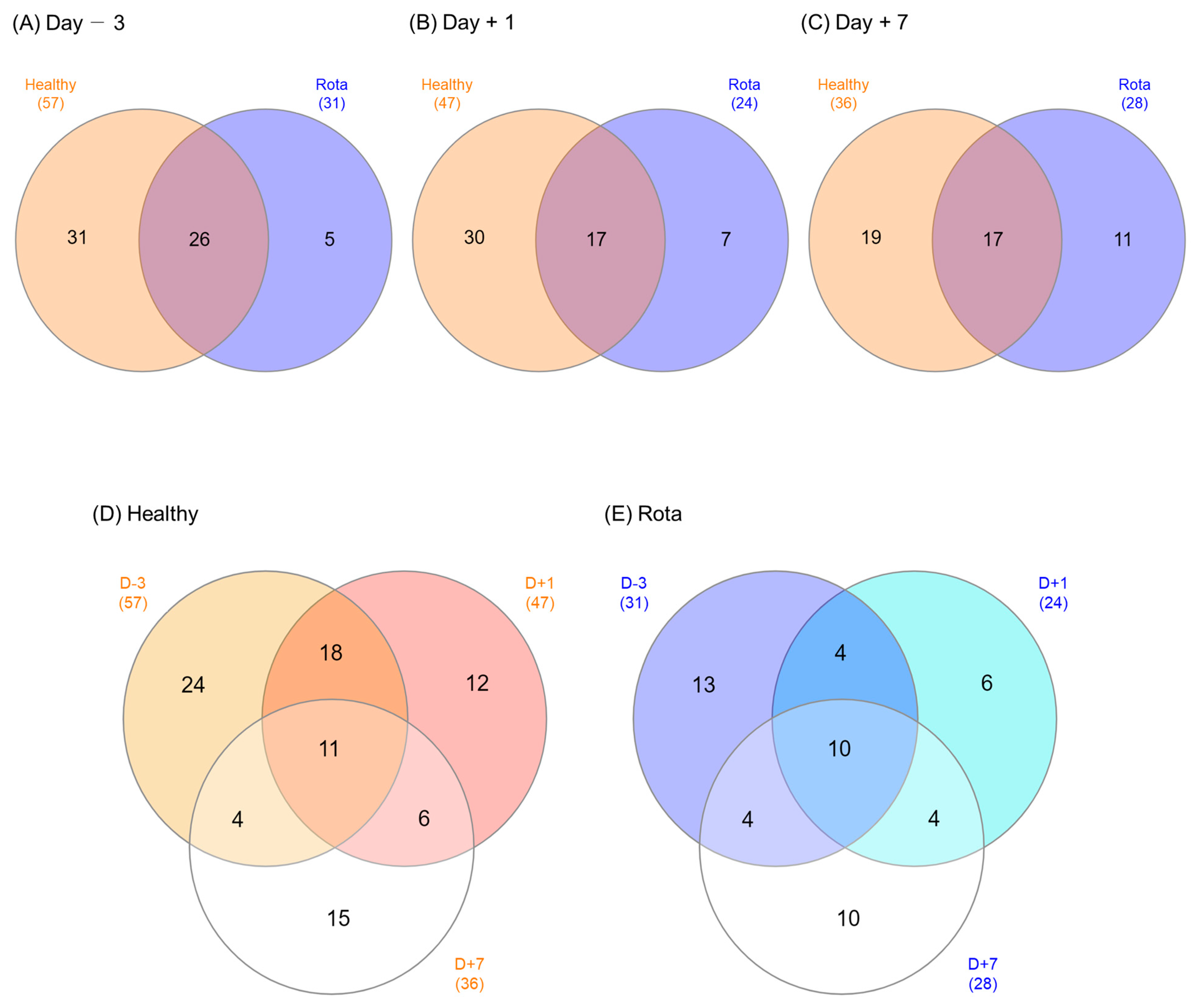
3.3. Diversity Analysis of Fecal Microbiota in Healthy and Rota Groups
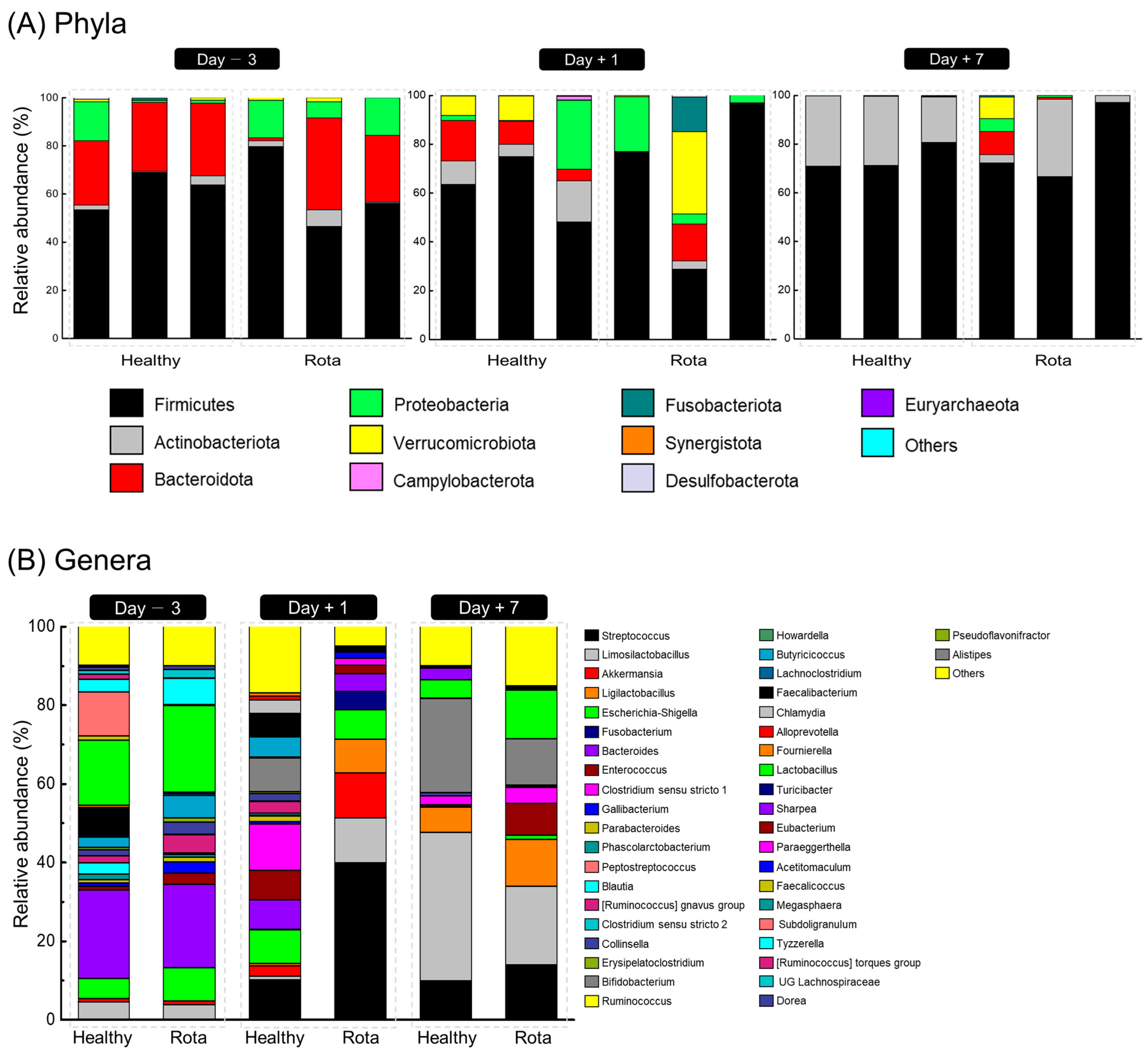
3.4. Taxonomic Composition of Fecal Microbiota in Healthy and Rota Groups
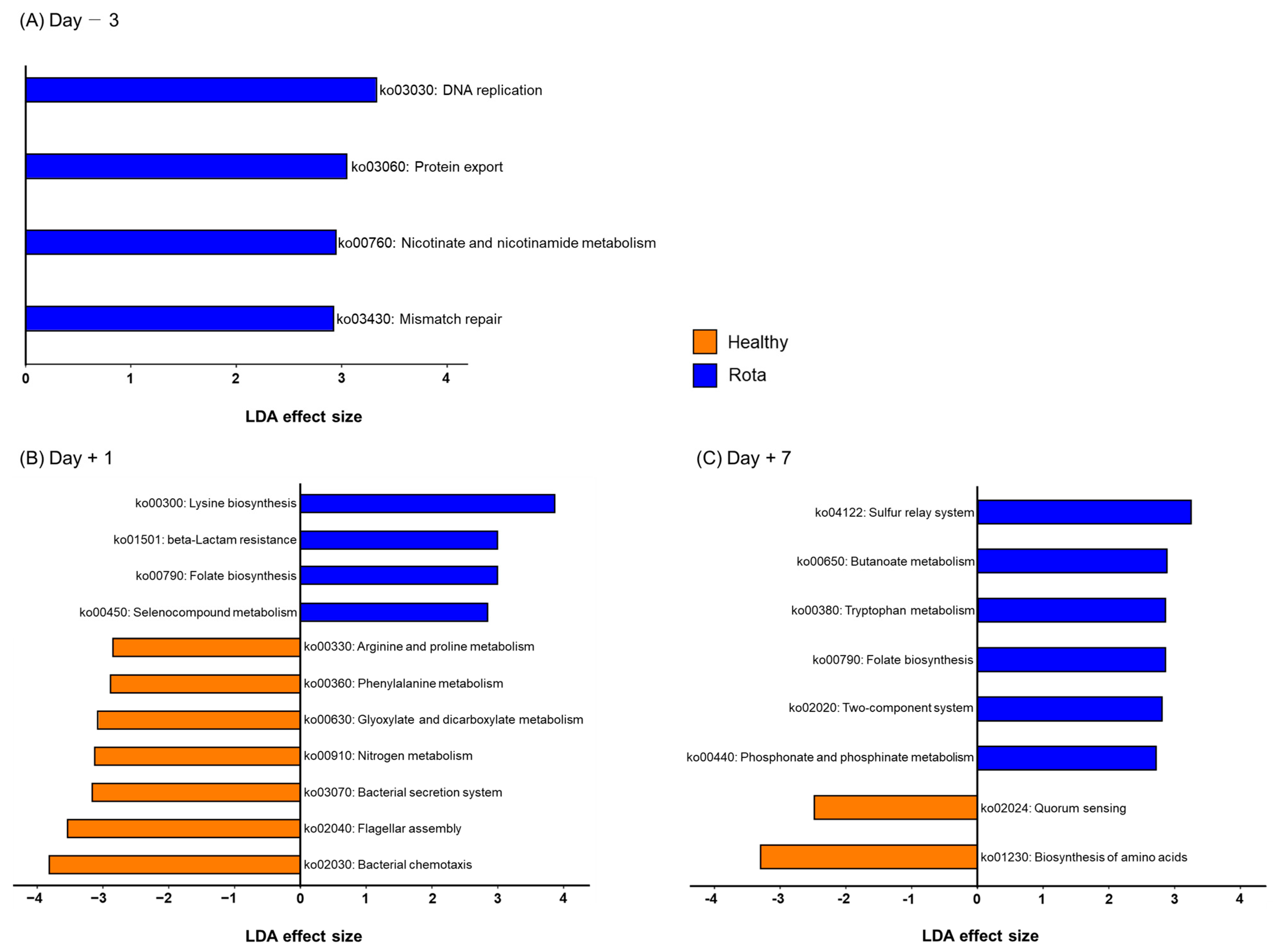
3.5. Predicted Kyoto Encyclopedia of Genes and Functional Pathways in the Fecal Microbiota in Healthy and Rota Groups
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Cho, Y.-I.; Yoon, K.J. An Overview of Calf Diarrhea—Infectious Etiology, Diagnosis, and Intervention. J. Vet. Sci. 2014, 15, 1–17. [Google Scholar] [CrossRef] [PubMed]
- Windeyer, M.C.; Leslie, K.E.; Godden, S.M.; Hodgins, D.C.; Lissemore, K.D.; LeBlanc, S.J. Factors Associated with Morbidity, Mortality, and Growth of Dairy Heifer Calves up to 3 Months of Age. Prev. Vet. Med. 2014, 113, 231–240. [Google Scholar] [CrossRef] [PubMed]
- Svensson, C.; Linder, A.; Olsson, S.O. Mortality in Swedish Dairy Calves and Replacement Heifers. J. Dairy Sci. 2006, 89, 4769–4777. [Google Scholar] [CrossRef] [PubMed]
- Malmuthuge, N.; Griebel, P.J.; Guan, L.L. The Gut Microbiome and Its Potential Role in the Development and Function of Newborn Calf Gastrointestinal Tract. Front. Vet. Sci. 2015, 2, 36. [Google Scholar] [CrossRef]
- Ma, T.; Villot, C.; Renaud, D.; Skidmore, A.; Chevaux, E.; Steele, M.; Guan, L.L. Linking Perturbations to Temporal Changes in Diversity, Stability, and Compositions of Neonatal Calf Gut Microbiota: Prediction of Diarrhea. ISME J. 2020, 14, 2223–2235. [Google Scholar] [CrossRef]
- Alipour, M.J.; Jalanka, J.; Pessa-Morikawa, T.; Kokkonen, T.; Satokari, R.; Hynönen, U.; Iivanainen, A.; Niku, M. The Composition of the Perinatal Intestinal Microbiota in Cattle. Sci. Rep. 2018, 8, 10437. [Google Scholar] [CrossRef]
- Guzman, C.E.; Wood, J.L.; Egidi, E.; White-Monsant, A.C.; Semenec, L.; Grommen, S.V.H.; Hill-Yardin, E.L.; De Groef, B.; Franks, A.E. A Pioneer Calf Foetus Microbiome. Sci. Rep. 2020, 10, 17712. [Google Scholar] [CrossRef] [PubMed]
