Dose-Dependent Genetic Resistance to Azole Fungicides Found in the Apple Scab Pathogen
Abstract
:1. Introduction
2. Materials and Methods
2.1. Generation of a Mapping Population and Measurement of Sensitivity
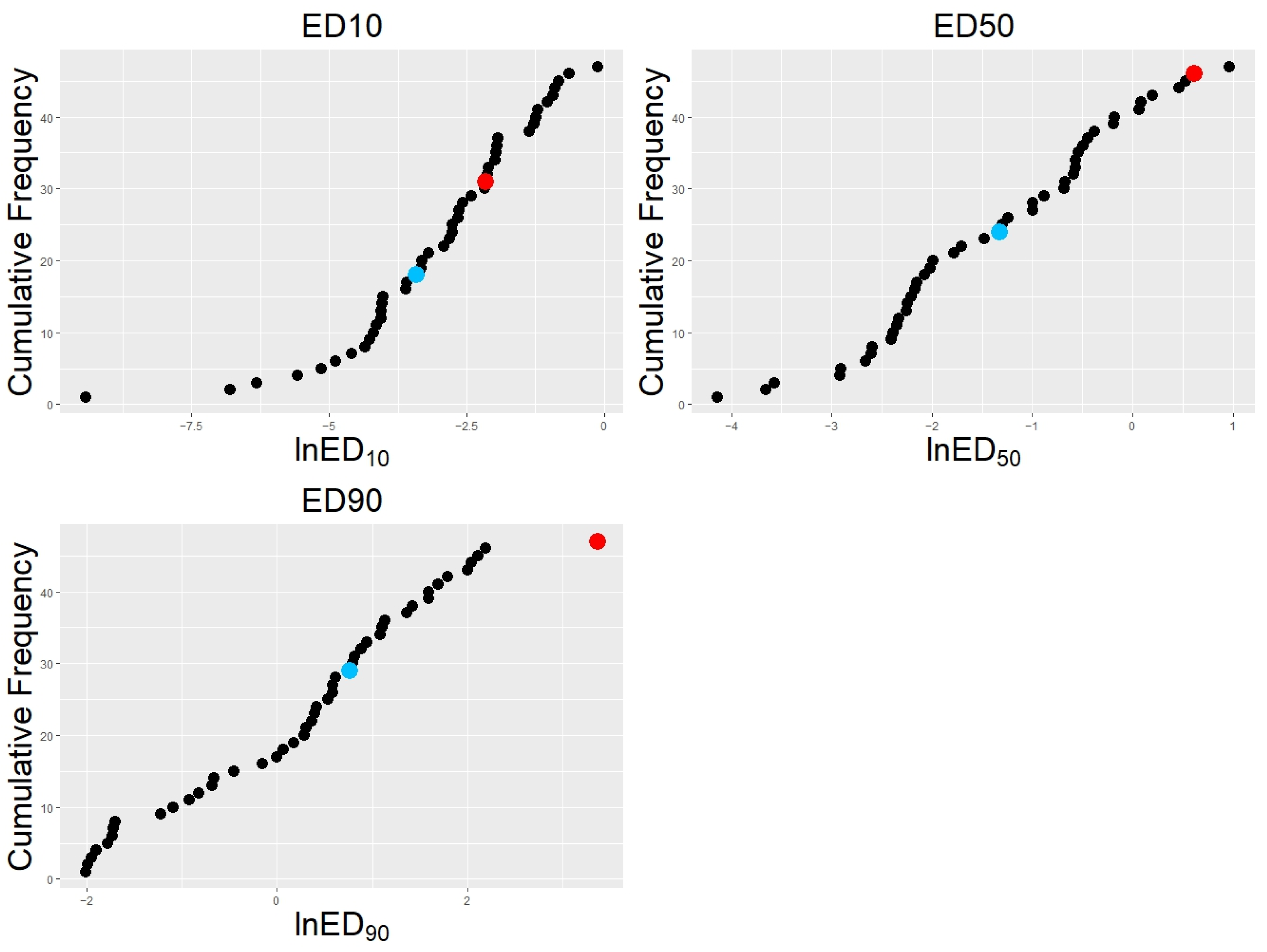
2.2. Calculation of Effective Dosages
2.3. DNA Extraction and Sequencing
2.4. Calling Single Nucleotide Polymorphism Markers
2.5. Linkage Map Generation
2.6. Mapping of Quantitative Trait Loci
2.7. Candidate Gene Characterisation
3. Results
3.1. Progeny Isolates Segregated by Tebuconazole Sensitivity
3.2. Whole-Genome Resequencing Identified Single Nucleotide Polymorphisms across the Mapping Population
3.3. Single Nucleotide Polymorphisms Mapped to Nine Linkage Groups
3.4. Two Dose-Dependent Quantitative Trait Loci for Tebuconazole Sensitivity Were Identified
3.5. Quantitative Trait Loci Location and Gene Content
4. Discussion
4.1. Multiple Loci Contribute to the Level of Tebuconazole Sensitivity in the Apple Scab Pathogen
4.2. Quantitative Trait Loci Indicate a Novel Mechanism of Resistance
4.3. Dose-Dependent Resistance May Result from Variant Efflux Transporter Genes
4.4. Alternative Resistance Mechanisms
4.5. Lab-Based vs. In-Field Assessments
4.6. Implications for Practical Management
4.7. Summary
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Carisse, O.; Bernier, J. Effect of Environmental Factors on Growth, Pycnidial Production and Spore Germination of Microsphaeropsis Isolates with Biocontrol Potential against Apple Scab. Mycol. Res. 2002, 106, 1455–1462. [Google Scholar] [CrossRef]
