Development and Evaluation of a Semi-Nested PCR Method Based on the 18S ribosomal RNA Gene for the Detection of Babesia aktasi Infections in Goats
Abstract
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Ethics Statement
2.2. Primer Design
2.3. Standard Positive Reference DNA Samples Used in This Study
2.4. Semi-Nested PCR Amplification of 18S rRNA Gene
2.5. Efficiency and Detection Threshold of the Semi-Nested PCR
2.6. Field Application of Semi-Nested PCR in Genomic DNAs Isolated from Goats
3. Results
3.1. Primer Selection for Semi-Nested PCR
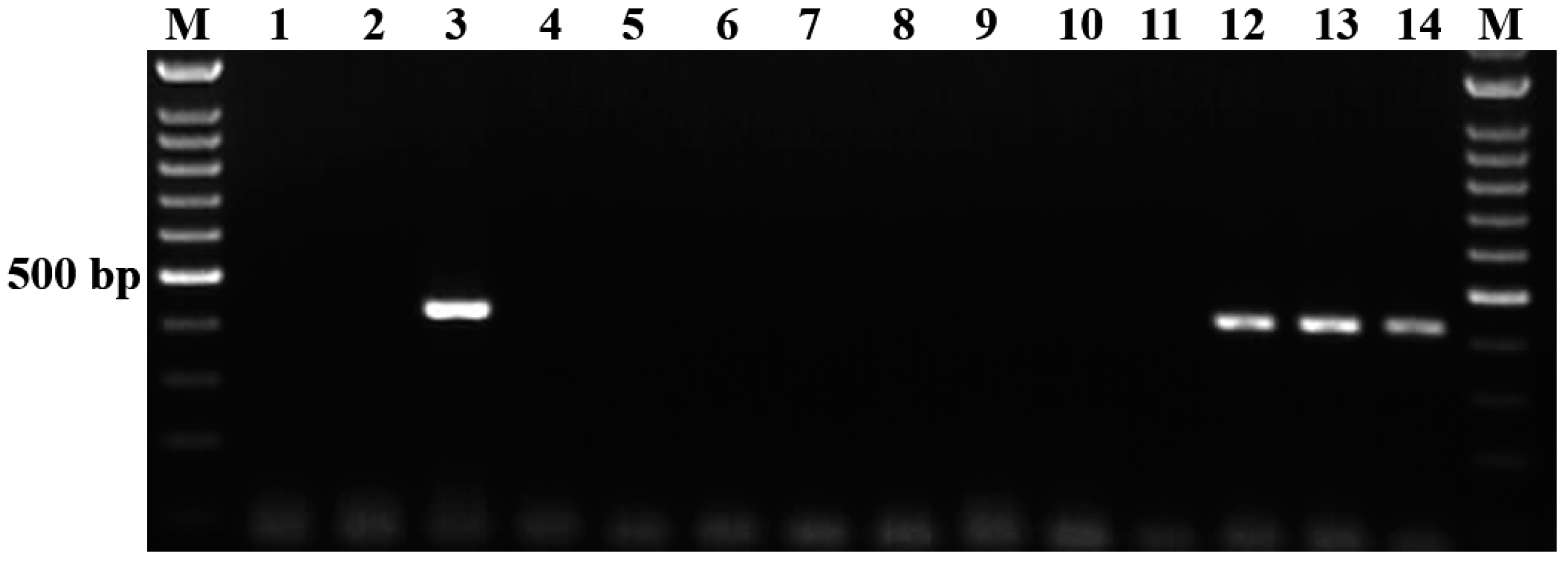
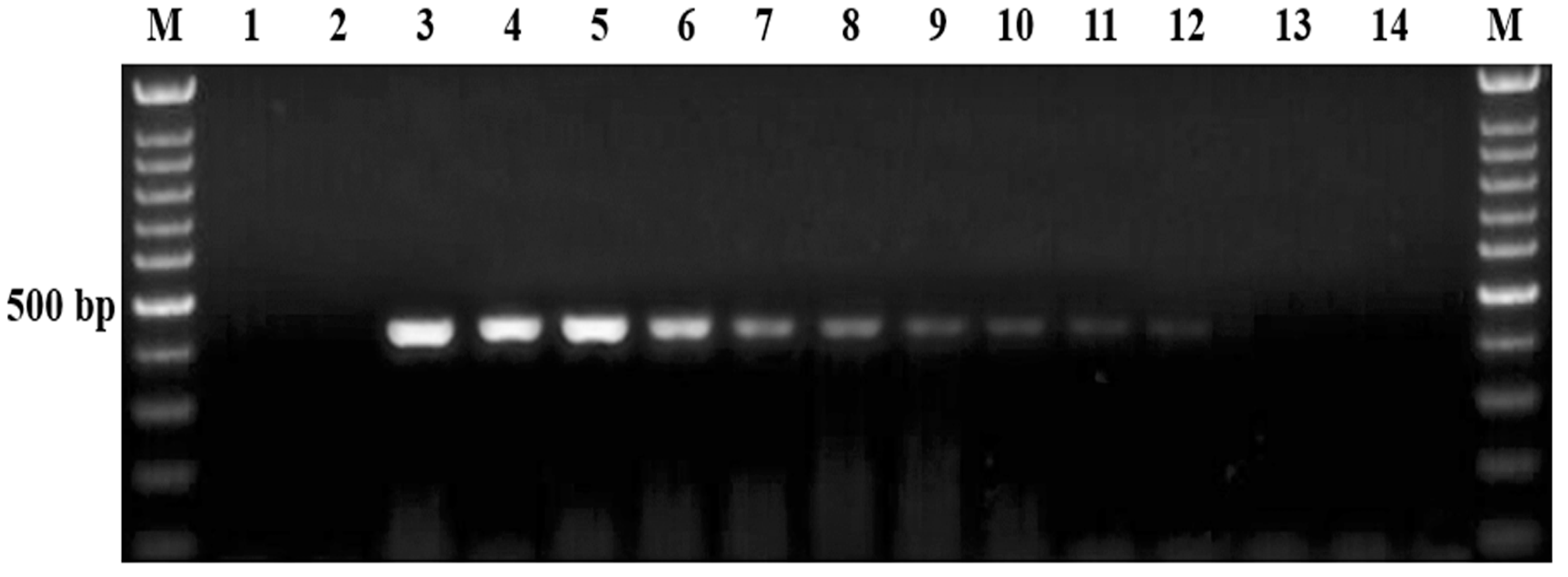
3.2. Evaluation of Analytical Specificity of the Semi-Nested PCR
3.3. Analytical Sensitivity of the Semi-Nested PCR
3.4. Field Application and Detecting Performance of Semi-Nested PCR on Field Blood Samples
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Gray, J.S.; Estrada-Peña, A.; Zintl, A. Vectors of Babesiosis. Annu. Rev. Entomol. 2019, 64, 149–165. [Google Scholar] [CrossRef] [PubMed]
- Martínez-García, G.; Santamaría-Espinosa, R.M.; Lira-Amaya, J.J.; Figueroa, J.V. Challenges in tick-borne pathogen detection: The case for Babesia spp. identification in the tick vector. Pathogens 2021, 10, 92. [Google Scholar] [CrossRef] [PubMed]
- Schnittger, L.; Ganzinelli, S.; Bhoora, R.; Omondi, D.; Nijhof, A.M.; Florin-Christensen, M. The Piroplasmida Babesia, Cytauxzoon, and Theileria in farm and companion animals: Species compilation, molecular phylogeny, and evolutionary insights. Parasitol Res. 2022, 121, 1207–1245. [Google Scholar] [CrossRef] [PubMed]
- Iqbal, F.; Fatima, M.; Shahnawaz, S.; Naeem, M.; Shaikh, R.S.; Ali, M.; Shaikh, A.S.; Aktas, M. A Study on the determination of risk factors associated with babesiosis and prevalence of Babesia sp., by PCR amplification, in small ruminants from Southern Punjab (Pakistan). Parasite J. Société Française Parasitol. 2011, 18, 229. [Google Scholar]
- Ozubek, S.; Bastos, R.G.; Alzan, H.F.; Inci, A.; Aktas, M.; Suarez, C.E. Bovine babesiosis in Turkey: Impact, current gaps, and opportunities for intervention. Pathogens 2020, 9, 1041. [Google Scholar] [CrossRef]
- Kappes, A.; Tozooneyi, T.; Shakil, G.; Railey, A.F.; McIntyre, K.M.; Mayberry, D.E.; Rushton, J.; Pendell, D.L.; Marsh, T.L. Livestock health and disease economics: A scoping review of selected literature. Front. Vet. Sci. 2023, 10, 1168649. [Google Scholar] [CrossRef]
