Activating Fc Gamma Receptors and Viral Receptors Are Required for Antibody-Dependent Enhancement of Porcine Reproductive and Respiratory Syndrome Virus Infection
Abstract
:Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Cells and Virus
2.2. Antibodies
2.3. Formation of PRRSV-Antibody ICs
2.4. Detection of PRRSV-ADE Infection
2.5. RNA Interfering Assay
2.6. Antibody Blocking Assay
2.7. RNA Extraction and Quantification RT-PCR
2.8. Immunoblot Assay
2.9. Flow Cytometry
2.10. Statistical Analysis
3. Results
3.1. ADE Activity of PRRSV Infection Exists in PRRSV-Specific Porcine IgG in PAMs
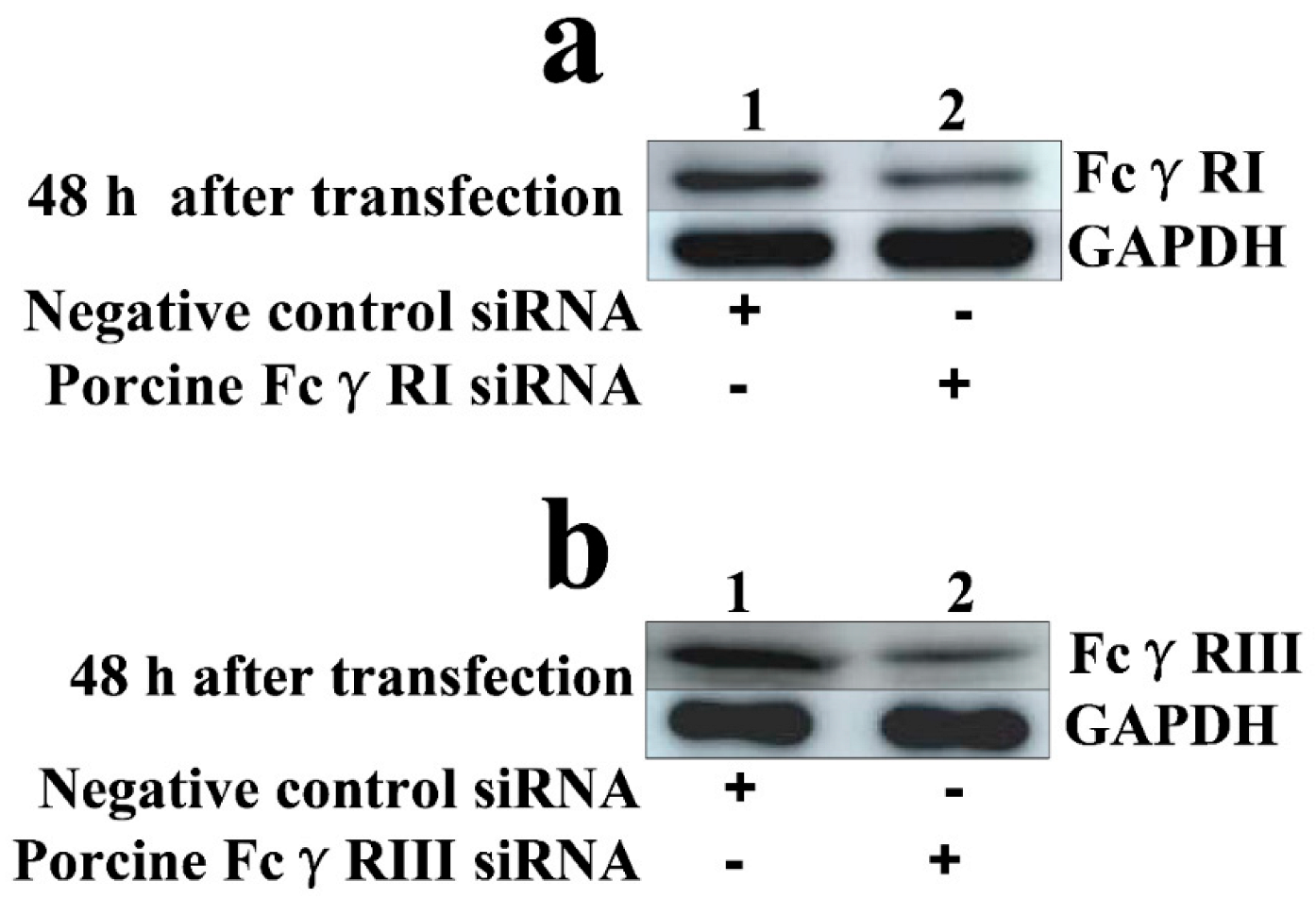
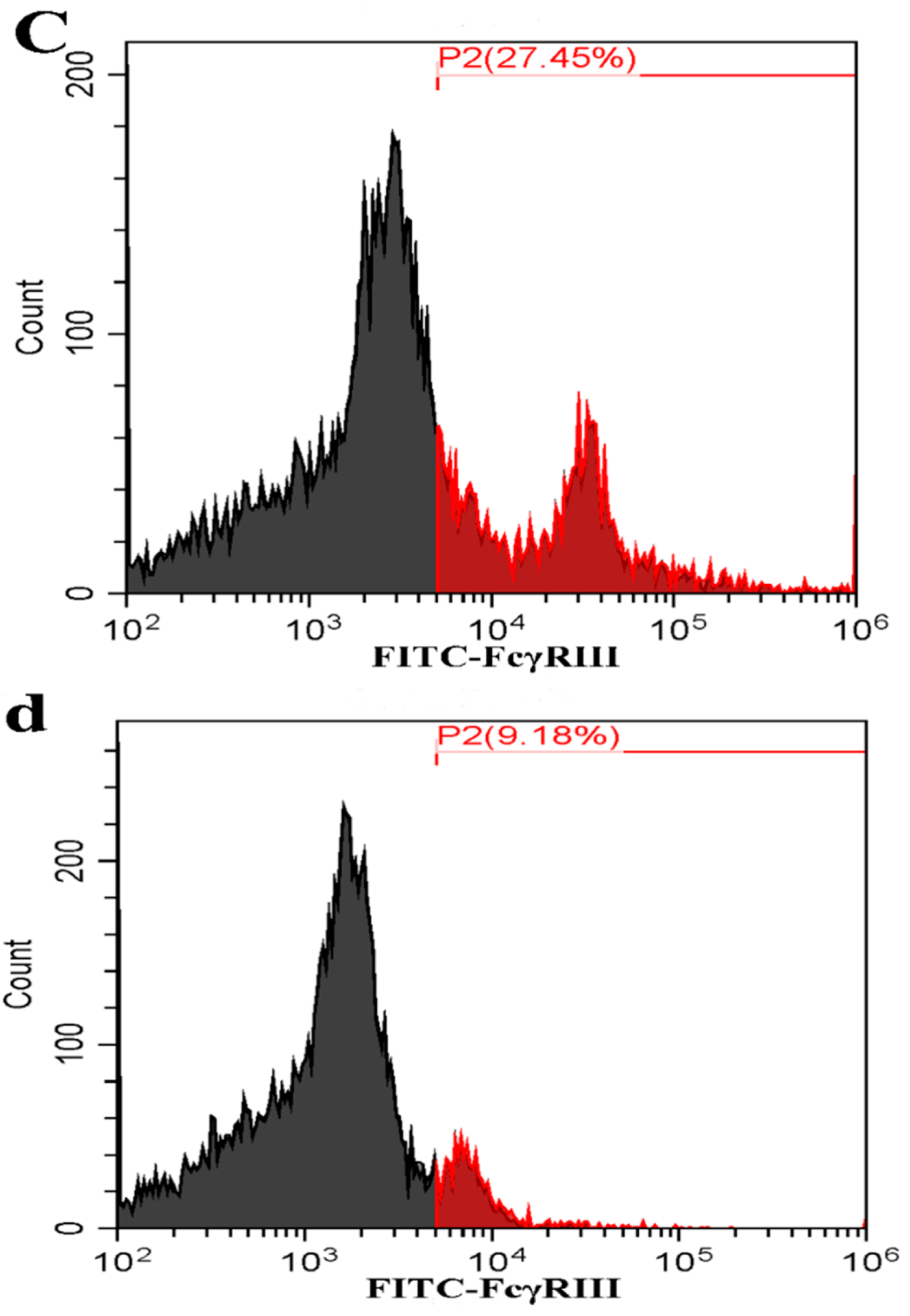
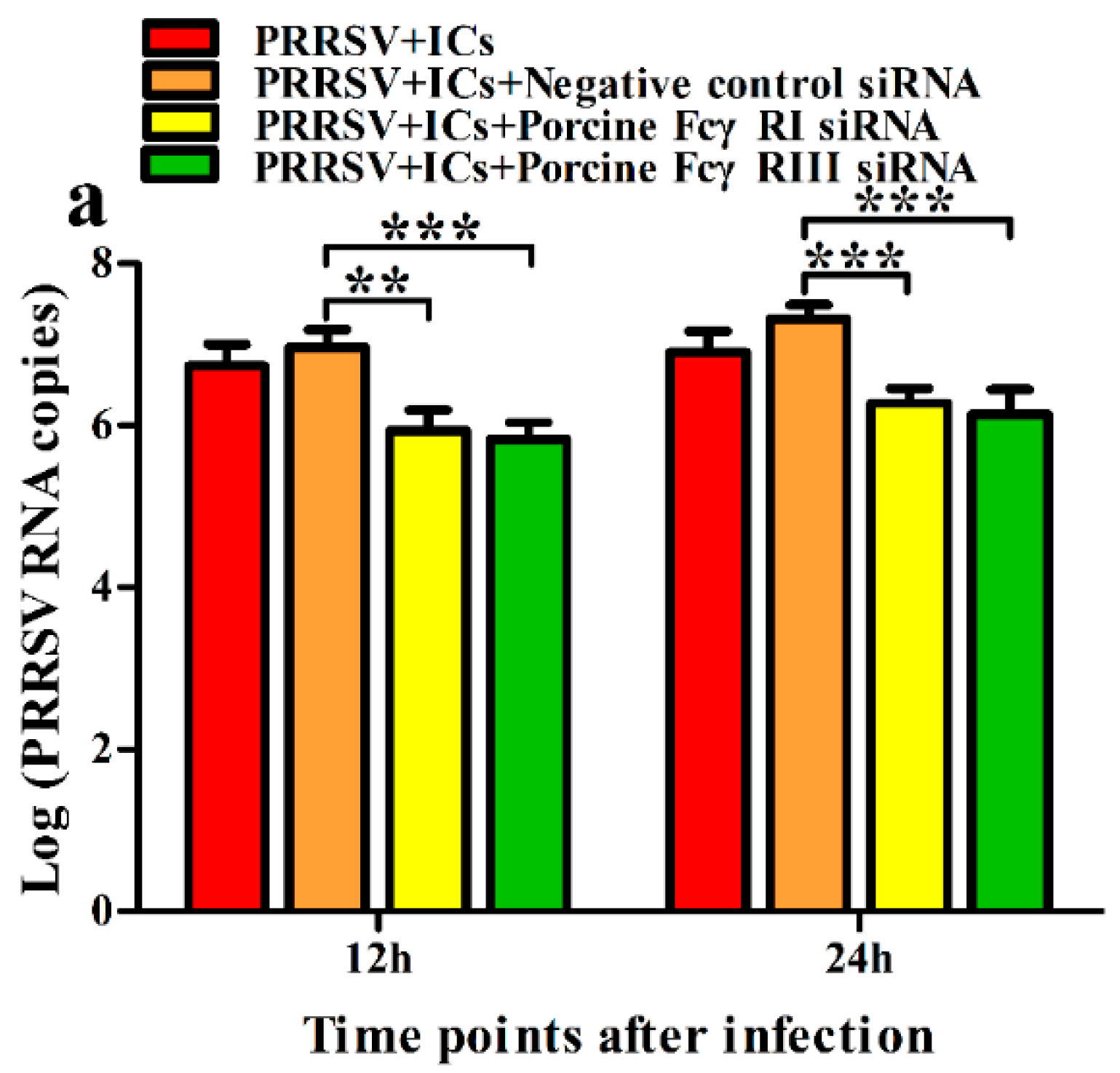
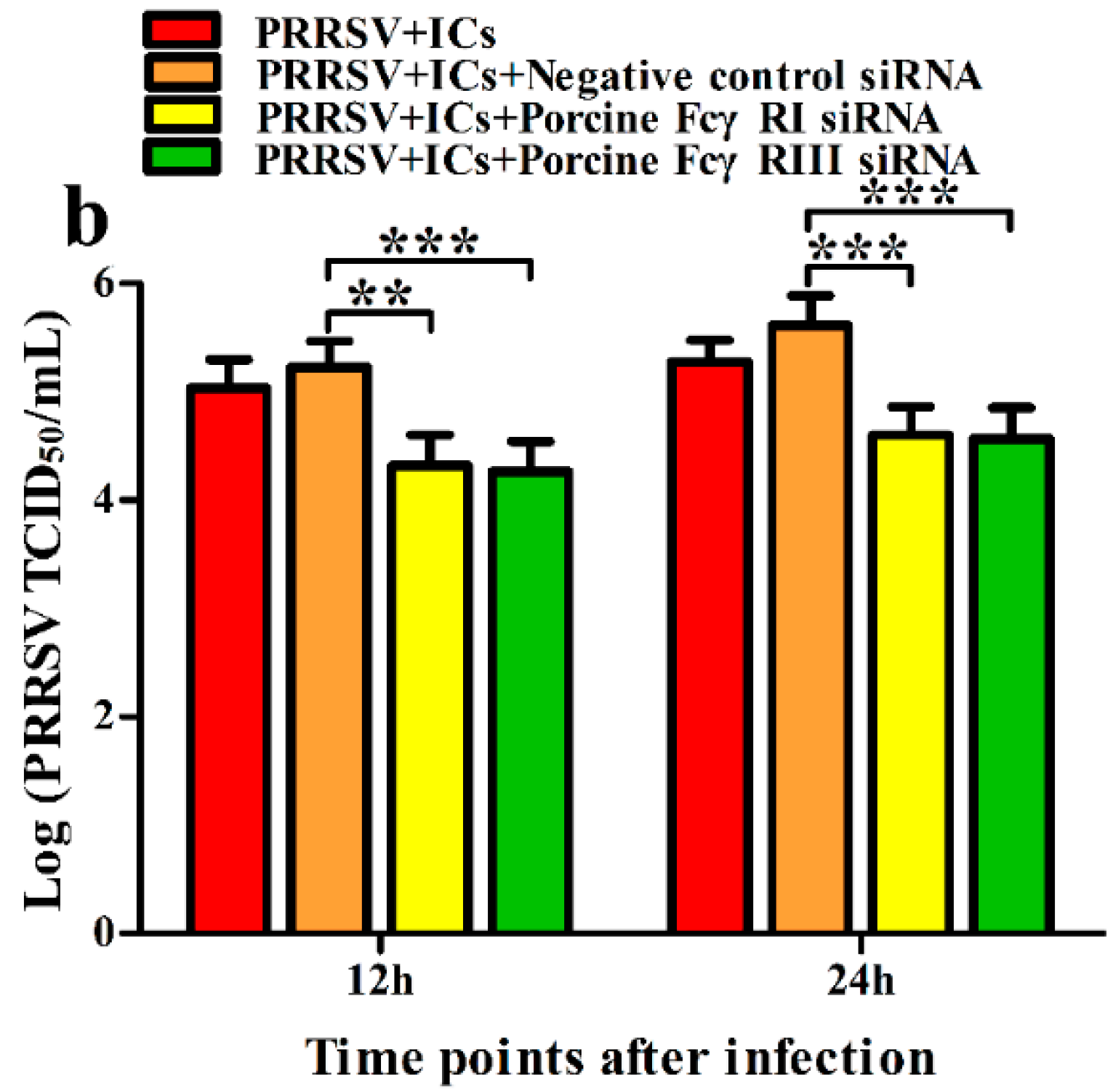
3.2. FcγRI and FcγRIII Are Responsible for Mediating ADE of PRRSV Infection in PAMs
3.3. Sn and CD163 Are Required for ADE of PRRSV Infection in PAMs
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Rowland, R.R.R.; Lunney, J.K. Alternative strategies for the control and elimination of PRRS. Vet. Microbiol. 2017, 209, 1–4. [Google Scholar] [CrossRef] [PubMed]
- Dokland, T. The structural biology of PRRSV. Virus Res. 2010, 154, 86–97. [Google Scholar] [CrossRef]
- Xie, C.Z.; Wang, Z.; Ha, Z.; Zhang, Y.; Xie, Y.B.; Zhang, H.; Nan, F.L.; Zhang, J.Y.; Zhao, G.Y.; Li, Z.X.; et al. Genetic characterization of a new NSP2-deletion porcine reproductive and respiratory syndrome virus in China. Microb. Pathog. 2021, 150, 104729. [Google Scholar] [CrossRef]
- Ruedas-Torres, I.; Rodríguez-Gómez, I.M.; Sánchez-Carvajal, J.M.; Larenas-Muñoz, F.; Pallarés, F.J.; Carrasco, L.; Gómez-Laguna, J. The jigsaw of PRRSV virulence. Vet. Microbiol. 2021, 260, 109168. [Google Scholar] [CrossRef]
- Lunney, J.K.; Fang, Y.; Ladinig, A.; Chen, N.; Li, Y.; Rowland, B.; Renukaradhya, G.J. Porcine reproductive and respiratory syndrome virus (PRRSV): Pathogenesis and interaction with the immune system. Annu. Rev. Anim. Biosci. 2016, 4, 129–154. [Google Scholar] [CrossRef]
