Toolbox for Genetic Transformation of Non-Conventional Saccharomycotina Yeasts: High Efficiency Transformation of Yeasts Belonging to the Schwanniomyces Genus
Abstract
:1. Introduction
2. Materials and Methods
2.1. Growth Conditions
2.2. Yeast Strains, Plasmids and Oligonucleotides
2.3. Gene Optimisation, Synthesis and Testing
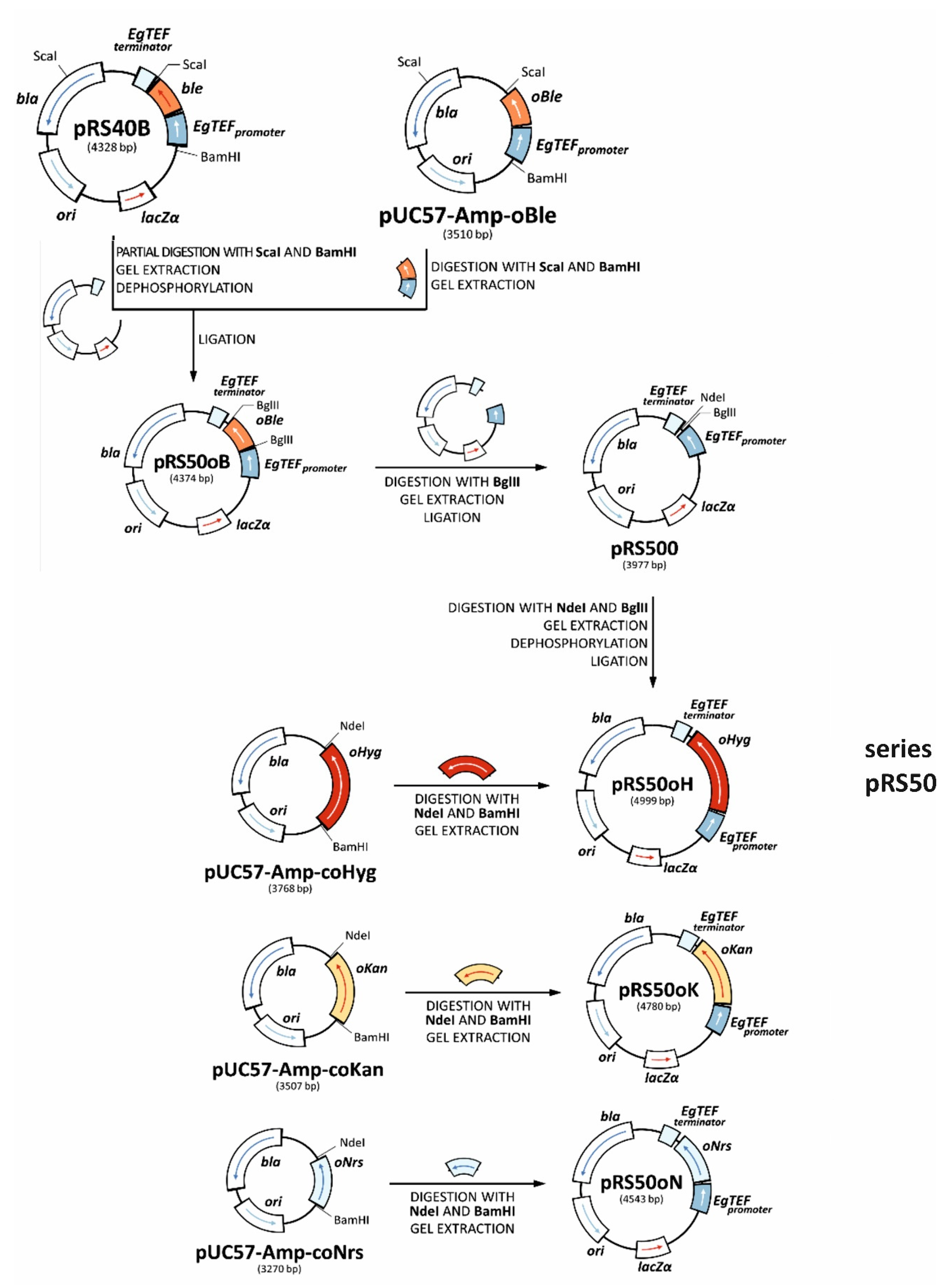
2.4. Plasmid Construction
2.4.1. Construction of Integrative Plasmids
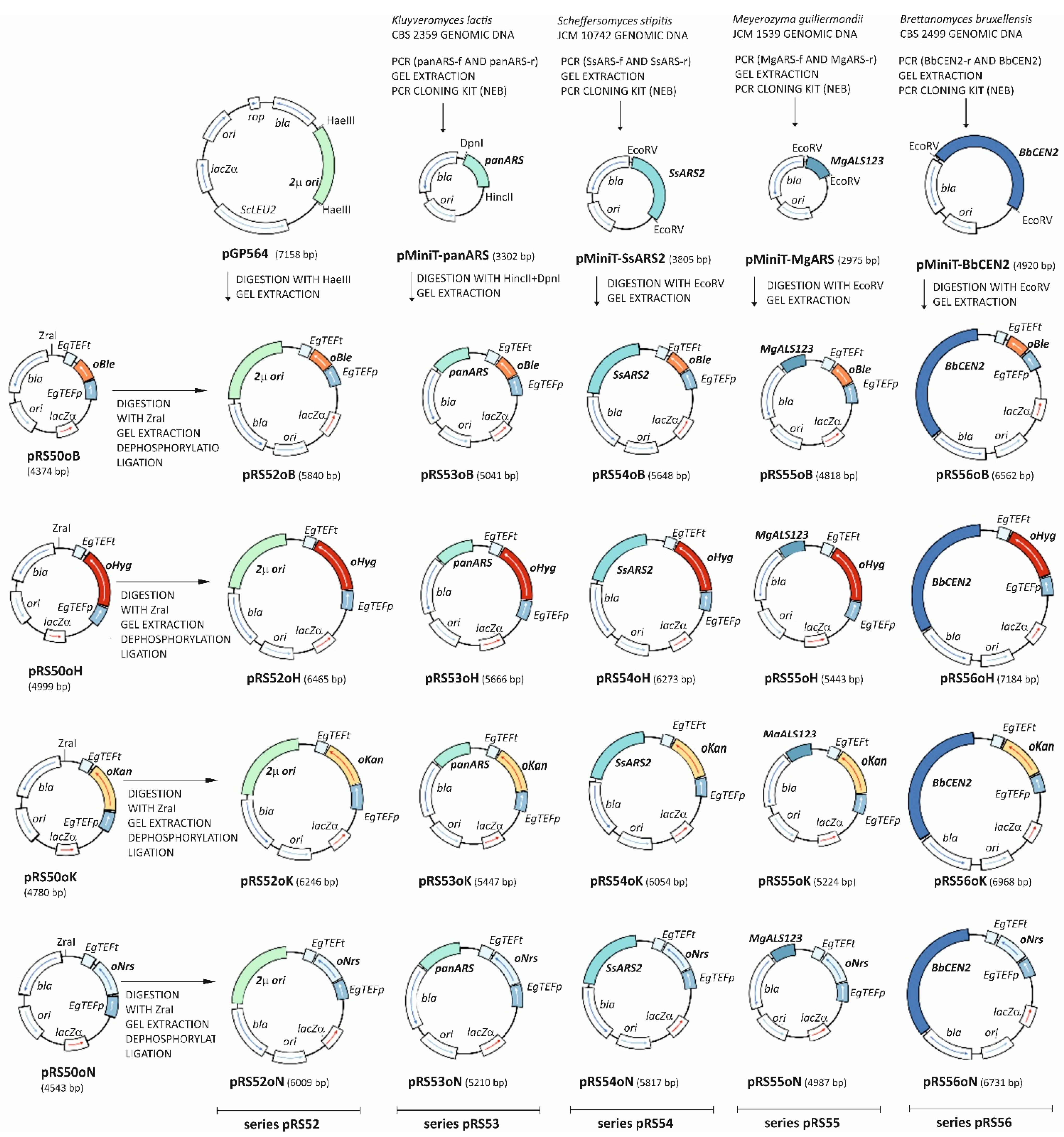
2.4.2. Construction of Replicative Plasmids
2.5. Determination of Yeast Antibiotic Resistance
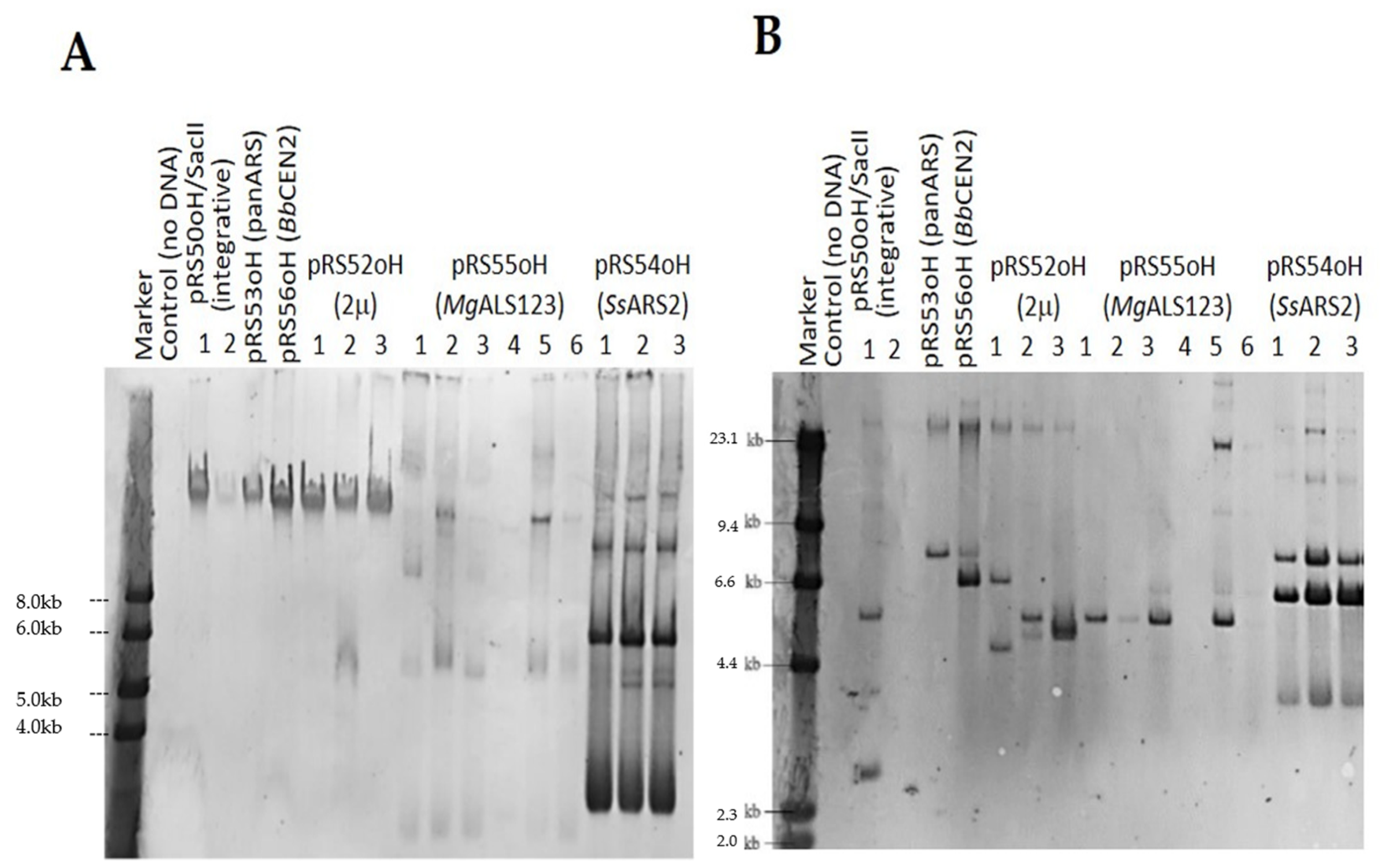
2.6. Yeast Transformation and Molecular Analysis by Southern Blotting
3. Results and Discussion
3.1. Plasmid Set for Transformation of Yeasts Belonging to the Saccharomycotina Subphylum
3.1.1. Integrative Yeast Plasmids
3.1.2. Replicative Yeast Plasmids
3.2. Transformation of Yeasts Belonging to the Schwanniomyces Genus
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Wendland, J. Special Issue: Non-Conventional Yeasts: Genomics and Biotechnology. Microorganisms 2019, 8, 21. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kurtzman, C.P.; Sugiyama, J. Saccharomycotina and Taphrinomycotina: The Yeasts and Yeastlike Fungi of the Ascomycota. In Systematics and Evolution. The Mycota; McLaughlin, D., Spatafora, J., Eds.; Springer: Berlin/Heidelberg, Germany, 2015; Volume 7B, ISBN 978-3-662-46010-8. [Google Scholar]
- Miklenić, M.; Žunar, B.; Štafa, A.; Svetec, I.-K. Improved electroporation procedure for genetic transformation of Dekkera/Brettanomyces bruxellensis. FEMS Yeast Res. 2015, 15, fov096. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bischoff, F.; Giersberg, M.; Matthes, F.; Schwalenberg, T.; Worch, S.; Kunze, G. Selection of the Optimal Yeast Host for the Synthesis of Recombinant Enzymes. In Recombinant Protein Production in Yeast; Humana Press: New York, NY, USA, 2019. [Google Scholar] [CrossRef]