- Gomez, D.E.E.; Arroyo, L.G.G.; Costa, M.C.C.; Viel, L.; Weese, J.S.S. Characterization of the Fecal Bacterial Microbiota of Healthy and Diarrheic Dairy Calves. J. Vet. Intern. Med. 2017, 31, 928–939. [Google Scholar] [CrossRef]
- Oikonomou, G.; Teixeira, A.G.V.; Foditsch, C.; Bicalho, M.L.; Machado, V.S.; Bicalho, R.C. Fecal Microbial Diversity in Pre-Weaned Dairy Calves as Described by Pyrosequencing of Metagenomic 16S RDNA. Associations of Faecalibacterium Species with Health and Growth. PLoS ONE 2013, 8, e63157. [Google Scholar] [CrossRef]
- Slanzon, G.S.; Ridenhour, B.J.; Moore, D.A.; Sischo, W.M.; Parrish, L.M.; Trombetta, S.C.; McConnel, C.S. Fecal Microbiome Profiles of Neonatal Dairy Calves with Varying Severities of Gastrointestinal Disease. PLoS ONE 2022, 17, e0262317. [Google Scholar] [CrossRef]
- Gomez, D.E.; Galvão, K.N.; Rodriguez-Lecompte, J.C.; Costa, M.C. The Cattle Microbiota and the Immune System: An Evolving Field. Vet. Clin. N. Am. Food Anim. Pract. 2019, 35, 485–505. [Google Scholar] [CrossRef]
- Hennessy, M.; Indugu, N.; Vecchiarelli, B.; Redding, L.; Bender, J.; Pappalardo, C.; Leibstein, M.; Toth, J.; Stefanovski, D.; Katepalli, A.; et al. Short Communication: Comparison of the Fecal Bacterial Communities in Diarrheic and Nondiarrheic Dairy Calves from Multiple Farms in Southeastern Pennsylvania. J. Dairy Sci. 2021, 104, 7225–7232. [Google Scholar] [CrossRef]
- Hennessy, M.L.; Indugu, N.; Vecchiarelli, B.; Bender, J.; Pappalardo, C.; Leibstein, M.; Toth, J.; Katepalli, A.; Garapati, S.; Pitta, D. Temporal Changes in the Fecal Bacterial Community in Holstein Dairy Calves from Birth through the Transition to a Solid Diet. PLoS ONE 2020, 15, e0238882. [Google Scholar] [CrossRef] [PubMed]