- Tirado, S.M.; Yoon, K.J. Antibody-dependent enhancement of virus infection and disease. Viral Immunol. 2003, 16, 69–86. [Google Scholar] [CrossRef]
- Yoon, K.J.; Wu, L.L.; Zimmerman, J.J.; Hill, H.T.; Platt, K.B. Antibody-dependent enhancement (ADE) of porcine reproductive and respiratory syndrome virus (PRRSV) infection in pigs. Viral Immunol. 1996, 9, 51–63. [Google Scholar] [CrossRef]
- Bao, D.; Wang, R.; Qiao, S.; Wan, B.; Wang, Y.; Liu, M.; Shi, X.; Guo, J.; Zhang, G. Antibody-dependent enhancement of PRRSV infection down-modulates TNF-α and IFN-β transcription in macrophages. Vet. Immunol. Immunopathol. 2013, 156, 128–134. [Google Scholar] [CrossRef]
- Loving, C.L.; Osorio, F.A.; Murtaugh, M.P.; Zuckermann, F.A. Innate and adaptive immunity against porcine reproductive and respiratory syndrome virus. Vet. Immunol. Immunopathol. 2015, 167, 1–14. [Google Scholar] [CrossRef]
- Zhang, L.; Li, W.; Sun, Y.; Kong, L.; Xu, P.; Xia, P.; Zhang, G. Antibody-mediated porcine reproductive and respiratory syndrome virus infection downregulates the production of interferon-α and tumor necrosis factor-α in porcine alveolar macrophages via Fc gamma receptor I and III. Viruses 2020, 12, 187. [Google Scholar] [CrossRef] [Green Version]
- Nan, Y.; Wu, C.; Gu, G.; Sun, W.; Zhang, Y.J.; Zhou, E.M. Improved vaccine against PRRSV: Current progress and future perspective. Front. Microbiol. 2017, 8, 1635. [Google Scholar] [CrossRef]
- Ravetch, J.; Bolland, S. IgG Fc receptors. Annu. Rev. Immunol. 2001, 19, 275–290. [Google Scholar] [CrossRef] [PubMed]
- Bournazos, S.; Ravetch, J.V. Fcγ receptor function and the design of vaccination strategies. Immunity 2017, 47, 224–233. [Google Scholar] [CrossRef]
- Lu, J.; Sun, P.D. Structural mechanism of high affinity FcγRI recognition of immunoglobulin G. Immunol. Rev. 2015, 268, 192–200. [Google Scholar] [CrossRef]