- Kurtzman, C.P. Phylogeny of the ascomycetous yeasts and the renaming of Pichia anomala to Wickerhamomyces anomalus. Antonie Van Leeuwenhoek 2010, 99, 13–23. [Google Scholar] [CrossRef] [PubMed]
- Piontek, M.; Hagedorn, J.; Hollenberg, C.P.; Gellissen, G.; Strasser, A.W.M. Two novel gene expression systems based on the yeasts Schwanniomyces occidentalis and Pichia stipitis. Appl. Microbiol. Biotechnol. 1998, 50, 331–338. [Google Scholar] [CrossRef] [PubMed]
- Costaglioli, P.; Meilhoc, E.; Masson, J.M. High-efficiency electrotransformation of the yeast Schwanniomyces occidentalis. Curr. Genet. 1994, 27, 26–30. [Google Scholar] [CrossRef]
- Suthar, D.H.; Chattoo, B.B. Expression of Vitreoscilla hemoglobin enhances growth and levels of α-amylase in Schwanniomyces occidentalis. Appl. Microbiol. Biotechnol. 2006, 72, 94–102. [Google Scholar] [CrossRef]
- Álvaro-Benito, M.; Fernández-Lobato, M.; Baronian, K.; Kunze, G. Assessment of Schwanniomyces occidentalis as a host for protein production using the wide-range Xplor®2 expression platform. Appl. Microbiol. Biotechnol. 2012, 97, 4443–4456. [Google Scholar] [CrossRef] [Green Version]
- Utashima, Y.; Yamashita, S.; Arima, T.-H.; Masaki, K. Codon optimization enables the Zeocin resistance marker’s use in the ascomycete yeast Debaryomyces occidentalis. J. Gen. Appl. Microbiol. 2017, 63, 254–257. [Google Scholar] [CrossRef] [Green Version]
- Brachmann, C.B.; Davies, A.; Cost, G.J.; Caputo, E.; Li, J.; Hieter, P.; Boeke, J.D. Designer deletion strains derived from Saccharomyces cerevisiae S288C: A useful set of strains and plasmids for PCR-mediated gene disruption and other applications. Yeast 1998, 14, 115–132. [Google Scholar] [CrossRef]
- Jones, G.M.; Stalker, J.; Humphray, S.; West, A.; Cox, T.; Rogers, J.; Dunham, I.; Prelich, G. A systematic library for comprehensive overexpression screens in Saccharomyces cerevisiae. Nat. Methods 2008, 5, 239–241. [Google Scholar] [CrossRef]