- Ma, T.; O’Hara, E.; Song, Y.; Fischer, A.J.; He, Z.; Steele, M.A.; Guan, L.L. Altered Mucosa-Associated Microbiota in the Ileum and Colon of Neonatal Calves in Response to Delayed First Colostrum Feeding. J. Dairy Sci. 2019, 102, 7073–7086. [Google Scholar] [CrossRef] [PubMed]
- Renaud, D.L.; Buss, L.; Wilms, J.N.; Steele, M.A. Technical Note: Is Fecal Consistency Scoring an Accurate Measure of Fecal Dry Matter in Dairy Calves? J. Dairy Sci. 2020, 103, 10709–10714. [Google Scholar] [CrossRef] [PubMed]
- Tsuchiaka, S.; Masuda, T.; Sugimura, S.; Kobayashi, S.; Komatsu, N.; Nagai, M.; Omatsu, T.; Furuya, T.; Oba, M.; Katayama, Y.; et al. Development of a Novel Detection System for Microbes from Bovine Diarrhea by Real-Time PCR. J. Vet. Med. Sci. 2016, 78, 383–389. [Google Scholar] [CrossRef]
- Herlemann, D.P.R.; Labrenz, M.; Jürgens, K.; Bertilsson, S.; Waniek, J.J.; Andersson, A.F. Transitions in Bacterial Communities along the 2000 Km Salinity Gradient of the Baltic Sea. ISME J. 2011, 5, 1571–1579. [Google Scholar] [CrossRef]
- Bolyen, E.; Rideout, J.R.; Dillon, M.R.; Bokulich, N.A.; Abnet, C.C.; Al-Ghalith, G.A.; Alexander, H.; Alm, E.J.; Arumugam, M.; Asnicar, F.; et al. Author Correction: Reproducible, Interactive, Scalable and Extensible Microbiome Data Science Using QIIME 2. Nat. Biotechnol. 2019, 37, 1091. [Google Scholar] [CrossRef]
- Martin, M. Cutadapt Removes Adapter Sequences from High-Throughput Sequencing Reads. EMBnet. J. 2011, 17, 10–12. [Google Scholar] [CrossRef]
- Callahan, B.J.; McMurdie, P.J.; Rosen, M.J.; Han, A.W.; Johnson, A.J.A.; Holmes, S.P. DADA2: High-Resolution Sample Inference from Illumina Amplicon Data. Nat. Methods 2016, 13, 581–583. [Google Scholar] [CrossRef]
- Quast, C.; Pruesse, E.; Yilmaz, P.; Gerken, J.; Schweer, T.; Yarza, P.; Peplies, J.; Glöckner, F.O. The SILVA Ribosomal RNA Gene Database Project: Improved Data Processing and Web-Based Tools. Nucleic Acids Res. 2013, 41, 590–596. [Google Scholar] [CrossRef]