- Nimmerjahn, F.; Bruhns, P.; Horiuchi, K.; Ravetch, J.V. FcgammaRIV: A novel FcR with distinct IgG subclass specificity. Immunity 2005, 23, 41–51. [Google Scholar] [CrossRef]
- Gu, W.; Guo, L.; Yu, H.; Niu, J.; Huang, M.; Luo, X.; Li, R.; Tian, Z.; Feng, L.; Wang, Y. Involvement of CD16 in antibody-dependent enhancement of porcine reproductive and respiratory syndrome virus infection. J. Gen. Virol. 2015, 96, 1712–1722. [Google Scholar] [CrossRef]
- Maemura, T.; Kuroda, M.; Armbrust, T.; Yamayoshi, S.; Halfmann, P.J.; Kawaoka, Y. Antibody-dependent enhancement of SARS-CoV-2 infection is mediated by the IgG receptors FcγRIIA and FcγRIIIA but does not contribute to aberrant cytokine production by macrophages. MBio 2021, 12, e0198721. [Google Scholar] [CrossRef]
- Pincetic, A.; Bournazos, S.; DiLillo, D.J.; Maamary, J.; Wang, T.T.; Dahan, R.; Fiebiger, B.M.; Ravetch, J.V. Type I and type II Fc receptors regulate innate and adaptive immunity. Nat. Immunol. 2014, 15, 707–716. [Google Scholar] [CrossRef]
- Halloran, P.J.; Sweeney, S.E.; Strohmeier, C.M.; Kim, Y.B. Molecular cloning and identification of the porcine cytolytic trigger molecule G7 as a Fc gamma RIII alpha (CD16) homologue. J. Immunol. 1994, 158, 400–413. [Google Scholar]
- Zhang, G.; Qiao, S.; Li, Q.; Wang, X.; Duan, Y.; Wang, L.; Xiao, Z.; Xia, C. Molecular cloning and expression of the porcine high-affinity immunoglobulin G Fc receptor (FcgammaRI). Immunogenetics 2006, 8, 845–849. [Google Scholar] [CrossRef]
- Crocker, P.R.; Mucklow, S.; Bouckson, V.; McWilliam, A.; Willis, A.C.; Gordon, S.; Milon, G.; Kelm, S.; Bradfield, P. Sialoadhesin, a macrophage sialic acid binding receptor for haemopoietic cells with 17 immunoglobulin-like domains. EMBO J. 1994, 13, 4490–4503. [Google Scholar] [CrossRef]
- Hartnell, A.; Steel, J.; Turley, H.; Jones, M.; Jackson, D.G.; Crocker, P.R. Characterization of human sialoadhesin, a sialic acid binding receptor expressed by resident and inflammatory macrophage populations. Blood 2001, 97, 288–296. [Google Scholar] [CrossRef]