- Chee, M.K.; Haase, S.B. New and Redesigned pRS Plasmid Shuttle Vectors for Genetic Manipulation of Saccharomyces cerevisiae. G3 Genes|Genomes|Genet. 2012, 2, 515–526. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Goldstein, A.L.; McCusker, J.H. Three new dominant drug resistance cassettes for gene disruption in Saccharomyces cerevisiae. Yeast 1999, 15, 1541–1553. [Google Scholar] [CrossRef]
- Christianson, T.W.; Sikorski, R.S.; Dante, M.; Shero, J.H.; Hieter, P. Multifunctional yeast high-copy-number shuttle vectors. Gene 1992, 110, 119–122. [Google Scholar] [CrossRef]
- Liachko, I.; Dunham, M.J. An autonomously replicating sequence for use in a wide range of budding yeasts. FEMS Yeast Res. 2013, 14, 364–367. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yang, V.W.; A Marks, J.; Davis, B.P.; Jeffries, T.W. High-efficiency transformation of Pichia stipitis based on its URA3 gene and a homologous autonomous replication sequence, ARS2. Appl. Environ. Microbiol. 1994, 60, 4245–4254. [Google Scholar] [CrossRef] [Green Version]
- Foureau, E.; Courdavault, V.; Gallón, S.M.N.; Besseau, S.; Simkin, A.J.; Crèche, J.; Atehortùa, L.; Giglioli-Guivarc’H, N.; Clastre, M.; Papon, N. Characterization of an autonomously replicating sequence in Candida guilliermondii. Microbiol. Res. 2013, 168, 580–588. [Google Scholar] [CrossRef]
- Ishchuk, O.P.; Zeljko, T.V.; Schifferdecker, A.J.; Wisén, S.M.; Hagström, K.; Rozpędowska, E.; Andersen, M.R.; Hellborg, L.; Ling, Z.; Sibirny, A.A.; et al. Novel Centromeric Loci of the Wine and Beer Yeast Dekkera bruxellensis CEN1 and CEN2. PLoS ONE 2016, 11, e0161741. [Google Scholar] [CrossRef] [Green Version]
- Winston, F.; Chumley, F.; Fink, G.R. [13] Eviction and transplacement of mutant genes in yeast. Methods Enzymol. 1983, 101, 211–228. [Google Scholar] [CrossRef]