- Sevinc, F.; Sevinc, M.; Ekici, O.D.; Yildiz, R.; Isik, N.; Aydogdu, U. Babesia ovis infections: Detailed clinical and laboratory observations in the pre-and post-treatment periods of 97 field cases. Vet. Parasitol. 2013, 191, 35–43. [Google Scholar] [CrossRef]
- Benitez, D.; Mesplet, M.; Echaide, I.; de Echaide, S.T.; Schnittger, L.; Florin-Christensen, M. Mitigated clinical disease in water buffaloes experimentally infected with Babesia bovis. Ticks Tick-Borne Dis. 2018, 9, 1358–1363. [Google Scholar] [CrossRef] [PubMed]
- Stuen, S. Haemoparasites—Challenging and wasting infections in small ruminants: A Review. Animals 2020, 10, 2179. [Google Scholar] [CrossRef]
- Galon, E.M.; Zafar, I.; Ji, S.; Li, H.; Ma, Z.; Xuan, X. Molecular reports of ruminant Babesia in Southeast Asia. Pathogens 2022, 11, 915. [Google Scholar] [CrossRef]
- Babeş, V. L’Étiologie d’une Enzootie des Moutons, Dénommée Carceag en Roumanie; Gauthier-Villars, 1892; Available online: https://www.biusante.parisdescartes.fr/histoire/medica/resultats/index.php?do=pdf&cote=90166x1892x29 (accessed on 24 September 2024).
- Liu, A.H.; Yin, H.; Guan, G.Q.; Schnittger, L.; Liu, Z.J.; Ma, M.L.; Dang, Z.S.; Liu, J.L.; Ren, Q.Y.; Bai, Q.; et al. At least two genetically distinct large Babesia species infective to sheep and goats in China. Vet. Parasitol. 2007, 147, 246–251. [Google Scholar] [CrossRef]
- Niu, Q.; Liu, Z.; Yang, J.; Yu, P.; Pan, Y.; Zhai, B.; Luo, J.; Yin, H. Genetic Diversity and molecular characterization of Babesia motasi-like in small ruminants and ixodid ticks from China. Infect. Genet. Evol. 2016, 41, 8–15. [Google Scholar] [CrossRef] [PubMed]
- Bosman, A.-M.; Oosthuizen, M.C.; Peirce, M.A.; Venter, E.H.; Penzhorn, B.L. Babesia lengau sp. nov., a novel Babesia species in cheetah (Acinonyx Jubatus, Schreber, 1775) populations in South Africa. J. Clin. Microbiol. 2010, 48, 2703–2708. [Google Scholar] [CrossRef]
- Ozubek, S.; Aktas, M. Molecular evidence of a new Babesia sp. in goats. Vet. Parasitol. 2017, 233, 1–8. [Google Scholar] [CrossRef] [PubMed]
- Ozubek, S.; Ulucesme, M.C.; Aktas, M. Discovery of a novel species infecting goats: Morphological and molecular characterization of Babesia aktasi n. sp. Pathogens 2023, 12, 113. [Google Scholar] [CrossRef] [PubMed]
- Ozubek, S.; Ulucesme, M.C.; Bastos, R.G.; Alzan, H.F.; Laughery, J.M.; Suarez, C.E.; Aktas, M. Experimental infection of non-immunosuppressed and immunosuppressed goats reveals differential pathogenesis of Babesia aktasi n. sp. Front. Cell. Infect. Microbiol. 2023, 13, 1277956. [Google Scholar] [CrossRef] [PubMed]