- Asano, K.; Nabeyama, A.; Miyake, Y.; Qiu, C.H.; Kurita, A.; Tomura, M.; Kanagawa, O.; Fujii, S.; Tanaka, M. CD169-positive macrophages dominate antitumor immunity by crosspresenting dead cell-associated antigens. Immunity 2011, 34, 85–95. [Google Scholar] [CrossRef]
- Kawasaki, N.; Vela, J.L.; Nycholat, C.M.; Rademacher, C.; Khurana, A.; van Rooijen, N.; Crocker, P.R.; Kronenberg, M.; Paulson, J.C. Targeted delivery of lipid antigen to macrophages via the CD169/sialoadhesin endocytic pathway induces robust invariant natural killer T cell activation. Proc. Natl. Acad. Sci. USA 2013, 110, 7826–7831. [Google Scholar] [CrossRef]
- Akiyama, H.; Ramirez, N.G.; Gudheti, M.V.; Gummuluru, S. CD169-mediated trafficking of HIV to plasma membrane invaginations in dendritic cells attenuates efficacy of anti-gp120 broadly neutralizing antibodies. PLoS Pathog. 2015, 11, e1004751. [Google Scholar] [CrossRef]
- Vanderheijden, N.; Delputte, P.L.; Favoree, H.W.; Vandekerckhove, J.; Van Damme, J.; van Woense, P.A.; Nauwynck, H.J. Involvement of sialoadhesin in entry of porcine reproductive and respiratory syndrome virus into porcine alveolar macrophags. J. Virol. 2003, 77, 8207–8215. [Google Scholar] [CrossRef]
- Zhang, L.; Xu, R.; Wei, F.; Li, W.; Li, X.; Zhang, G.; Xia, P. Activation of sialoadhesin down-regulates the levels of innate antiviral cytokines in porcine alveolar macrophages in vitro. Virus Res. 2020, 275, 197792. [Google Scholar] [CrossRef]
- Martínez, V.G.; Moestrup, S.K.; Holmskov, U.; Mollenhauer, J.; Lozano, F. The conserved scavenger receptor cysteine-rich superfamily in therapy and diagnosis. Pharmacol. Rev. 2011, 63, 967–1000. [Google Scholar] [CrossRef]
- Welch, S.K.; Calvert, J.G. A brief review of CD163 and its role in PRRSV infection. Virus Res. 2010, 154, 98–103. [Google Scholar] [CrossRef]
- Fujiwara, Y.; Ohnishi, K.; Horlad, H.; Saito, Y.; Shiraishi, D.; Takeya, H.; Yoshii, D.; Kaieda, S.; Hoshino, T.; Komohara, Y. CD163 deficiency facilitates lipopolysaccharide-induced inflammatory responses and endotoxin shock in mice. Clin. Transl. Immunol. 2020, 9, e1162. [Google Scholar] [CrossRef]