- Bowen, J.K.; Mesarich, C.H.; Bus, V.G.M.; Beresford, R.M.; Plummer, K.M.; Templeton, M.D. Venturia inaequalis: The Causal Agent of Apple Scab. Mol. Plant Pathol. 2011, 12, 105–122. [Google Scholar] [CrossRef] [PubMed]
- Chatzidimopoulos, M.; Lioliopoulou, F.; Sotiropoulos, T.; Vellios, E. Efficient Control of Apple Scab with Targeted Spray Applications. Agronomy 2020, 10, 217. [Google Scholar] [CrossRef]
- Cox, K.D. Fungicide Resistance in Venturia inaequalis, the Causal Agent of Apple Scab, in the United States. In Fungicide Resistance in Plant Pathogens: Principles and a Guide to Practical Management; Ishii, H., Hollomon, D.W., Eds.; Springer: Tokyo, Japan, 2015; pp. 433–448. ISBN 978-4-431-55641-1. [Google Scholar]
- FRAG Fungicide Resistance Action Group. Fungicide Resistance Management in Apple and Pear Pathogens. Available online: https://media.ahdb.org.uk/media/Default/Imported%20Publication%20Docs/AHDB%20Cereals%20&%20Oilseeds/Disease/FRAG%20Fungicide%20resistance%20management%20in%20apple%20and%20pear%20pathogens%20(March%202015).pdf (accessed on 10 February 2023).
- MacHardy, W.E. Apple Scab: Biology, Epidemiology, and Management; APS Press: St Paul, MN, USA, 1996; ISBN 0-89054-206-6. [Google Scholar]
- AHDB Agriculture and Horticulture Development Board. Apple Best Practice Guide: Apple Scab—Additional Information. Available online: https://applegrowing.niab.com/apple-scab-additional-information/ (accessed on 2 February 2023).
