Abstract
Karnal bunt of wheat is an internationally quarantined disease affecting trade, quality, and production of wheat. During 2015–2016, a severe outbreak of Karnal bunt disease occurred in north-western plain zone of India. The present study was undertaken to decipher genetic variations in Indian isolates of Tilletia indica collected from different locations. Seven multilocus sequence fragments were selected to differentiate and characterize these T. indica isolates. A phylogenetic tree constructed based on pooled sequences of actin-related protein 2 (ARP2), β-tubulin (TUB), eukaryotic translation initiation factor 3 subunit A (EIF3A), glyceraldehyde-3-phosphate dehydrogenase (GAPDH), histone 2B (H2B), phosphoglycerate kinase (PGK), and serine/threonine-protein kinase (STPK) showed that isolate KB-11 (Kaithal, Haryana) was highly conserved as it was located in cluster 1 and has the maximum sequence similarity with the reference strain. Other isolates in cluster 1 included KB-16 and KB-17, both from Uttar Pradesh, and KB-19 from Haryana. Isolates KB-07 (Jind, Haryana) and KB-18 (Mujaffar Nagar, Uttar Pradesh) were the most diverse and grouped in a subgroup of cluster 2. Maximum numbers of single nucleotide polymorphisms (SNPs) (675) were in the PGK gene across the T. indica isolates. The minimum numbers of SNPs (67) were in KB-11 (Kaithal, Haryana), while the maximum number of SNPs (165) was identified in KB-18, followed by 164 SNPs in KB-14. KB-18 isolate was found to be the most diverse amongst all T. indica isolates. This first study on multilocus sequence typing (MLST) revealed that the population of T. indica was highly diverse.
1. Introduction
Tilletia indica Mitra is a floret infecting fungal pathogen which causes Karnal bunt disease of wheat. It is a re-emerging disease in India. In 2014–2015, in the north-western plain zone of India, it occurred in epidemic form when up to 15% of disease incidence was reported [1]. The disease was first reported in Karnal district, Haryana, India [2]. Until now, several countries have had restrictions on importing wheat from countries where the disease has been reported [3]. Strict quarantine measures are imposed on wheat-importing countries that have zero tolerance limits [4]. The disease affects the quality and quantity of wheat grain. The fungus has a unique pathogenesis mechanism: it infects the host plants after dikaryotization between compatible mating-type secondary sporidia, colonizing only the endosperm [5,6], and infects the ear in a partial systemic manner [7]. Teliospores survive in the soil for many years in a dormant state, which is the main source of primary inoculum for the disease [8]. These teliospores are liberated during harvesting/threshing to the soil surface, and the disease cycle begins again. The secondary sporidia have been shown to be very durable and can remain dormant and then regenerate very rapidly under favorable conditions [9]. Moreover, the erratic occurrence of Karnal bunt disease is climate-dependent and requires certain conditions, further making it a complex phytopathogen. Seed- or soil-borne teliospores and their successive germination seem to play only a starting role in Karnal bunt epidemics [10].
The disease is difficult to manage effectively using cultural and fungicidal approaches because of its unique infection behavior. The best approach to manage the disease is through the use of resistant cultivars. Phenotyping the wheat genotypes using different pathogenic isolates of T. indica does not give a uniform disease reaction pattern as the pathogen is heterothallic, creating much variability. Efforts have been made by previous workers to study pathogenic and genetic diversity based on randomly amplified polymorphic DNA (RAPD)/Universal rice primers (URP)/ inter simple sequence repeats (ISSR) markers and ITS sequences [6,11,12], but genetic variability through multilocus sequence typing (MLST) has not been done. The MLST tool using partial sequence analysis of seven to ten housekeeping genes is the most popular typing approach for epidemiological investigations of phytopathogenic fungi and other microorganisms [13,14]. Among very few studies using MLST in fungal genotyping, multilocus molecular identification of Colletotrichum tamarilloi, Damm, P.F. Cannon and Crous revealed mutant isolates and a possible detection of a new pathogenic species affecting tamarillo fruits [15]. In this approach, for each locus studied, different genetic sequences present within a species are assigned as distinct alleles. The combination of the identified alleles at each of the loci defines the allelic profile or sequence type for each isolate. The sequence generated can be used to determine whether the fungal populations are clonal or have undergone recombination patterns. This sequence-based approach produces unambiguous, reproducible results and can be used to compare different isolates of a pathogen. Keeping in view the unique infection biology of the pathogen, and its recent severe outbreak, the study aimed to identify intraspecific variations among twenty T. indica isolates using multilocus sequence typing and single nucleotide polymorphisms (SNPs) approaches.