- Salih, D.A.; El Hussein, A.M.; Singla, L.D. Diagnostic approaches for tick-borne haemoparasitic diseases in livestock. J. Vet. Med. Anim. Health 2015, 7, 45–56. [Google Scholar]
- Almería, S.; Castella, J.; Ferrer, D.; Ortuno, A.; Estrada-Peña, A.; Gutierrez, J.F. Bovine piroplasms in Minorca (Balearic Islands, Spain): A comparison of PCR-based and light microscopy detection. Vet. Parasitol. 2001, 99, 249–259. [Google Scholar] [CrossRef] [PubMed]
- Mosqueda, J.; Olvera-Ramírez, A.; Aguilar-Tipacamú, G.; Cantó, G. Current advances in detection and treatment of babesiosis. Curr. Med. Chem. 2012, 19, 1504–1518. [Google Scholar] [CrossRef]
- Papadopoulos, B.; Brossard, M.; Perié, N.M. Piroplasms of domestic animals in the Macedonia Region of Greece 3. Piroplasms of Small Ruminants. Vet. Parasitol. 1996, 63, 67–74. [Google Scholar] [CrossRef]
- Alvarez, J.A.; Rojas, C.; Figueroa, J.V. Diagnostic tools for the identification of Babesia sp. in persistently infected cattle. Pathogens 2019, 8, 143. [Google Scholar] [CrossRef] [PubMed]
- Zhou, B.; Yang, G.; Hu, Z.; Chen, K.; Guo, W.; Wang, X.; Du, C. Development of a real-time quantitative PCR based on a taqman-mgb probe for the rapid detection of Theileria haneyi. Microorganisms 2023, 11, 2633. [Google Scholar] [CrossRef] [PubMed]
- Schnittger, L.; Yin, H.; Qi, B.; Gubbels, M.J.; Beyer, D.; Niemann, S.; Jongejan, F.; Ahmed, J.S. Simultaneous detection and differentiation of Theileria and Babesia parasites infecting small ruminants by reverse line blotting. Parasitol. Res. 2004, 92, 189–196. [Google Scholar] [CrossRef] [PubMed]
- Alhassan, A.; Pumidonming, W.; Okamura, M.; Hirata, H.; Battsetseg, B.; Fujisaki, K.; Yokoyama, N.; Igarashi, I. Development of a single-round and multiplex PCR method for the simultaneous detection of Babesia caballi and Babesia equi in horse blood. Vet. Parasitol. 2005, 129, 43–49. [Google Scholar] [CrossRef] [PubMed]
- Aktaş, M.; Altay, K.; Dumanli, N. Development of a polymerase chain reaction method for diagnosis of Babesia ovis infection in sheep and goats. Vet. Parasitol. 2005, 133, 277–281. [Google Scholar] [CrossRef] [PubMed]
- Razmi, G.; Pourhosseini, M.; Yaghfouri, S.; Rashidi, A.; Seidabadi, M. Molecular detection of Theileria spp. and Babesia spp. in sheep and ixodid ticks from the northeast of Iran. J. Parasitol. 2013, 99, 77–81. [Google Scholar] [CrossRef] [PubMed]