- Zhang, J.; Li, L.; Lv, Q.; Yan, L.; Wang, Y.; Jiang, Y. The Fungal CYP51s: Their Functions, Structures, Related Drug Resistance, and Inhibitors. Front. Microbiol. 2019, 10, 691. [Google Scholar] [CrossRef] [PubMed]
- Parker, J.E.; Warrilow, A.G.S.; Price, C.L.; Mullins, J.G.L.; Kelly, D.E.; Kelly, S.L. Resistance to Antifungals That Target CYP51. J. Chem. Biol. 2014, 7, 143–161. [Google Scholar] [CrossRef] [PubMed]
- Schwabe, W.F.S.; Jones, A.L.; Jonker, J.P. Changes in the Susceptibility of Developing Apple Fruit to Venturia inaequalis. Phytopathology 1984, 74, 118–121. [Google Scholar] [CrossRef]
- O’Leary, A.L.; Sutton, T.B. The Influence of Temperature and Moisture on the Quantitative Production of Pseudothecia of Venturia inaequalis. Phytopathology 1986, 76, 199–204. [Google Scholar] [CrossRef]
- Poblete, J.A.; Latorre, B.A. Efecto preventivo y curativo de los fungicidas inhibidores de esteroles en el control de Venturia inaequalis del manzano. Cienc. E Investig. Agrar. 2001, 28, 145–150. [Google Scholar] [CrossRef]
- Cordero-Limon, L. Genetics behind the Variability in Sensitivity to the Demethylation Inhibitor (DMI) Fungicides Myclobutanil and Tebuconazole in Venturia inaequalis. PhD Thesis, University of Reading, Reading, UK, 2018. [Google Scholar]
- Chapman, K.S.; Sundin, G.W.; Beckerman, J.L. Identification of Resistance to Multiple Fungicides in Field Populations of Venturia inaequalis. Plant Dis. 2011, 95, 921–926. [Google Scholar] [CrossRef]
- Stanis, V.F.; Jones, A.L. Reduced Sensitivity to Sterol-Inhibiting Fungicides in Field Isolates of Venturia inaequalis. Phytopathology 1985, 75, 1098–1101. [Google Scholar] [CrossRef]
- Köller, W.; Smith, F.D.; Reynolds, K.L. Phenotypic Instability of Flusilazole Sensitivity in Venturia inaequalis. Plant Pathol. 1991, 40, 608–611. [Google Scholar] [CrossRef]
- Jobin, T.; Carisse, O. Incidence of Myclobutanil- and Kresoxim-Methyl-Insensitive Isolates of Venturia inaequalis in Quebec Orchards. Plant Dis. 2007, 91, 1351–1358. [Google Scholar] [CrossRef] [PubMed]
- Gao, L.; Berrie, A.; Yang, J.; Xu, X. Within- and between-Orchard Variability in the Sensitivity of Venturia inaequalis to Myclobutanil, a DMI Fungicide, in the UK. Pest. Manag. Sci. 2009, 65, 1241–1249. [Google Scholar] [CrossRef]
- Sombardier, A.; Dufour, M.-C.; Blancard, D.; Corio-Costet, M.-F. Sensitivity of Podosphaera aphanis Isolates to DMI Fungicides: Distribution and Reduced Cross-Sensitivity: Sensitivity of P. Aphanis to DMI Fungicides. Pest. Manag. Sci. 2010, 66, 35–43. [Google Scholar] [CrossRef] [PubMed]
- Stević, M.; Vukša, P.; Elezović, I. Resistance of Venturia inaequalis to Demethylation Inhibiting (DMI) Fungicides. Žemdir-bystė-Agric. 2010, 97, 65–72. [Google Scholar]
- Henríquez, S.J.L.; Sarmiento, V.O.; Alarcón, C.P. Sensitivity of Venturia inaequalis Chilean Isolates to Difenoconazole, Fenarimol, Mancozeb, and Pyrimethanil. Chil. J. Agric. Res. 2011, 71, 39–44. [Google Scholar] [CrossRef]
- Beresford, R.; Wright, P.; Wood, P.; Park, N. Sensitivity of Venturia inaequalis to Myclobutanil Penconazole and Dodine in Relation to Fungicide Use in Hawkes Bay Apple Orchards. N. Z. Plant Prot. 2012, 65, 106–113. [Google Scholar] [CrossRef]