- Philippidis, P.; Mason, J.C.; Evans, B.J.; Nadra, I.; Taylor, K.M.; Haskard, D.O.; Landis, R.C. Hemoglobin scavenger receptor CD163 mediates interleukin-10 release and heme oxygenase-1 synthesis: Antiinflammatory monocyte-macrophage responses in vitro, in resolving skin blisters in vivo, and after cardiopulmonary bypass surgery. Circ. Res. 2004, 94, 119–126. [Google Scholar] [CrossRef] [Green Version]
- Calvert, J.G.; Slade, D.E.; Shields, S.L.; Jolie, R.; Mannan, R.M.; Ankenbauer, R.G.; Welch, S.K. CD163 expression confers susceptibility to porcine reproductive and respiratory syndrome viruses. J. Virol. 2007, 81, 7371–7379. [Google Scholar] [CrossRef]
- Burkard, C.; Opriessnig, T.; Mileham, A.J.; Stadejek, T.; Ait-Ali, T.; Lillico, S.G.; Whitelaw, C.B.A.; Archibald, A.L. Pigs lacking the scavenger receptor cysteine-rich domain 5 of CD163 are resistant to porcine reproductive and respiratory syndrome virus 1 infection. J. Virol. 2018, 92, e00415-18. [Google Scholar] [CrossRef]
- Zhang, Y.; Zhou, Y.; Yang, Q.; Mu, C.; Duan, E.; Chen, J.; Yang, M.; Xia, P.; Cui, B. Ligation of Fc gamma receptor IIB enhances levels of antiviral cytokine in response to PRRSV infection in vitro. Vet. Microbiol. 2012, 160, 473–480. [Google Scholar] [CrossRef]
- Kronimus, Y.; Dodel, R.; Galuska, S.P.; Neumann, S. IgG Fc N-glycosylation: Alterations in neurologic diseases and potential therapeutic target? J. Autoimmun. 2019, 96, 14–23. [Google Scholar] [CrossRef]
- Jefferis, R.; Lund, J.; Pound, J.D. IgG-Fc-mediated effector functions: Molecular definition of interaction sites for effector ligands and the role of glycosylation. Immunol. Rev. 1998, 163, 59–76. [Google Scholar] [CrossRef]
- Taylor, A.; Foo, S.S.; Bruzzone, R.; Dinh, L.V.; King, N.J.; Mahalingam, S. Fc receptors in antibody-dependent enhancement of viral infections. Immunol. Rev. 2015, 268, 340–364. [Google Scholar] [CrossRef]