2. Materials and Methods
2.1. Collection, Isolation and Maintenance of T. indica Isolates
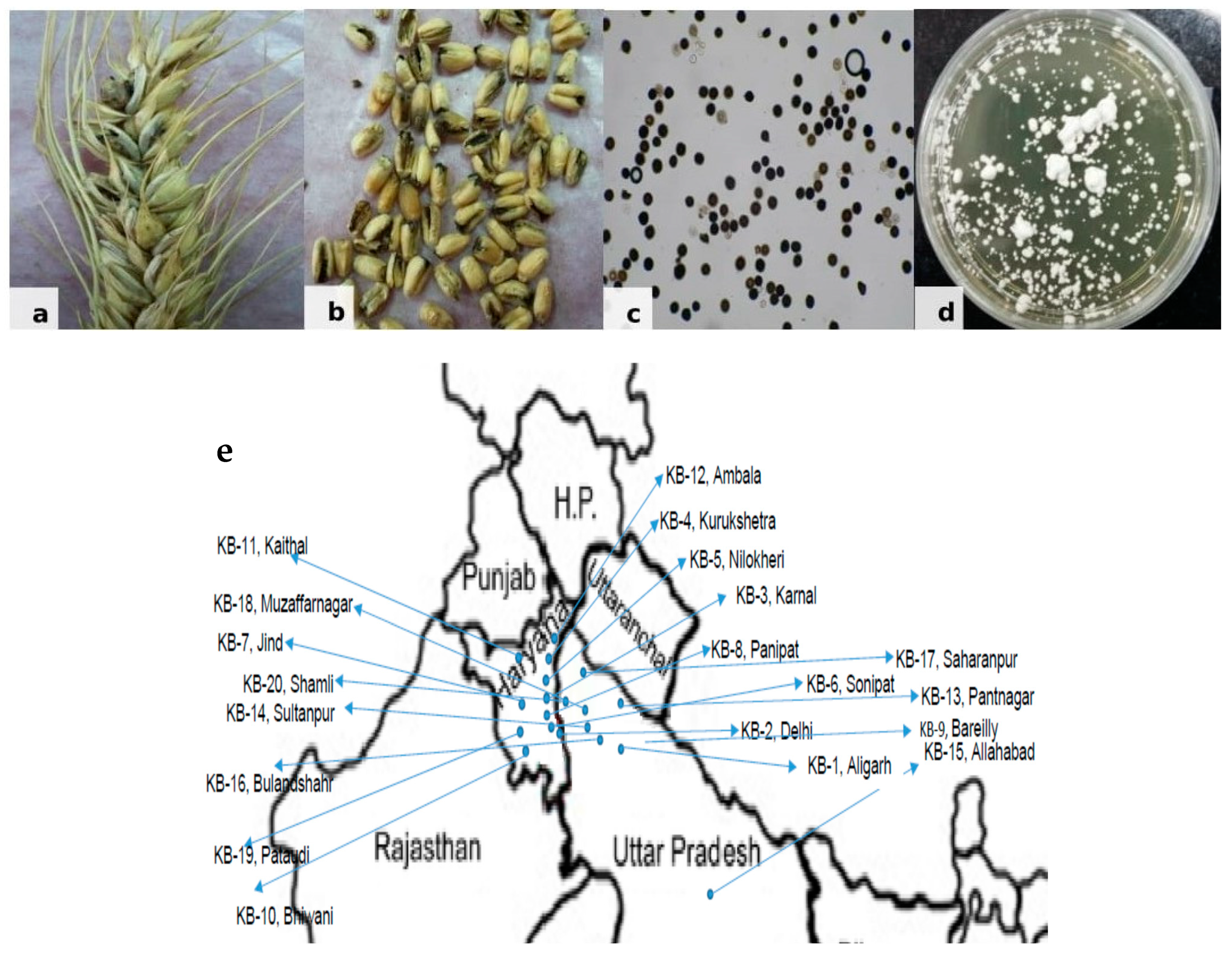
Tilletia indica samples were collected from different regions of the north-western plain zone of India from farmers’ field during 2014–2015 where recommended varieties, PBW 343, WH1105 and HD2967, are grown by the farmers. Infected wheat grain samples (Figure 1) brought to the Fungal Molecular Biology Laboratory, Division of Plant Pathology, ICAR-IARI, Pusa Campus, New Delhi, India, were surface sterilized using 70% ethanol. Cultures of T. indica isolates were raised from teliospores using the earlier standardized technique by [16]. For the separation of teliospores, bunted grains were vortexed in a sterile, capped vial containing 10 mL sterile distilled water. Each teliospore suspension was filtered through a 45-µm mesh sieve to remove grain chaff, and the filtrate was collected in 15-mL sterile, screw-capped centrifuge tube. To pellet down teliospores, tubes were centrifuged at 10,000 rpm for 2 min, and the supernatant was discarded. The teliospore pellet was re-suspended in 7 mL of 4% sodium hypochlorite and incubated for 2 min for sterilization. After incubation, tubes were centrifuged, the supernatant discarded, and the pellet was washed in a similar way twice using sterile distilled water. Finally, the pellet was re-suspended in 7 mL sterile distilled water and kept overnight in the refrigerator. About 0.5 mL of teliospore suspension was poured on water agar plates (1.5%) and incubated at 16 ± 2 °C in an incubator with exposure to alternate light and dark periods of 12 h. Plates were observed microscopically on a regular basis for the germination of teliospores. A small disc of water agar bearing a single germinating teliospore was transferred to potato-dextrose agar (PDA) test tube slants and incubated at 16 ± 2 °C with alternate light and dark periods. After 3–4 days, a creamy white powdery growth of fungus appeared (Figure 1) showering sporidia downward from the disc, which covered the entire slant within a few days. These T. indica isolates were further maintained at 16 ± 2 °C in the incubator by regular sub-culturing.