- Mondino, P.; Casanova, L.; Celio, A.; Bentancur, O.; Leoni, C.; Alaniz, S. Sensitivity of Venturia inaequalis to Trifloxystrobin and Difenoconazole in Uruguay. J. Phytopathol. 2014, 163, 1–10. [Google Scholar] [CrossRef]
- Villani, S.M.; Biggs, A.R.; Cooley, D.R.; Raes, J.J.; Cox, K.D. Prevalence of Myclobutanil Resistance and Difenoconazole Insensitivity in Populations of Venturia inaequalis. Plant Dis. 2015, 99, 1526–1536. [Google Scholar] [CrossRef]
- Li, X.; Li, H.; Yu, Z.; Gao, L.; Yang, J. Investigating the Sensitivity of Venturia inaequalis Isolates to Difenoconazole and Pyraclostrobin in Apple Orchards in China. Eur. J. Plant Pathol. 2021, 161, 207–217. [Google Scholar] [CrossRef]
- Chatzidimopoulos, M.; Zambounis, A.; Lioliopoulou, F.; Vellios, E. Detection of Venturia inaequalis Isolates with Multiple Resistance in Greece. Microorganisms 2022, 10, 2354. [Google Scholar] [CrossRef] [PubMed]
- Nasonov, A.; Yakuba, G.; Marchenko, N.; Lobodina, E.; Astapchuk, I. Evaluation of Sensitivity of Apple Scab Pathogen to Difenoconazole Using the Discriminatory Dose Technique. BIO Web Conf. 2022, 47, 10002. [Google Scholar] [CrossRef]
- Hildebrand, P.D.; Lockhart, C.L.; Newbery, R.J.; Ross, R.G. Resistance of Venturia inaequalis to Bitertanol and Other Demethylation-Inhibiting Fungicides. Can. J. Plant Pathol. 1988, 10, 311–316. [Google Scholar] [CrossRef]
- Köller, W.; Wilcox, W.F.; Barnard, J.; Jones, A.L.; Braun, P.G. Detection and Quantification of Resistance of Venturia inaequalis Populations to Sterol Demethylation Inhibitors. Phytopathology 1997, 87, 184–190. [Google Scholar] [CrossRef] [PubMed]
- Köller, W.; Wilcox, W.F. Interactive Effects of Dodine and the DMI Fungicide Fenarimol in the Control of Apple Scab. Plant Dis. 2000, 84, 863–870. [Google Scholar] [CrossRef] [PubMed]
- Stammler, G.; Cordero, J.; Koch, A.; Semar, M.; Schlehuber, S. Role of the Y134F Mutation in CYP51 and Overexpression of CYP51 in the Sensitivity Response of Puccinia triticina to Epoxiconazole. Crop. Prot. 2009, 28, 891–897. [Google Scholar] [CrossRef]
- Becher, R.; Wirsel, S.G.R. Fungal Cytochrome P450 Sterol 14α-Demethylase (CYP51) and Azole Resistance in Plant and Human Pathogens. Appl. Microbiol. Biotechnol. 2012, 95, 825–840. [Google Scholar] [CrossRef] [PubMed]
- Schmitz, H.K.; Medeiros, C.-A.; Craig, I.R.; Stammler, G. Sensitivity of Phakopsora pachyrhizi towards Quinone-Outside-Inhibitors and Demethylation-Inhibitors, and Corresponding Resistance Mechanisms: Sensitivity and Resistance Mechanisms of P. pachyrhizi towards Fungicides. Pest. Manag. Sci. 2014, 70, 378–388. [Google Scholar] [CrossRef]
- Frenkel, O.; Cadle-Davidson, L.; Wilcox, W.F.; Milgroom, M.G. Mechanisms of Resistance to an Azole Fungicide in the Grapevine Powdery Mildew Fungus, Erysiphe necator. Phytopathology 2015, 105, 370–377. [Google Scholar] [CrossRef]
- Mair, W.; Lopez-Ruiz, F.; Stammler, G.; Clark, W.; Burnett, F.; Hollomon, D.; Ishii, H.; Thind, T.S.; Brown, J.K.; Fraaije, B.; et al. Proposal for a Unified Nomenclature for Target-site Mutations Associated with Resistance to Fungicides. Pest Manag. Sci. 2016, 72, 1449–1459. [Google Scholar] [CrossRef]