- Xia, Y. Q2-Repeat-Rarefy: QIIME2 Plugin for Generating the Average Rarefied Table for Library Size Normalization Using Repeated Rarefaction. Available online: https://github.com/yxia0125/q2-repeat-rarefy (accessed on 2 February 2023).
- Douglas, G.M.; Maffei, V.J.; Zaneveld, J.; Yurgel, S.N.; Brown, J.R.; Taylor, C.M.; Huttenhower, C.; Langille, M.G.I. PICRUSt2: An Improved and Customizable Approach for Metagenome Inference. BioRxiv 2020, 672295. [Google Scholar] [CrossRef]
- Segata, N.; Izard, J.; Waldron, L.; Gevers, D.; Miropolsky, L.; Garrett, W.S.; Huttenhower, C. Metagenomic Biomarker Discovery and Explanation. Genome Biol. 2011, 12, R60. [Google Scholar] [CrossRef] [PubMed]
- Zeineldin, M.; Aldridge, B.; Lowe, J. Dysbiosis of the Fecal Microbiota in Feedlot Cattle with Hemorrhagic Diarrhea. Microb. Pathog. 2018, 115, 123–130. [Google Scholar] [CrossRef] [PubMed]
- Jang, J.Y.; Kim, S.; Kwon, M.S.; Lee, J.; Yu, D.H.; Song, R.H.; Choi, H.J.; Park, J. Rotavirus-Mediated Alteration of Gut Microbiota and Its Correlation with Physiological Characteristics in Neonatal Calves. J. Microbiol. 2019, 57, 113–121. [Google Scholar] [CrossRef]
- Chuang, S.T.; Chen, C.T.; Hsieh, J.C.; Li, K.Y.; Ho, S.T.; Chen, M.J. Development of Next-Generation Probiotics by Investigating the Interrelationships between Gastrointestinal Microbiota and Diarrhea in Preruminant Holstein Calves. Animals 2022, 12, 695. [Google Scholar] [CrossRef]
- Portincasa, P.; Bonfrate, L.; Vacca, M.; De Angelis, M.; Farella, I.; Lanza, E.; Khalil, M.; Wang, D.Q.H.; Sperandio, M.; Di Ciaula, A. Gut Microbiota and Short Chain Fatty Acids: Implications in Glucose Homeostasis. Int. J. Mol. Sci. 2022, 23, 1105. [Google Scholar] [CrossRef]
- Hang, B.P.T.; Wredle, E.; Dicksved, J. Analysis of the Developing Gut Microbiota in Young Dairy Calves—Impact of Colostrum Microbiota and Gut Disturbances. Trop. Anim. Health Prod. 2021, 53, 50. [Google Scholar] [CrossRef]