Figure 1.
Karnal Bunt disease. (a) Infected wheat spike. (b) Infected wheat grains. (c) Mass of teliospores. (d) Mycelial culture of Tilletia indica. (e) Map showing collected KB samples from north western plain zone of India.
2.2. DNA Isolation
Fungal DNA was isolated from 10–15-day-old mycelium of T. indica isolates which were grown on a shaker incubator at 16 ± 2 °C in potato dextrose broth (PDB) using the CTAB (cetyl trimethylammonium bromide) method [17]. Fungal mycelium was collected by filtering from the broth through autoclaved Whatman paper in aseptic conditions and used immediately for isolation of DNA. Approximately 5 g of mycelium was ground in liquid nitrogen in a mortar and transferred to a sterile 30-mL polypropylene tube. In total, 10 mL preheated CTAB extraction buffer (100 mM Tris pH 8.0, 5 M NaCl, 0.5 EDTA (Ethylenediamine tetraacetic acid), pH 8.0, 2% CTAB, 0.1% mercaptoethenol) was added to the ground mycelium powder, mixed gently and incubated at 65 °C for 1 h. After incubation, an equal volume of chloroform–isoamyl alcohol solution (24:1) was added, mixed gently and centrifuged at 10,000 rpm for 10 min. The upper aqueous phase was transferred to a new, pre-sterilized 30-mL polypropylene tube. DNA was precipitated with an equal volume of pre-chilled isopropanol for 2 h at −20 °C. After incubation, DNA was pelleted down by centrifuging at 10,000 rpm for 20 min at 4 °C. DNA pellets were washed twice with 75 % ethanol, air-dried and dissolved in TE buffer (10 mM Tris HCl (pH 8.0) and 1 mM EDTA (pH 8.0)). The concentration of DNA was measured on a nanodrop spectrophotometer (Nanodrop 2000, Thermo Scientific). Qualitative assessment of the DNA was performed by running on 0.8% agarose gel stained with ethidium bromide (0.5 µg/L) in TAE (Tris-acetate-EDTA) buffer (40 Mm Tris-acetate, 1 mM EDTA (pH 8.0)) along with the 1 Kb DNA ladder (MBI, Fermentas). Electrophoresis was carried out in 1× TAE buffer at constant voltage (75 V) for 1 h and DNA bands were observed using a gel documentation system (UVITECH, UK, New Fire Reader).
2.3. Selection of MLST Loci and Amplification
For the multilocus sequence typing (MLST), the seven genes selected after BLAST analysis with T. indica were: actin-related protein 2 (ARP2; AOA177TN81), β-tubulin (TUB; A0A177TPV8), eukaryotic translation initiation factor 3 subunit A (EIF3A; A0A177TP83), glyceraldehyde-3-phosphate dehydrogenase (GAPDH; A0A177TT56), histone 2 B (H2B; A0A177TGR2), phosphoglycerate kinase (PGK; A0A177TT19) and serine/threonine-protein kinase (STPK; A0A177TLFO). These genes are highly conserved in fungi. Housekeeping genes are typically constitutive genes that are required for the maintenance of basal cellular functions that are essential for the existence of a cell, regardless of its specific role in the tissue or organism. The primers (Table 1) were designed using IDT Oligo Analyzer from sequences taken from NCBI GenBank (http://www.ncbi.nlm.nih.gov/). The designed primers were synthesized by GCC Biotech Pvt. India. The PCR amplifications were carried out in 25µL reaction volume, consisting of 200 ng of genomic DNA, 200 µmol/L dNTP mix (dATP, dGTP, dCTP, dTTP), 0.1 µmol/L each primer, 3.5 mmol/L MgCl2, 1.5 U Taq DNA polymerase, 9.5 µL water, and 1× Taq buffer in a thermal cycler (Bio-Rad Laboratories India Pvt Ltd.). The PCR conditions used were: 95 °C for 5 min—initial denaturation; 95 °C for 1 min—denaturation for 35 cycles at 55–59 °C for 30 s—annealing (gradient); 72 °C for 1 min—extension; and 72 °C for 7 min—final extension with three replications.

Table 1.
List of genes and primers used for multilocus sequence typing in Tilletia indica.