- Huf, A.; Rehfus, A.; Lorenz, K.H.; Bryson, R.; Voegele, R.T.; Stammler, G. Proposal for a New Nomenclature for CYP51 Haplotypes in Zymoseptoria tritici and Analysis of Their Distribution in Europe. Plant Pathol. 2018, 67, 1706–1712. [Google Scholar] [CrossRef]
- Muellender, M.M.; Mahlein, A.; Stammler, G.; Varrelmann, M. Evidence for the Association of Target-site Resistance in Cyp51 with Reduced DMI Sensitivity in European Cercospora beticola Field Isolates. Pest Manag. Sci. 2021, 77, 1765–1774. [Google Scholar] [CrossRef] [PubMed]
- Cools, H.J.; Hawkins, N.J.; Fraaije, B.A. Constraints on the Evolution of Azole Resistance in Plant Pathogenic Fungi. Plant Pathol. 2013, 62, 36–42. [Google Scholar] [CrossRef]
- Yaegashi, H.; Hirayama, K.; Akahira, T.; Ito, T. Point Mutation in CYP51A1 of Venturia inaequalis Is Associated with Low Sensitivity to Sterol Demethylation Inhibitors. J. Gen. Plant Pathol. 2020, 86, 245–249. [Google Scholar] [CrossRef]
- Hoffmeister, M.; Zito, R.; Böhm, J.; Stammler, G. Mutations in CYP51 of Venturia inaequalis and Their Effects on DMI Sensitivity. J. Plant Dis. Prot. 2021, 128, 1467–1478. [Google Scholar] [CrossRef]
- Schnabel, G.; Jones, A.L. The 14α-Demethylasse (CYP51A1) Gene Is Overexpressed in Venturia inaequalis Strains Resistant to Myclobutanil. Phytopathology 2001, 91, 102–110. [Google Scholar] [CrossRef]
- Villani, S.M.; Hulvey, J.; Hily, J.-M.; Cox, K.D. Overexpression of the CYP51A1 Gene and Repeated Elements Are Associated with Differential Sensitivity to DMI Fungicides in Venturia inaequalis. Phytopathology 2016, 106, 562–571. [Google Scholar] [CrossRef]
- Cordero-Limon, L.; Shaw, M.W.; Passey, T.A.; Robinson, J.D.; Xu, X. Cross-resistance between Myclobutanil and Tebuconazole and the Genetic Basis of Tebuconazole Resistance in Venturia inaequalis. Pest Manag. Sci. 2021, 77, 844–850. [Google Scholar] [CrossRef]
- Xu, X.-M.; Gao, L.-Q.; Yang, J.-R. Are Insensitivities of Venturia inaequalis to Myclobutanil and Fenbuconazole Correlated? Crop. Prot. 2010, 29, 183–189. [Google Scholar] [CrossRef]
- Errampalli, D. Distribution of Myclobutanil Fungicide Sensitivities among Populations of Venturia inaequalis, the Causal Agent of Apple Scab, in Ontario. Acta Hortic. 2004, 638, 157–162. [Google Scholar] [CrossRef]
- Beresford, R.M.; Wright, P.J.; Wood, P.N.; Park, N.M.; Larsen, N.J.; Fisher, B.M. Resistance of Venturia inaequalis to Demethylation Inhibitor and Dodine Fungicides in Four New Zealand Applegrowing Regions. NZPP 2013, 66, 274–283. [Google Scholar] [CrossRef]
- Koller, W. Baseline Sensitivities of Venturia inaequalis to Sterol Demethylation Inhibitors. Plant Dis. 1991, 75, 726. [Google Scholar] [CrossRef]
- Pfeufer, E.E.; Ngugi, H.K. Orchard Factors Associated with Resistance and Cross Resistance to Sterol Demethylation Inhibitor Fungicides in Populations of Venturia inaequalis from Pennsylvania. Phytopathology 2012, 102, 272–282. [Google Scholar] [CrossRef] [PubMed]
- Ritz, C.; Baty, F.; Streibig, J.C.; Gerhard, D. Dose-Response Analysis Using R. PLoS ONE 2015, 10, e0146021. [Google Scholar] [CrossRef] [PubMed]