- Esmaeilnejad, B.; Tavassoli, M.; Asri-Rezaei, S.; Dalir-Naghadeh, B.; Mardani, K.; Golabi, M.; Arjmand, J.; Kazemnia, A.; Jalilzadeh, G. Determination of prevalence and risk factors of infection with Babesia ovis in small ruminants from West Azerbaijan Province, Iran by Polymerase Chain Reaction. J. Arthropod-Borne Dis. 2015, 9, 246. [Google Scholar]
- Bazmani, A.; Abolhooshyar, A.; Imani-Baran, A.; Akbari, H. Semi-nested polymerase chain reaction-based detection of Babesia spp. in small ruminants from Northwest of Iran. Vet. World 2018, 11, 268. [Google Scholar] [CrossRef]
- Tu, H.L.C.; Nugraheni, Y.R.; Tiawsirisup, S.; Saiwichai, T.; Thiptara, A.; Kaewthamasorn, M. Development of a Novel multiplex PCR assay for the detection and differentiation of Plasmodium caprae from Theileria luwenshuni and Babesia spp. in goats. Acta Trop. 2021, 220, 105957. [Google Scholar] [CrossRef]
- Erster, O.; Roth, A.; Wollkomirsky, R.; Leibovich, B.; Savitzky, I.; Zamir, S.; Molad, T.; Shkap, V. Quantitative analysis of Babesia ovis infection in sheep and ticks. Vet. Parasitol. 2016, 221, 39–45. [Google Scholar] [CrossRef]
- Guan, G.; Chauvin, A.; Luo, J.; Inoue, N.; Moreau, E.; Liu, Z.; Gao, J.; Thekisoe, O.M.; Ma, M.; Liu, A. The development and evaluation of a loop-mediated isothermal amplification (LAMP) method for detection of Babesia spp. infective to sheep and goats in China. Exp. Parasitol. 2008, 120, 39–44. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Gao, S.; Zhang, S.; He, X.; Liu, J.; Liu, A.; Li, Y.; Liu, G.; Luo, J.; Guan, G.; et al. Rapid Detection of Babesia motasi responsible for human babesiosis by cross-priming amplification combined with a vertical flow. Parasites Vectors 2020, 13, 377. [Google Scholar] [CrossRef]
- Nagore, D.; García-Sanmartín, J.; Garcıa-Pérez, A.L.; Juste, R.A.; Hurtado, A. Identification, Genetic diversity and prevalence of Theileria and Babesia species in a sheep population from Northern Spain. Int. J. Parasitol. 2004, 34, 1059–1067. [Google Scholar] [CrossRef] [PubMed]
- Niu, Q.; Luo, J.; Guan, G.; Ma, M.; Liu, Z.; Liu, A.; Dang, Z.; Gao, J.; Ren, Q.; Li, Y.; et al. Detection and differentiation of ovine Theileria and Babesia by reverse line blotting in China. Parasitol Res. 2009, 104, 1417–1423. [Google Scholar] [CrossRef]
- Altay, K.; Aktas, M.; Dumanli, N. Detection of Babesia ovis by PCR in Rhipicephalus bursa collected from naturally infested sheep and goats. Res. Vet. Sci. 2008, 85, 116–119. [Google Scholar] [CrossRef]