- Lee, W.S.; Wheatley, A.K.; Kent, S.J.; DeKosky, B.J. Antibody-dependent enhancement and SARS-CoV-2 vaccines and therapies. Nat. Microbiol. 2020, 5, 1185–1191. [Google Scholar] [CrossRef]
- Shukla, R.; Ramasamy, V.; Shanmugam, R.K.; Ahuja, R.; Khanna, N. Antibody-dependent enhancement: A challenge for developing a safe dengue vaccine. Front. Cell. Infect. Microbiol. 2020, 10, 572681. [Google Scholar] [CrossRef]
- Dejnirattisai, W.; Jumnainsong, A.; Onsirisakul, N.; Fitton, P.; Vasanawathana, S.; Limpitikul, W.; Puttikhunt, C.; Edwards, C.; Duangchinda, T.; Supasa, S.; et al. Cross-reacting antibodies enhance dengue virus infection in humans. Science 2010, 328, 745–748. [Google Scholar] [CrossRef]
- Takada, A.; Kawaoka, Y. Antibody-dependent enhancement of viral infection: Molecular mechanisms and in vivo implications. Rev. Med. Virol. 2003, 13, 387–398. [Google Scholar] [CrossRef] [PubMed]
- Shi, P.; Su, Y.; Li, Y.; Zhang, L.; Lu, D.; Li, R.; Zhang, L.; Huang, J. The alternatively spliced porcine FcγRI regulated PRRSV-ADE infection and proinflammatory cytokine production. Dev. Comp. Immunol. 2019, 90, 186–198. [Google Scholar] [CrossRef] [PubMed]
- Takeda, A.; Sweet, R.W.; Ennis, F.A. Two receptors are required for antibody-dependent enhancement of human immunodeficiency virus type 1 infection: CD4 and Fc gamma R. J. Virol. 1990, 64, 5605–5610. [Google Scholar] [CrossRef]
- Wang, Z.; Deng, T.; Zhang, Y.; Niu, W.; Nie, Q.; Yang, S.; Liu, P.; Pei, P.; Chen, L.; Li, H.; et al. ACE2 can act as the secondary receptor in the FcγR-dependent ADE of SARS-CoV-2 infection. iScience 2022, 25, 103720. [Google Scholar] [CrossRef] [PubMed]
- Van Breedam, W.; Van Gorp, H.; Zhang, J.Q.; Crocker, P.R.; Delputte, P.L.; Nauwynck, H.J. The M/GP5 glycoprotein complex of porcine reproductive and respiratory syndrome virus binds the sialoadhesin receptor in a sialic acid-dependent manner. PLoS Pathog. 2010, 6, e1000730. [Google Scholar] [CrossRef]