- Štafa, A.; Miklenić, M.S.; Zandona, A.; Žunar, B.; Čadež, N.; Petković, H.; Svetec, I.K. In Saccharomyces cerevisiae gene targeting fidelity depends on a transformation method and proportion of the overall length of the transforming and targeted DNA. FEMS Yeast Res. 2017, 17, fox041. [Google Scholar] [CrossRef]
- Sikorski, R.S.; Hieter, P. A system of shuttle vectors and yeast host strains designed for efficient manipulation of DNA in Saccharomyces cerevisiae. Genetics 1989, 122, 19–27. [Google Scholar] [CrossRef]
- Gnügge, R.; Rudolf, F. Saccharomyces cerevisiae Shuttle vectors. Yeast 2017, 34, 205–221. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Santos, M.; Gomes, A.C.; Santos, M.C.; Carreto, L.C.; Moura, G. The genetic code of the fungal CTG clade. Comptes Rendus Biol. 2011, 334, 607–611. [Google Scholar] [CrossRef] [PubMed]
- Papon, N.; Courdavault, V.; Clastre, M.; Simkin, A.J.; Crèche, J.; Giglioli-Guivarc’H, N. Deus ex Candida genetics: Overcoming the hurdles for the development of a molecular toolbox in the CTG clade. Microbiology 2012, 158, 585–600. [Google Scholar] [CrossRef] [PubMed]
- Letzring, D.P.; Dean, K.M.; Grayhack, E.J. Control of translation efficiency in yeast by codon–anticodon interactions. RNA 2010, 16, 2516–2528. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dvir, S.; Velten, L.; Sharon, E.; Zeevi, D.; Carey, L.B.; Weinberger, A.; Segal, E. Deciphering the rules by which 5′-UTR sequences affect protein expression in yeast. Proc. Natl. Acad. Sci. USA 2013, 110, E2792–E2801. [Google Scholar] [CrossRef] [Green Version]
- Wang, T.-T.; Choi, Y.-J.; Lee, B.H. Transformation Systems of non-Saccharomyces Yeasts. Crit. Rev. Biotechnol. 2001, 21, 177–218. [Google Scholar] [CrossRef]
- Wright, M.C.; Philippsen, P. Replicative transformation of the filamentous fungus Ashbya gossypii with plasmids containing Saccharomyces cerevisiae ARS elements. Gene 1991, 109, 99–105. [Google Scholar] [CrossRef]
- Bijlani, S.; Thevandavakkam, M.A.; Tsai, H.-J.; Berman, J. Autonomously Replicating Linear Plasmids That Facilitate the Analysis of Replication Origin Function in Candida albicans. MSphere 2019, 4, e00103-19. [Google Scholar] [CrossRef] [Green Version]