The PCR products were separated on 1.2% agarose gel stained with ethidium bromide (0.5 µg/L) in TAE buffer (pH 8.0) along with the 1 Kb DNA ladder (MBI, Fermentas). Electrophoresis was carried out in 1× TAE buffer at constant voltage (75 V) for 1 h. Amplified PCR products were visualized using the gel documentation system.
2.4. Sequencing and Phylogenetic Analysis
The amplified PCR products of each gene, viz. actin-related protein 2 (ARP2), β-tubulin (TUB), eukaryotic translation initiation factor 3 subunit A (EIF3A), glyceraldehyde-3-phosphate dehydrogenase (GAPDH), histone 2 B (H2B), phosphoglycerate kinase (PGK), and serine/threonine-protein kinase (STPK) were sequenced by Eurofins Pvt. Ltd. Bangaluru, India. Sequences were trimmed for phylogenetic analysis. These sequences were used to determine a final panel of seven gene fragments that gave the highest discrimination for MLST. The evolutionary history was inferred by using the maximum likelihood method based on the Kimura 2-parameter model. The bootstrap consensus tree inferred from 1000 replicates was taken to represent the evolutionary history of the isolates analyzed. Branches corresponding to partitions reproduced in less than 50% of bootstrap replicates are collapsed. The percentage of replicate trees in which the associated isolates clustered together in the bootstrap test (1000 replicates) is shown next to the branches. Initial tree(s) for the heuristic search were obtained automatically by applying Neighbor-Join and BioNJ algorithms to a matrix of pairwise distances, estimated using the maximum composite likelihood (MCL) approach, and then selecting the topology with superior log-likelihood value. For phylogenetic analysis, 20 amplified nucleotide gene sequences, including the reference sequence (pooled selected gene sequences), were taken. Evolutionary analysis was conducted in CLC Genomics Workbench 9.0 (http://www.clcbio.com) and a phylogenetic tree was constructed [18].
2.5. Single Nucleotide Polymorphism Analysis
Single nucleotide polymorphism analysis was conducted with the reference genome (LWDF00000000.1) containing seven gene sequences, namely ENA|OAJ04384|OAJ04384.1 T. indica actin-related protein 2, ENA|OAJ05357|OAJ05357.1 T. indica tubulin, A|OAJ00852|OAJ00852.1 T. indica hypothetical protein, ENA|OAJ06275|OAJ06275.1 T. indica glyceraldehyde-3-phosphate dehydrogenase, ENA|OAJ03744|OAJ03744.1 T. indica hypothetical protein, ENA|OAJ06449|OAJ06449.1 T. indica phosphoglycerate kinase and ENA|OAJ01891 |OAJ01891.1 T. indica hypothetical protein. Single nucleotide polymorphisms (SNPs) were identified using the Single Nucleotide Variant tool in CLC Genomics Workbench 9.0.
3. Results
3.1. PCR Amplification and BLAST Search
PCR amplification using template DNA from each isolate showed a sharp clear band of the size with respect to each specific gene, viz. actin-related protein 2, ARP2 (1200 bp); β-tubulin, TUB (1100 bp); eukaryotic translation initiation factor 3 subunit A, EIF3A (1500 bp); glyceraldehyde-3-phosphate dehydrogenase, GAPDH (800 bp); histone 2 B, H2B (300 bp); phosphoglycerate kinase, PGK (1000 bp); and serine/threonine-protein kinase, STPK (1200 bp). Upon homology analysis using the BLAST tool, amplified sequences showed maximum similarity with the respective reference gene in the whole genome sequence of T. indica DAOM236416 (NCBI database). After confirmation by BLAST homology, sequences were submitted to GenBank and accession numbers were obtained (Table 2).

Table 2.
Tilletia indica isolates GenBank accession numbers of generated sequences of different genes.