- Xin, H.; Ma, T.; Xu, Y.; Chen, G.; Chen, Y.; Villot, C.; Renaud, D.L.; Steele, M.A.; Guan, L.L. Characterization of Fecal Branched-Chain Fatty Acid Profiles and Their Associations with Fecal Microbiota in Diarrheic and Healthy Dairy Calves. J. Dairy Sci. 2021, 104, 2290–2301. [Google Scholar] [CrossRef]
- Furusawa, Y.; Obata, Y.; Fukuda, S.; Endo, T.A.; Nakato, G.; Takahashi, D.; Nakanishi, Y.; Uetake, C.; Kato, K.; Kato, T. Commensal Microbe-Derived Butyrate Induces the Differentiation of Colonic Regulatory T Cells. Nature 2013, 504, 446–450. [Google Scholar] [CrossRef] [PubMed]
- Huang, S.; Ji, S.; Yan, H.; Hao, Y.; Zhang, J.; Wang, Y.; Cao, Z.; Li, S. The Day-to-Day Stability of the Ruminal and Fecal Microbiota in Lactating Dairy Cows. Microbiologyopen 2020, 9, e990. [Google Scholar] [CrossRef] [PubMed]
- Lynn, D.J.; Benson, S.C.; Lynn, M.A.; Pulendran, B. Modulation of Immune Responses to Vaccination by the Microbiota: Implications and Potential Mechanisms. Nat. Rev. Immunol. 2022, 22, 33–46. [Google Scholar] [CrossRef] [PubMed]
- Foster, D.M.; Smith, G.W. Pathophysiology of Diarrhea in Calves. Vet. Clin. N. Am. Food Anim. Pract. 2009, 25, 13–36. [Google Scholar] [CrossRef] [PubMed]
- Gao, W.; Howden, B.P.; Stinear, T.P. Evolution of Virulence in Enterococcus Faecium, a Hospital-Adapted Opportunistic Pathogen. Curr. Opin. Microbiol. 2018, 41, 76–82. [Google Scholar] [CrossRef]
- Balikci, E.; Al, M. Some Serum Acute Phase Proteins and Immunoglobulins Concentrations in Calves with Rotavirus, Coronavirus, E. coli F5 and Eimeria Species. Iran. J. Vet. Res. 2014, 15, 397–401. [Google Scholar] [PubMed]
- Hou, Q.; Kwok, L.Y.; Zheng, Y.; Wang, L.; Guo, Z.; Zhang, J.; Huang, W.; Wang, Y.; Leng, L.; Li, H.; et al. Differential Fecal Microbiota Are Retained in Broiler Chicken Lines Divergently Selected for Fatness Traits. Sci. Rep. 2016, 6, 37376. [Google Scholar] [CrossRef]
- Sudo, N. Biogenic Amines: Signals between Commensal Microbiota and Gut Physiology. Front. Endocrinol. 2019, 10, 504. [Google Scholar] [CrossRef]