- Miklenić, M. Genetic Transformation of the Yeast Dekkera/Brettanomyces bruxellensis with Non-Homologous DNA. J. Microbiol. Biotechnol. 2013, 23, 674–680. [Google Scholar] [CrossRef] [Green Version]

| Geneticin G418 [μg/mL] | Hygromycin B [μg/mL] | cloNAT [μg/mL] | Phleomycin [μg/mL] | |
|---|---|---|---|---|
| S. cerevisiae | 200 | 300 | 100 | 10 |
| K. lactis | 150 | 200 | 20 | 100 |
| S. stipitis | selection not possible | 600 | 50 | 100 |
| M. guilliermondii | selection not possible | 500 | 200 | 200 |
| B. bruxellensis | 250 | 150 | 35 | 150 |
| S. pseudopolymorpshus | selection not possible | 200 | 50 | Not determined |
| S. polymorphus var. polymorphus | selection not possible | 200 | 50 | Not determined |
| S. polymorphus var. africanus | selection not possible | 400 | 50 | Not determined |
| Yeast Strains | Genotype | Reference |
|---|---|---|
| Saccharomyces cerevisiae BY 4742 | MATα his3Δ1 leu2Δ0 lys2Δ0 ura3Δ0 | [11] |
| Kluyveromyces lactis CBS 2359T | Type strain | Westerdijk Fungal Biodiversity Institute, The Netherlands |
| Scheffersomyces stipitis JCM 10742T | Type strain | Japan Collection of Microorganisms, Japan |
| Meyerozyma guilliermondii JCM 1539T | Type strain | Japan Collection of Microorganisms, Japan |
| Brettanomyces bruxellensis CBS 2499 | Westerdijk Fungal Biodiversity Institute, The Netherlands | |
| Schwanniomyces pseudopolymorphus JCM3652T | Type strain | Japan Collection of Microorganisms, Japan |
| Schwanniomyces polymorphus var. polymorphus JCM3647T | Type strain | Japan Collection of Microorganisms, Japan |
| Schwanniomyces polymorphus var. africanus JCM7443T | Type strain | Japan Collection of Microorganisms, Japan |
| Plasmids | Feature Important for This Study | Reference |
| pGP564 | contains 2µ replication origin | [12] |
| pRS40B * | source of the pRS40 backbone | [13] |