3.2. Multilocus Phylogenetic Analysis
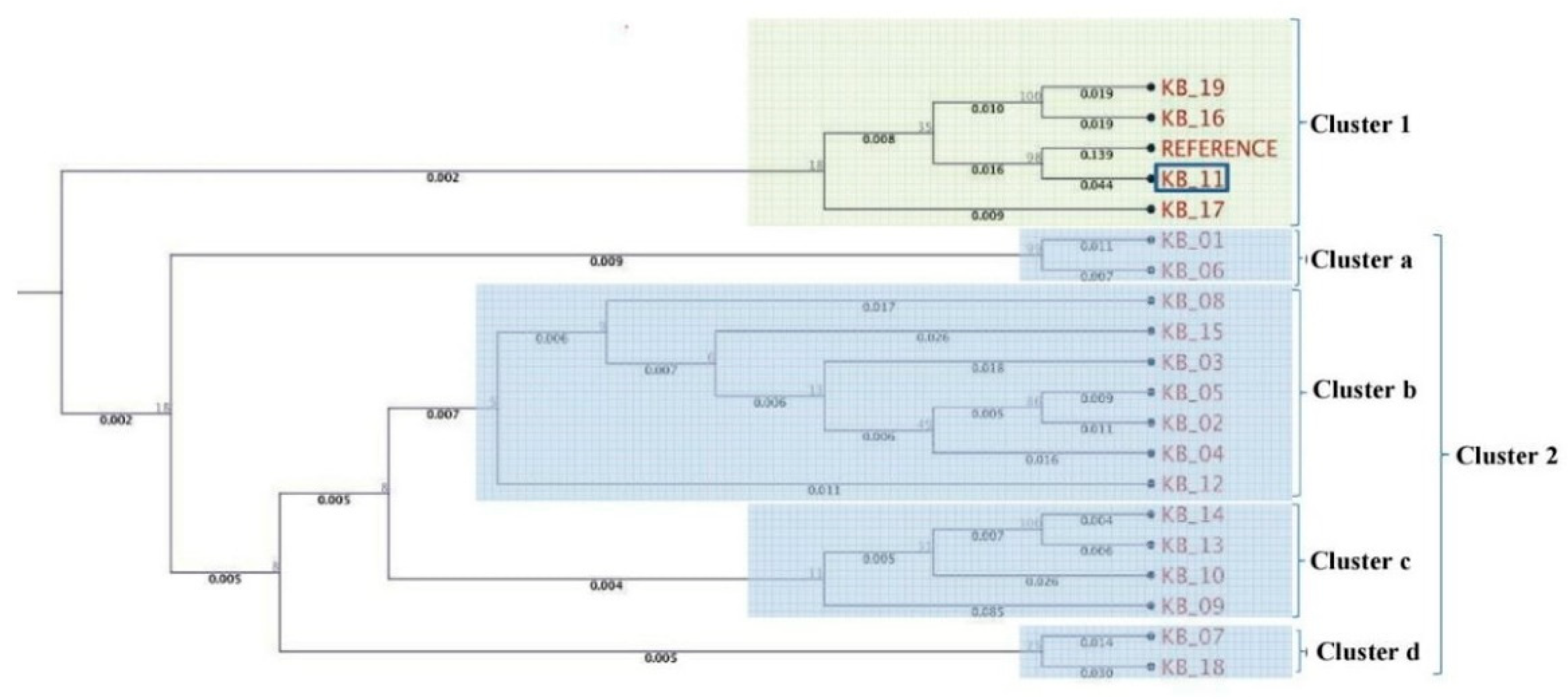
A phylogenetic tree was constructed based on the pooled amplified gene sequences, namely ARP2, TUB EIF3A, GAPDH, H2B, PGK and STPK, and with the reference gene sequence. The phylogenetic tree grouped the isolates into two major clusters (Figure 2). The reference sequence was positioned in cluster 1. Isolate KB-11 (Kaithal, Haryana) showed the highest similarity to the reference sequence and is therefore located in cluster 1. Among cluster 2, only two isolates, viz. KB-01(Aligarh, Uttar Pradesh) and KB-06 (Sonipat, Haryana), were grouped in sub cluster a. KB-12 (Ambala, Haryana) is an outlier of sub-cluster b, which exhibited genetic diversity from other isolates (KB-08, KB-15, KB-03, KB-05, KB-02, KB-04) present in the same cluster. KB-09 (Bareilly, Uttar Pradesh) was in the outlier of sub-cluster c, indicating that it shares very limited similarity with KB-10, KB-13 and KB-14 isolates. Isolates KB-07 (Jind, Haryana) and KB-18 (Mujaffar Nagar, Uttar Pradesh) were the most diverse and were grouped in sub-cluster d. In further pooled-gene phylogenetic analysis, T. indica isolates clustered with Tilletia walkeri Cast l. and Carris but not with Tilletia caries (DC.) Tul. and C. Tul. and Ustilago maydis (DC.) Corda (Figure 3).
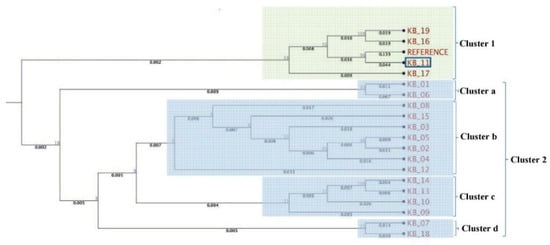
Figure 2.
Molecular phylogenetic analysis of T. indica isolates using seven gene sequences.

Figure 3.
Comparative phylogeny with Tilletia caries, Tilletia walkeri and Ustilago maydis.