- Ranjbar-Bahadori, S.; Eckert, B.; Omidian, Z.; Shirazi, N.S.; Shayan, P. Babesia ovis as the main causative agent of sheep babesiosis in Iran. Parasitol Res. 2012, 110, 1531–1536. [Google Scholar] [CrossRef] [PubMed]
- Aydin, M.F.; Aktas, M.; Dumanli, N. Molecular identification of Theileria and Babesia in sheep and goats in the Black Sea Region in Turkey. Parasitol Res. 2013, 112, 2817–2824. [Google Scholar] [CrossRef] [PubMed]
- Gökpinar, S.; Gazyağci, A.N.; Aydenizöz, M.; Kaya, U. A research on Babesia and Theileria species in sheep and goats of Kırıkkale Province through Molecular Methods. Turk. J. Vet. Anim. Sci. 2021, 45, 912–919. [Google Scholar] [CrossRef]
- Gubbels, J.M.; De Vos, A.P.; Van Der Weide, M.; Viseras, J.; Schouls, L.M.; De Vries, E.; Jongejan, F. Simultaneous detection of bovine Theileria and Babesia species by reverse line blot hybridization. J. Clin. Microbiol. 1999, 37, 1782–1789. [Google Scholar] [CrossRef]
- Bekker, C.P.; De Vos, S.; Taoufik, A.; Sparagano, O.A.; Jongejan, F. Simultaneous detection of Anaplasma and Ehrlichia species in ruminants and detection of Ehrlichia ruminantium in Amblyomma variegatum ticks by reverse line blot hybridization. Vet. Microbiol. 2002, 89, 223–238. [Google Scholar] [CrossRef]
- Ulucesme, M.C.; Ozubek, S.; Karoglu, A.; Turk, Z.I.; Olmus, I.; Irehan, B.; Aktas, M. Small ruminant piroplasmosis: High prevalence of Babesia aktasi n. sp. in goats in Türkiye. Pathogens 2023, 12, 514. [Google Scholar] [CrossRef] [PubMed]
- Tirosh-Levy, S.; Gottlieb, Y.; Fry, L.M.; Knowles, D.P.; Steinman, A. Twenty years of equine piroplasmosis research: Global distribution, molecular diagnosis, and phylogeny. Pathogens 2020, 9, 926. [Google Scholar] [CrossRef] [PubMed]
- Katoh, K.; Standley, D.M. MAFFT Multiple Sequence Alignment Software Version 7: Improvements in Performance and Usability. Mol. Biol. Evol. 2013, 30, 772–780. [Google Scholar] [CrossRef] [PubMed]
- AbouLaila, M.; Yokoyama, N.; Igarashi, I. Development and evaluation of two nested PCR assays for the detection of Babesia bovis from cattle blood. Vet. Parasitol. 2010, 172, 65–70. [Google Scholar] [CrossRef] [PubMed]
- Ybañez, A.P.; Arrabis, O.V.; Alvarez, D.J.M.; Galon, E.M.S.; Jayag, R.M.P.; Delan, E.S.; Ybañez, R.H.D.; Xuan, X. Evaluation on the presence of Anaplasma, Ehrlichia, and Babesia spp. in goats (Capra hircus) in Cebu, the Philippines. Vet. World 2019, 12, 774. [Google Scholar] [CrossRef]