| pRS50 series (pRS50oK, pRS50oH, pRS50oN, pRS50oB) | Integrative plasmids with codon-optimized selectable markers (KanR, HygR, NarR, PhlR) | This study |
| pRS52 series (pRS52oK, pRS52oH, pRS52oN, pRS52oB) | Contains 2μ replication origin from S. cerevisiae and codon-optimized selectable markers (KanR, HygR, NarR, PhlR) | This study |
| pRS53 series (pRS53oK, pRS53oH, pRS53oN, pRS53oB) | Contains panARS replication origin from K. lactis and codon-optimized selectable markers (KanR, HygR, NarR, PhlR) | This study |
| pRS54 series (pRS54oK, pRS54oH, pRS54oN, pRS54oB) | Contains SsARS replication origin from S. stipitis and codon-optimized selectable markers (KanR, HygR, NarR, PhlR) | This study |
| pRS55 series (pRS55oK, pRS55oH, pRS55oN, pRS55oB) | Contains MgALS123 replication origin from M. guilliermondii and codon-optimized selectable markers (KanR, HygR, NarR, PhlR) | This study |
| pRS56 series (pRS56oK, pRS56oH, pRS56oN, pRS56oB) | Contains BbCEN2 replication origin from B. bruxellensis and codon-optimized selectable markers (KanR, HygR, NarR, PhlR) | This study |
| Oligonucleotides | Sequence | Reference |
| panARS-f | gtgaggtaccgaaggaatttgctgttatggag | This study |
| panARS-r | gtgaggtaccactgacactgttgactctg | This study |
| SsARS2-f | gatatccagaataattgatggtccgc | This study |
| SsARS2-r | gatatctggattgttgtgctctcg | This study |
| MgARS-f | gatatcagatgacaagcccaaacac | This study |
| MgARS-r | gatatccatatgtccttgccagttgaacca | This study |
| DbCEN2-f | gatatcctgaggttgctaagcccc | This study |
| DbCEN2-r | gatatcgtgaatagtgaagccaactggt | This study |
| AgTEF-f | aggcctcccgggacatggaggcccagaat | This study |
| AgTEF-r | aggcctcccgggcagtatagcgaccagcattc | This study |
| Transformation Efficiency (Transformants/μg) | ||||||
|---|---|---|---|---|---|---|
| pRS50oH/SacII (Linear) | pRS52oH (2μ Origin) | pRS53oH (panARS Origin) | pRS54oH (SsARS2 Origin) | pRS55oH (MgALS123 Origin) | pRS56oH (BbCEN2 Origin) | |