- Pratt, J.; Hromadkova, J.; Malmuthuge, N.; Guan, L.L. Gut Microbiota and the Gut–Brain Axis in Neonatal Calves: Implications for Psychobiotic Usage for Stress Regulation. In Gut Microbiota, Immunity, and Health in Production Animals; Springer: Berlin/Heidelberg, Germany, 2022; pp. 299–325. [Google Scholar]
- Yatsunenko, T.; Rey, F.E.; Manary, M.J.; Trehan, I.; Dominguez-Bello, M.G.; Contreras, M.; Magris, M.; Hidalgo, G.; Baldassano, R.N.; Anokhin, A.P.; et al. Human Gut Microbiome Viewed across Age and Geography. Nature 2012, 486, 222–227. [Google Scholar] [CrossRef]
- LeBlanc, J.G.; Milani, C.; de Giori, G.S.; Sesma, F.; van Sinderen, D.; Ventura, M. Bacteria as Vitamin Suppliers to Their Host: A Gut Microbiota Perspective. Curr. Opin. Biotechnol. 2013, 24, 160–168. [Google Scholar] [CrossRef]
- Coleman, D.N.; Alharthi, A.S.; Liang, Y.; Lopes, M.G.; Lopreiato, V.; Vailati-Riboni, M.; Loor, J.J. Multifaceted Role of One-Carbon Metabolism on Immunometabolic Control and Growth during Pregnancy, Lactation and the Neonatal Period in Dairy Cattle. J. Anim. Sci. Biotechnol. 2021, 12, 27. [Google Scholar] [CrossRef]
- Zhuang, Y.; Cui, K.; Huang, W.; Han, Y.; Diao, Q.; Zhang, N. Early Solid Diet Supplementation Influences Proteomic of Rumen Epithelium in Goat Kids. preprint 2020. [Google Scholar] [CrossRef] [PubMed]
- Kim, E.T.; Lee, S.J.; Kim, T.Y.; Lee, H.G.; Atikur, R.M.; Gu, B.H.; Kim, D.H.; Park, B.Y.; Son, J.K.; Kim, M. Dynamic Changes in Fecal Microbial Communities of Neonatal Dairy Calves by Aging and Diarrhea. Animals 2021, 11, 1113. [Google Scholar] [CrossRef] [PubMed]
- Hudault, S.; Guignot, J.; Servin, A.L. Escherichia Coli Strains Colonising the Gastrointestinal Tract Protect Germfree Mice against Salmonella Typhimurium Infection. Gut 2001, 49, 47–55. [Google Scholar] [CrossRef] [PubMed]


| Item | Diet |
|---|---|
| Crude protein | 20.0% or more |