- Ceylan, O.; Sevinc, F. Endemic Instability of ovine babesiosis in Turkey: A country-wide sero-epidemiological study. Vet. Parasitol. 2020, 278, 109034. [Google Scholar] [CrossRef]
- Udonsom, R.; Mahittikorn, A.; Jirapattharasate, C. Molecular detection and genetic diversity of tick-borne pathogens in goats from the Southern Part of Thailand. Pathogens 2022, 11, 477. [Google Scholar] [CrossRef] [PubMed]
- Yeruham, I.; Hadani, A.; Galker, F. Some epizootiological and clinical aspects of ovine babesiosis caused by Babesia ovis—A review. Vet. Parasitol. 1998, 74, 153–163. [Google Scholar] [CrossRef] [PubMed]
- Yeruham, I.; Hadani, A.; Galker, F. The Effect of the Ovine Host Parasitaemia on the development of Babesia ovis (Babes, 1892) in the tick Rhipicephalus bursa (Canestrini and Fanzago, 1877). Vet. Parasitol. 2001, 96, 195–202. [Google Scholar] [CrossRef]
- Rampersad, J.; Cesar, E.; Campbell, M.D.; Samlal, M.; Ammons, D. A field evaluation of PCR for the routine detection of Babesia equi in horses. Vet. Parasitol. 2003, 114, 81–87. [Google Scholar] [CrossRef] [PubMed]
- Lempereur, L.; Beck, R.; Fonseca, I.; Marques, C.; Duarte, A.; Santos, M.; Zúquete, S.; Gomes, J.; Walder, G.; Domingos, A.; et al. Guidelines for the detection of Babesia and Theileria parasites. Vector-Borne Zoonotic Dis. 2017, 17, 51–65. [Google Scholar] [CrossRef] [PubMed]
- Lv, K.; Zhang, Y.; Yang, Y.; Liu, Z.; Deng, L. Development of nested PCR and duplex real-time fluorescence quantitative PCR assay for the simultaneous detection of Theileria equi and Babesia caballi. Front. Vet. Sci. 2022, 9, 873190. [Google Scholar] [CrossRef] [PubMed]
- Liu, I.-L.; Chi, N.-Y.; Chang, C.-L.; Hung, M.-L.; Chiu, C.-T.; Chen, H.-W. A novel pcr-based point-of-care method enables rapid, sensitive and reliable diagnosis of Babesia gibsoni infection in dogs. BMC Vet. Res. 2019, 15, 428. [Google Scholar] [CrossRef] [PubMed]
- Inokuma, H.; Okuda, M.; Yoshizaki, Y.; Hiraoka, H.; Miyama, T.; Itamoto, K.; Une, S.; Nakaichi, M.; Taura, Y. Clinical observations of Babesia gibsoni infection with low parasitaemia confirmed by PCR in dogs. Vet. Rec. 2005, 156, 116–118. [Google Scholar] [CrossRef]
- Horta, S.; Barreto, M.C.; Pepe, A.; Campos, J.; Oliva, A. Highly sensitive method for diagnosis of subclinical B. ovis infection. Ticks Tick Borne Dis. 2014, 5, 902–906. [Google Scholar] [CrossRef] [PubMed]
- Bilgiç, H.B.; Karagenç, T.; Simuunza, M.; Shiels, B.; Tait, A.; Eren, H.; Weir, W. development of a multiplex PCR assay for simultaneous detection of Theileria annulata, Babesia bovis and Anaplasma marginale in cattle. Exp. Parasitol. 2013, 133, 222–229. [Google Scholar] [CrossRef] [PubMed]