3.3. Single Nucleotide Polymorphism (SNP) Analysis
A large number of SNPs were identified after comparison of T. indica DAOM, LWDF010001 sequences with the MLST locus sequences (Table 3). Phosphoglycerate kinase gene sequences showed the maximum numbers (675) of SNPs. This was followed by 640 SNPs in actin-related protein across T. indica isolates. According to SNPs in each gene, the maximum number of SNPs were identified in actin-related protein gene sequences of both KB-13 (Pant Nagar, Uttar Pradesh) and KB-14 (Sultanpur, Uttar Pradesh) (58 SNPs), while histone 2B (H2B) gene sequences were found most conserved among all the genes since either no SNP or very few SNPs (1–2) were observed in different T. indica isolates. Pooled SNP data analysis showed that minimum numbers of SNPs were identified in KB-11 isolate (Kaithal, Haryana), which had 67 SNPs, whereas the highest number of SNPs was found in KB-18 (Mujaffar Nagar, Uttar Pradesh), which had 165 SNPs. Isolate KB-18 was found to be the most diverse among all the T. indica isolates studied.

Table 3.
Single nucleotide polymorphism (SNP) with respect to housekeeping genes in different isolates of Tilletia indica.
3.4. Discussion
India is the second largest producer of wheat in the world. Wheat production has reached 103.60 million tons in the country, and India can export surplus wheat to other countries, but Karnal bunt disease is a major limitation in wheat export, causing huge monetary loss in wheat trade. Considering the international quarantine policy of disease [19] and the unique pathogenesis mechanism of T. indica, efforts have been made to generate whole genome data of a virulent isolate (RAKB_UP_1) of T. indica (NCBI accession no: MBSW00000000) [20] (Gurjar et al. 2019). Sequence typing methods help in understanding the epidemiology, evolution of pathogens and disease outbreaks [21]. The present study supplemented new data on genotypic variation in T. indica isolates collected from the north-western plain zone of India with respect to multilocus sequence typing (MLST) and single nucleotide polymorphisms (SNPs).
MLST is a preferred method for the genotyping of strains of a particular species, and it is more reliable and informative. This method identifies major phylogenetic clades and molecular groups in sub-populations of a species. It has been utilized to identify variations among the isolates of pathogenic microbes, especially in evolution, pathogenesis and ecology [22,23]. In the present study, the phylogenetic analysis, based on seven gene sequences, showed that isolates of T. indica did not form clusters according to their geographical preferences. Haryana (KB-07) and Uttar Pradesh (KB18) isolates were most diverse amongst all T. indica isolates. According to their geographical preferences, there is a high genetic variation among the isolates of T. indica. The pathogenic and genetic variability studies in T. indica done earlier were based on RAPD and URP markers [6,24,25]. In T. indica–wheat interaction, the dikaryotization of the haploid allantoid sporidia takes place before infection on or within the host tissues. A comparison of nucleotide sequences of microbes and pathogens is the most unambiguous way to differentiate strains/isolates [26]. We, therefore, used a seven gene-based MLST scheme to genotype different isolates of T. indica. MLST was earlier successfully used to study epidemiology and population structures in many fungal species [27]. These genes taken for the study are all housekeeping genes, still showing high number of polymorphisms in sequences in this species, and were therefore considered useful for designing the MLST scheme.
Compared to the reference sequences, the highest single nucleotide polymorphism was recorded in the ARP gene (6–58 nucleotides/isolate, 58 nucleotides in KB-13 and KB-14) and PKG gene (0–42 nucleotides/isolate, 42 nucleotides in KB-13 and KB-14). The lowest single nucleotide polymorphism was recorded in the H2B gene (0–2 nucleotides/isolate). Pooled SNP data analysis showed that the maximum and minimum polymorphisms occurred in KB-18 and KB-11 isolates (165 and 67 nucleotides, respectively). Earlier studies reported that isolates of T. indica undergo sexual reproduction after teliospore germination when compatible mating-type secondary sporidia fuse to form diakaryon and thus increase chances of variation [28]. In earlier studies, the genetic and pathogenic nature of the fungus was found variable due to the recombination of secondary allantoid sporidia. Two types of secondary sporidia are produced like allantoid sporidia and filiform sporidia, of which only the allantoid type is thought to be able to infect and cause the disease. Allantoid secondary sporidia are ballistospores. T. indica, being a heterothallic fungus, demands fusion between secondary sporidia of opposite mating types, resulting in high variation [29]. Therefore, the MLST approach was found useful in understanding genetic variation in T. indica and, potentially, in epidemiological investigations. It is simpler, faster, and less expensive than whole-genome sequencing of fungal pathogens [30]. By analyzing SNPs, a diagnostic marker for differentiating isolates revealing intraspecific population diversity of T. indica can be developed. This is the first study to reveal information on single nucleotide polymorphism in T. indica isolates based on MLST loci.
4. Conclusions
MLST has great potential as a tool to interpret the population structures of pathogenic fungi which can be utilized in crop improvement programs. The present study revealed that the population of T. indica in the north-western plain zone of India was highly diverse. MLST was found to be useful in understanding genetic variations in T. indica. Large numbers of putative SNPs were also detected in T. indica isolates. This genetic information will be helpful in understanding the epidemiology and in devising management strategies for Karnal bunt disease.