- An, T.Q.; Tian, Z.J.; He, Y.X.; Xiao, Y.; Jiang, Y.F.; Peng, J.M.; Zhou, Y.J.; Liu, D.; Tong, G.Z. Porcine reproductive and respiratory syndrome virus attachment is mediated by the N terminal domain of the sialoadhesin receptor. Vet. Microbiol. 2010, 143, 371–378. [Google Scholar] [CrossRef]
- Delputte, P.L.; Van Breedam, W.; Delrue, I.; Oetke, C.; Crocker, P.R.; Nauwynck, H.J. Porcine arterivirus attachment to the macrophage-specific receptor sialoadhesin is dependent on the sialic acid-binding activity of the N-terminal immunoglobulin domain of sialoadhesin. J. Virol. 2007, 81, 9546–9550. [Google Scholar] [CrossRef]
- Chen, Y.; Guo, R.; He, S.; Zhang, X.; Xia, X.; Sun, H. Additive inhibition of porcine reproductive and respiratory syndrome virus infection with the soluble sialoadhesin and CD163 receptors. Virus Res. 2014, 179, 85–92. [Google Scholar] [CrossRef]
- Zhang, Q.; Yoo, D. PRRS virus receptors and their role for pathogenesis. Vet. Microbiol. 2015, 177, 229–241. [Google Scholar] [CrossRef]
- Tirado, S.M.; Evans, R.B.; Yoon, K.J. Monoclonal antibody analysis of porcine reproductive and respiratory syndrome virus epitopes associated with antibody-dependent enhancement and neutralization of virus infection. Vet. Immunol. Immunopathol. 2004, 102, 249–262. [Google Scholar] [CrossRef]

| Number | Start Position | End Position |
|---|---|---|
| 1 | 26 | 136 |
| 2 | 145 | 235 |
| 3 | 253 | 312 |
| 4 | 337 | 397 |
| 5 | 418 | 509 |
| 6 | 522 | 582 |
| 7 | 609 | 707 |
| 8 | 714 | 792 |
| 9 | 807 | 882 |
| 10 | 906 | 966 |
| 11 | 995 | 1073 |
| 12 | 1091 | 1169 |
| 13 | 1184 | 1248 |
| 14 | 1272 | 1331 |
| 15 | 1358 | 1433 |
| 16 | 1457 | 1519 |
| 17 | 1562 | 1637 |
| Name | Start Position | End Position |
|---|---|---|
| SRCR1 | 51 | 151 |
| SRCR2 | 158 | 258 |
| SRCR3 | 265 | 365 |
| SRCR4 | 372 | 472 |
| SRCR5 | 477 | 577 |
| SRCR6 | 582 | 682 |
| SRCR7 | 718 | 818 |
| SRCR8 | 823 | 925 |
| SRCR9 | 928 | 1028 |