- AbouLaila, M.; Yokoyama, N.; Igarashi, I. Development and evaluation of a nested PCR based on spherical body protein 2 gene for the diagnosis of Babesia bovis infection. Vet. Parasitol. 2010, 169, 45–50. [Google Scholar] [CrossRef] [PubMed]
- Troskie, M.; de Villiers, L.; Leisewitz, A.; Oosthuizen, M.C.; Quan, M. Development and validation of a multiplex, real-time PCR assay for Babesia rossi and Babesia vogeli. Ticks Tick-Borne Dis. 2019, 10, 421–432. [Google Scholar] [CrossRef] [PubMed]
- Stoltsz, H.; Byaruhanga, C.; Troskie, M.; Makgabo, M.; Oosthuizen, M.C.; Collins, N.E.; Neves, L. Improved detection of Babesia bigemina from various geographical areas in Africa using quantitative PCR and reverse line blot hybridisation. Ticks Tick-Borne Dis. 2020, 11, 101415. [Google Scholar] [CrossRef] [PubMed]
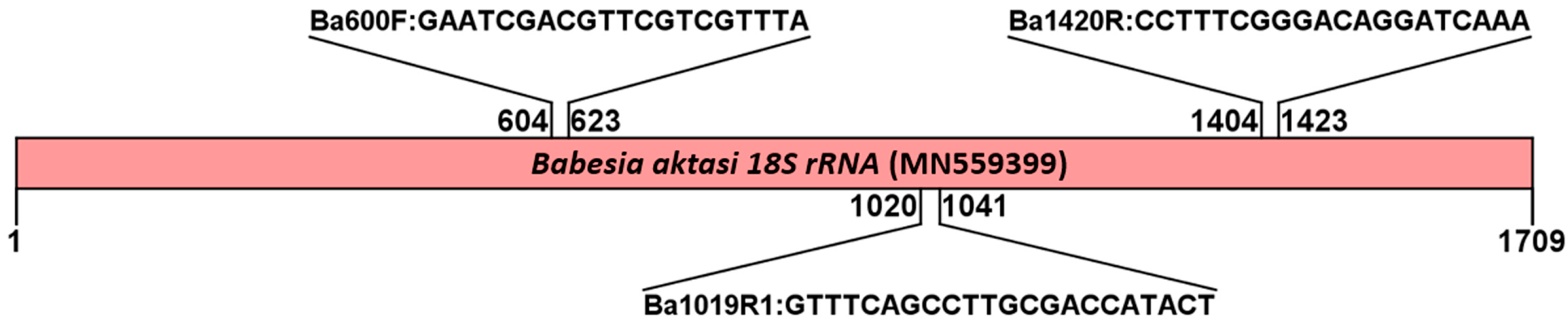

| Gene | Primer Name | Primer Sequences (5’→3’) | Reaction and/or Use | Amplicon Size |
|---|---|---|---|---|
| 18S rRNA | Ba600F Ba1420R | GAATCGACGTTCGTCGTTTA CCTTTCGGGACAGGATCAAA | First round PCR forward F First round PCR reverse R | 820 |
| Ba600F Ba1019R1 | GAATCGACGTTCGTCGTTTA GTTTCAGCCTTGCGACCATACT | Semi-nested PCR forward F Semi-nested PCR reverse R1 | 438 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ulucesme, M.C.; Ozubek, S.; Aktas, M. Development and Evaluation of a Semi-Nested PCR Method Based on the 18S ribosomal RNA Gene for the Detection of Babesia aktasi Infections in Goats. Vet. Sci. 2024, 11, 466. https://doi.org/10.3390/vetsci11100466
Ulucesme MC, Ozubek S, Aktas M. Development and Evaluation of a Semi-Nested PCR Method Based on the 18S ribosomal RNA Gene for the Detection of Babesia aktasi Infections in Goats. Veterinary Sciences. 2024; 11(10):466. https://doi.org/10.3390/vetsci11100466
Chicago/Turabian StyleUlucesme, Mehmet Can, Sezayi Ozubek, and Munir Aktas. 2024. "Development and Evaluation of a Semi-Nested PCR Method Based on the 18S ribosomal RNA Gene for the Detection of Babesia aktasi Infections in Goats" Veterinary Sciences 11, no. 10: 466. https://doi.org/10.3390/vetsci11100466
APA StyleUlucesme, M. C., Ozubek, S., & Aktas, M. (2024). Development and Evaluation of a Semi-Nested PCR Method Based on the 18S ribosomal RNA Gene for the Detection of Babesia aktasi Infections in Goats. Veterinary Sciences, 11(10), 466. https://doi.org/10.3390/vetsci11100466