Author Contributions
M.S.G. and R.A. conceived and designed the experiments; S.J. and M.S.S. strains and materials important for the experiments; S.J. and S.S. performed the experiments; M.S.G., J.S., S.A. and S.G. analysed the data and made the figures; M.S.G. & R.A. wrote the manuscript. All authors have read and agreed to the published version of the manuscript.
Funding
This work is financially supported by the Indian Council of Agricultural Research—Consortium Research Platform (CRP) on Genomics (ICAR-G/CRP-Genomics/2015-2720).
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
Data available in a publicly accessible repository. The data presented in this study are openly available in https://www.ncbi.nlm.nih.gov.
Acknowledgments
We are highly thankful to the Director, Joint Director (Research) and Head, Division of Plant Pathology, ICAR—Indian Agricultural Research Institute, New Delhi, India, for providing guidance and facilities for investigations.
Conflicts of Interest
The authors declare no conflict of interest.
Abbreviations
| ARP2 | Actin-related protein 2 |
| EIF3A | Eukaryotic translation initiation factor 3 subunit A |
| GAPDH | Glyceraldehyde-3-phosphate dehydrogenase |
| H2B | Histone 2B |
| KB | Karnal bunt |
| MLST | Multilocus sequence typing |
| PGK | Phosphoglycerate kinase |
| SNP | Single nucleotide polymorphism |
| STPK | serine/threonine-protein kinase |
| TUB | β-tubulin |
References
- Anonymous. IARI Annual Report (2015–2016); Indian Agricultural Research Institute: New Delhi, India, 2015; p. 83. Available online: www.iari.res.in (accessed on 3 January 2021).
- Mitra, M. A new bunt of wheat in India. Ann. Appl. Biol. 1931, 18, 178–179. [Google Scholar] [CrossRef]
- Bonde, M.R.; Nester, S.E.; Olsen, M.W.; Berner, D.K. Survival of teliospores of Tilletia indica in Arizona field soils. Plant Dis. 2004, 88, 804–810. [Google Scholar] [CrossRef]
- Singh, D.V.; Gogoi, R. Karnal bunt of wheat (Triticum sp.): A global scenario. Indian J. Agric. Sci. 2011, 81, 3–14. [Google Scholar]
- Aggarwal, R.; Singh, D.V.; Srivastava, K.D. Host pathogen interaction in Karnal bunt of wheat. Indian Phytopathol. 1994, 47, 381–386. [Google Scholar]
- Aggarwal, R.; Tripathi, A.; Yadav, A. Pathogenic and genetic variability in Tilletia indica monosporidial culture lines using universal rice primer-PCR. Eur. J. Plant Pathol. 2010, 128, 333–342. [Google Scholar] [CrossRef]
- Carris, L.M.; Castlebury, L.A.; Goates, B.J. Nonsystemic Bunt Fungi-Tilletia indica and T. horrida: A Review of History, Systematics, and Biology. Annu. Rev. Phytopathol. 2006, 44, 113–133. [Google Scholar] [CrossRef] [PubMed]
- Babadoost, M.; Mathre, D.E.; Johnston, R.H.; Bonde, M.R. Survival of teliospores of Tilletia indica in soil. Plant Dis. 2004, 88, 56–62. [Google Scholar] [CrossRef]
- Goates, B.J. Survival of secondary sporidia of floret infecting Tilletia species: Implications for epidemiology. Phytopathology 2010, 100, 655–662. [Google Scholar] [CrossRef]
- Dhaliwal, H.S. Multiplication of secondary sporidia of Tilletia indica on soil and wheat leaves and spikes and incidence of Karnal bunt. Can. J. Bot. 1989, 67, 2387–2390. [Google Scholar] [CrossRef]
- Thirumalaisamy, P.P.; Singh, D.V. Variability of Indian isolates of Tilletia indica assessed by pathogenecity and molecular marker. J. Phytopathol. 2012, 160, 525–531. [Google Scholar] [CrossRef]
- Shabana, P.; Saharan, M.S.; Verma, A.; Sharma, I. Comparative analysis of RAPD and ISSR marker assays for detecting genetic polymorphism in Tilletia indica. Eur. J. Exp. Biol. 2013, 3, 380–387. [Google Scholar]
- Taylor, J.W.; Fisher, M.C. Fungal multilocus sequence typing—It’s not just for bacteria. Curr. Opin. Microbiol. 2003, 6, 351–356. [Google Scholar] [CrossRef]
- Bain, J.M.; Tavanti, A.; Davidson, A.D.; Jacobsen, M.D.; Shaw, D.; Gow, N.A.R.; Odds, F.C. Multilocus Sequence Typing of the Pathogenic Fungus Aspergillus fumigates. J. Clin. Microbiol. 2007, 5, 1469–1477. [Google Scholar] [CrossRef] [PubMed]