| Gene Name | Sequence (5′-3′) |
|---|---|
| Porcine FcγRI | Forward: GCCUUGAGGUGUCAUGGAUTT Reverse: AUCCAUGACACCUCAAGGCTT |
| Porcine FcγRIII | Forward: GUGGAGAAUACACGUGUAATT Reverse: UUACACGUGUAUUCUCCACTT |
| Negative control | Forward: UUCUCCGAACGUGUCACGUTT Reverse: ACGUGACACGUUCGGAGAATT |
| Name | Sequence (5′-3′) |
|---|---|
| Porcine FcγRI forward Porcine FcγRI reverse | TGAAACAAAGTTGCTCCCA GCTGCGCTTGATGACCT |
| Porcine FcγRIII forward Porcine FcγRIII reverse | CTGCTGCTTCTGGTTTCA CCATTCCACCTCCACTC |
| β-actin forward β-actin reverse | CGGGACATCAAGGAGAAGC CTCGTTGCCGATGGTGATG |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhang, L.; Wang, H.; Li, W.; Feng, X.; Han, F.; Zhang, Y.; Chen, J.; Liu, D.; Xia, P. Activating Fc Gamma Receptors and Viral Receptors Are Required for Antibody-Dependent Enhancement of Porcine Reproductive and Respiratory Syndrome Virus Infection. Vet. Sci. 2022, 9, 470. https://doi.org/10.3390/vetsci9090470
Zhang L, Wang H, Li W, Feng X, Han F, Zhang Y, Chen J, Liu D, Xia P. Activating Fc Gamma Receptors and Viral Receptors Are Required for Antibody-Dependent Enhancement of Porcine Reproductive and Respiratory Syndrome Virus Infection. Veterinary Sciences. 2022; 9(9):470. https://doi.org/10.3390/vetsci9090470
Chicago/Turabian StyleZhang, Liujun, Huandi Wang, Wen Li, Xing Feng, Fangfang Han, Yina Zhang, Jing Chen, Deyi Liu, and Pingan Xia. 2022. "Activating Fc Gamma Receptors and Viral Receptors Are Required for Antibody-Dependent Enhancement of Porcine Reproductive and Respiratory Syndrome Virus Infection" Veterinary Sciences 9, no. 9: 470. https://doi.org/10.3390/vetsci9090470
APA StyleZhang, L., Wang, H., Li, W., Feng, X., Han, F., Zhang, Y., Chen, J., Liu, D., & Xia, P. (2022). Activating Fc Gamma Receptors and Viral Receptors Are Required for Antibody-Dependent Enhancement of Porcine Reproductive and Respiratory Syndrome Virus Infection. Veterinary Sciences, 9(9), 470. https://doi.org/10.3390/vetsci9090470