| S. pseudopolymorphus | 8.1 × 103 | 424 | 21 | 4.7 × 105 | 32 | 1.1 × 103 |
| S. polymorphus var. polymorphus | 952 | 360 | 117 | 3.7 × 105 | More than 2 × 104 | 714 |
| S. polymorphus var. africanus | 476 | 88 | 149 | 2.7 × 105 | More than 2 × 104 | 0 |
| Plasmid Stability | |||
|---|---|---|---|
| pRS50oH Integrated in the Genome | pRS54oH (SsARS2 Origin) | pRS55oH (MgALS123 Origin) | |
| S. pseudopolymorphus | 100% | 5.9% | Not replicative |
| S. polymorphus var. polymorphus | 100% | 28% | 1% |
| S. polymorphus var. africanus | 100% | 100% | 0.2% |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Matanović, A.; Arambašić, K.; Žunar, B.; Štafa, A.; Svetec Miklenić, M.; Šantek, B.; Svetec, I.-K. Toolbox for Genetic Transformation of Non-Conventional Saccharomycotina Yeasts: High Efficiency Transformation of Yeasts Belonging to the Schwanniomyces Genus. J. Fungi 2022, 8, 531. https://doi.org/10.3390/jof8050531
Matanović A, Arambašić K, Žunar B, Štafa A, Svetec Miklenić M, Šantek B, Svetec I-K. Toolbox for Genetic Transformation of Non-Conventional Saccharomycotina Yeasts: High Efficiency Transformation of Yeasts Belonging to the Schwanniomyces Genus. Journal of Fungi. 2022; 8(5):531. https://doi.org/10.3390/jof8050531
Chicago/Turabian StyleMatanović, Angela, Kristian Arambašić, Bojan Žunar, Anamarija Štafa, Marina Svetec Miklenić, Božidar Šantek, and Ivan-Krešimir Svetec. 2022. "Toolbox for Genetic Transformation of Non-Conventional Saccharomycotina Yeasts: High Efficiency Transformation of Yeasts Belonging to the Schwanniomyces Genus" Journal of Fungi 8, no. 5: 531. https://doi.org/10.3390/jof8050531
APA StyleMatanović, A., Arambašić, K., Žunar, B., Štafa, A., Svetec Miklenić, M., Šantek, B., & Svetec, I. -K. (2022). Toolbox for Genetic Transformation of Non-Conventional Saccharomycotina Yeasts: High Efficiency Transformation of Yeasts Belonging to the Schwanniomyces Genus. Journal of Fungi, 8(5), 531. https://doi.org/10.3390/jof8050531