- Caicedo, J.D.; Karina, P.; Lalangui, A.N.; Pozo, P.A.; Cevallos, V.; Arahana, S.; Mendez, K.S. Multilocus molecular identification and phylogenetic analysis of Colletotrichum tamarilloi as the causal agent of Tamarillo (Solanum betaceum) anthracnose in the Ecuadorian highlands. Eur. J. Plant Pathol. 2017, 148, 983–996. [Google Scholar] [CrossRef]
- Warham, E.J. Studies on Karnal Bunt of Wheat. Ph.D. Thesis, University of Wales, Aberystwyth, UK, 1987. [Google Scholar]
- Murray, M.G.; Thompson, W.F. Rapid isolation of high molecular weight plant DNA. Nucleic Acids Res. 1980, 8, 4321–4325. [Google Scholar] [CrossRef]
- Tamura, K.; Stecher, G.; Peterson, D.; Filipski, A.; Kumar, S. MEGA6: Molecular evolutionary genetics analysis version 6.0. Mol. Biol. Evol. 2013, 30, 2725–2729. [Google Scholar] [CrossRef]
- Rush, C.M.; Riemenschneider, R.; Stein, J.M.; Boratynski, T.; Bowden, R.L.; Matthew, H.R. Status of Karnal Bunt of Wheat in the Unites States 1996 to 2004. Plant Dis. 2005, 89, 212–223. [Google Scholar] [CrossRef]
- Gurjar, M.S.; Aggarwal, R.; Jogawat, A.; Kulshreshtha, D.; Sharma, S.; Solanke, A.U.; Dubey, H.; Jain, R.K. Genome Sequencing and Secretome analysis of Tilletia indica inciting Karnal bunt of wheat Provides Pathogenesis-related genes. 3 Biotech 2019, 9, 219. [Google Scholar] [CrossRef]
- Debourgogne, A.; Gueidan, C.; Henneguin, C.; Contet-Audonneau, N.; de Hoog, S.; Machouart, M. Development of a new MLST scheme for differentiation of Fusarium solani Species Complex (FSSC) isolates. J. Microbiol. Methods 2010, 82, 319–323. [Google Scholar] [CrossRef]
- Byrnes, E.J.; Bildfell, R.J.; Frank, S.A.; Mitchell, T.G.; Marr, K.A.; Heitman, J. Molecular evidence that the range of the Vancouver Island outbreak of Cryptococcus gattii infection has expanded into the Pacific Northwest in the United States. J. Infect. Dis. 2009, 199, 1081–1086. [Google Scholar] [CrossRef]
- Litvintseva, A.P.; Mitchell, T.G. Population Genetic Analyses Reveal the African origin and strain variation of Cryptococcus neoformans var. grubii. PLoS Pathog 2012, 8, e1002495. [Google Scholar] [CrossRef] [PubMed]
- Datta, R.; Rajebhosale, M.D.; Dhaliwal, H.S.; Singh, H.; Ranjekar, P.K.; Gupta, V.S. Intraspecific genetic variability analysis of Neovossia indica causing Karnal bunt of wheat using repetitive elements. Theor. Appl. Genet. 2000, 100, 569–575. [Google Scholar] [CrossRef]
- Tripathi, A.; Aggarwal, R.; Yadav, A. Determination of variability in monosporidial lines of Tilletia indica by RAPD analysis. Arch. Phytopathol. Plant Prot. 2011, 44, 1312–1321. [Google Scholar] [CrossRef]
- Urwin, R.; Maiden, M.C.J. Multi-locus sequence typing: A tool for global epidemiology. Trends Microbiol. 2003, 11, 479–487. [Google Scholar] [CrossRef]
- Meyer, W.; Aanensen, D.M.; Boekhout, T.; Cogliati, M.; Diaz, M.R.; Esposto, M.C.; Fisher, M.; Gilgado, F.; Hagen, F.; Kaocharoen, S.; et al. Consensus multi-locus sequence typing scheme for Cryptococcus neoformans and Cryptococcus gattii. Med. Mycol. 2009, 12, 1–14. [Google Scholar]
- Dhaliwal, H.S.; Singh, D.V. Up-to-date life cycle of Nevossia indica (Mitra) Mundkur. Curr. Sci. 1988, 57, 675–677. [Google Scholar]
- Duran, R.; Cromarty, R. Tilletia indica: A heterothallic wheat bunt 8 fungus with multiple alleles controlling incompatibility. Phytopathology 1977, 67, 812–815. [Google Scholar] [CrossRef]
- Chen, Y.; Frazzitta, A.E.; Litvintseva, A.P.; Fang, C.; Mitchell, T.G.; Springer, D.J.; Ding, Y.; Yuan, G.; Perfect, J.R. Next generation multilocus sequence typing (NGMLST) and the analytical software program MLSTEZ enable efficient, cost-effective, high-throughput, multilocus sequencing typing. Fungal Genet. Biol. 2015, 75, 64–71. [Google Scholar] [CrossRef]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).