| Crude fat | 10.0% or more |
| Calcium | 0.7% or more |
| Phosphorus | 1.5% or less |
| Crude fiber | 3.0% or less |
| Crude Ash | 12.0% or less |
| Vitamin A | More than 25,000 IU/kg |
| Measurements | Group | SEM | p Value | |
|---|---|---|---|---|
| Healthy | Rota | |||
| Day − 3 | ||||
| Chao1 estimate | 160 | 79.0 | 28.5 | 0.247 |
| Evenness | 0.62 | 0.60 | 0.04 | 0.800 |
| Shannon’s index | 4.42 | 3.76 | 0.25 | 0.147 |
| Simpson’s index | 0.89 | 0.87 | 0.03 | 0.713 |
| Day + 1 | ||||
| Chao1 estimate | 136 | 87.0 | 24.2 | 0.234 |
| Evenness | 0.63 | 0.48 | 0.04 | 0.074 |
| Shannon’s index | 4.43 | 3.03 | 0.35 | 0.055 |
| Simpson’s index | 0.92 | 0.77 | 0.04 | 0.139 |
| Day + 7 | ||||
| Chao1 estimate | 105 | 99.3 | 47.7 | 0.937 |
| Evenness | 0.55 | 0.59 | 0.03 | 0.447 |
| Shannon’s index | 3.56 | 3.68 | 0.62 | 0.895 |
| Simpson’s index | 0.85 | 0.87 | 0.04 | 0.795 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kim, S.-H.; Choi, Y.; Miguel, M.A.; Lee, S.-J.; Lee, S.-S.; Lee, S.-S. Analysis of Fecal Microbial Changes in Young Calves Following Bovine Rotavirus Infection. Vet. Sci. 2023, 10, 496. https://doi.org/10.3390/vetsci10080496
Kim S-H, Choi Y, Miguel MA, Lee S-J, Lee S-S, Lee S-S. Analysis of Fecal Microbial Changes in Young Calves Following Bovine Rotavirus Infection. Veterinary Sciences. 2023; 10(8):496. https://doi.org/10.3390/vetsci10080496
Chicago/Turabian StyleKim, Seon-Ho, Youyoung Choi, Michelle A. Miguel, Shin-Ja Lee, Sung-Sill Lee, and Sang-Suk Lee. 2023. "Analysis of Fecal Microbial Changes in Young Calves Following Bovine Rotavirus Infection" Veterinary Sciences 10, no. 8: 496. https://doi.org/10.3390/vetsci10080496
APA StyleKim, S.-H., Choi, Y., Miguel, M. A., Lee, S.-J., Lee, S.-S., & Lee, S.-S. (2023). Analysis of Fecal Microbial Changes in Young Calves Following Bovine Rotavirus Infection. Veterinary Sciences, 10(8), 496. https://doi.org/10.3390/vetsci10080496