- McKenna, A.; Hanna, M.; Banks, E.; Sivachenko, A.; Cibulskis, K.; Kernytsky, A.; Garimella, K.; Altshuler, D.; Gabriel, S.; Daly, M.; et al. The Genome Analysis Toolkit: A MapReduce Framework for Analyzing next-Generation DNA Sequencing Data. Genome Res. 2010, 20, 1297–1303. [Google Scholar] [CrossRef] [PubMed]
- Bolger, A.M.; Lohse, M.; Usadel, B. Trimmomatic: A Flexible Trimmer for Illumina Sequence Data. Bioinformatics 2014, 30, 2114–2120. [Google Scholar] [CrossRef] [PubMed]
- Mapleson, D.; Garcia Accinelli, G.; Kettleborough, G.; Wright, J.; Clavijo, B.J. KAT: A K-Mer Analysis Toolkit to Quality Control NGS Datasets and Genome Assemblies. Bioinformatics 2017, 33, 574–576. [Google Scholar] [CrossRef]
- Langmead, B.; Salzberg, S.L. Fast Gapped-Read Alignment with Bowtie 2. Nat. Methods 2012, 9, 357–359. [Google Scholar] [CrossRef]
- Passey, T.A.J.; Armitage, A.D.; Xu, X. Annotated Draft Genome Sequence of the Apple Scab Pathogen Venturia inaequalis. Microbiol. Resour. Announc. 2018, 7, e01062-18. [Google Scholar] [CrossRef]
- Li, H.; Handsaker, B.; Wysoker, A.; Fennell, T.; Ruan, J.; Homer, N.; Marth, G.; Abecasis, G.; Durbin, R.; 1000 Genome Project Data Processing Subgroup. The Sequence Alignment/Map Format and SAMtools. Bioinformatics 2009, 25, 2078–2079. [Google Scholar] [CrossRef]
- R Core Team. R: A Language and Environment for Statistical Computing; R Foundation for Statistical Computing: Vienna, Austria, 2022. [Google Scholar]
- Stam, P. Construction of Integrated Genetic Linkage Maps by Means of a New Computer Package: Join Map. Plant J. 1993, 3, 739–744. [Google Scholar] [CrossRef]
- Camacho, C.; Coulouris, G.; Avagyan, V.; Ma, N.; Papadopoulos, J.; Bealer, K.; Madden, T.L. BLAST+: Architecture and Applications. BMC Bioinform. 2009, 10, 421. [Google Scholar] [CrossRef]
- Paysan-Lafosse, T.; Blum, M.; Chuguransky, S.; Grego, T.; Pinto, B.L.; Salazar, G.A.; Bileschi, M.L.; Bork, P.; Bridge, A.; Colwell, L.; et al. InterPro in 2022. Nucleic Acids Res. 2023, 51, D418–D427. [Google Scholar] [CrossRef] [PubMed]
- Mistry, J.; Chuguransky, S.; Williams, L.; Qureshi, M.; Salazar, G.A.; Sonnhammer, E.L.L.; Tosatto, S.C.E.; Paladin, L.; Raj, S.; Richardson, L.J.; et al. Pfam: The Protein Families Database in 2021. Nucleic Acids Res. 2021, 49, D412–D419. [Google Scholar] [CrossRef] [PubMed]
- Shaw, M.W. A Model of the Evolution of Polygenically Controlled Fungicide Resistance. Plant Pathol. 1989, 38, 44–55. [Google Scholar] [CrossRef]
- Georgopoulos, S.G.; Skylakakis, G. Genetic Variability in the Fungi and the Problem of Fungicide Resistance. Crop. Prot. 1986, 5, 299–305. [Google Scholar] [CrossRef]
- Lendenmann, M.H.; Croll, D.; McDonald, B.A. QTL Mapping of Fungicide Sensitivity Reveals Novel Genes and Pleiotropy with Melanization in the Pathogen Zymoseptoria tritici. Fungal Genet. Biol. 2015, 80, 53–67. [Google Scholar] [CrossRef] [PubMed]
- Nakaune, R.; Adachi, K.; Nawata, O.; Tomiyama, M.; Akutsu, K.; Hibi, T. A Novel ATP-Binding Cassette Transporter Involved in Multidrug Resistance in the Phytopathogenic Fungus Penicillium digitatum. Appl. Environ. Microbiol. 1998, 64, 3983–3988. [Google Scholar] [CrossRef] [PubMed]
- Cowen, L.E.; Anderson, J.B.; Kohn, L.M. Evolution of Drug Resistance in Candida albicans. Annu. Rev. Microbiol. 2002, 56, 139–165. [Google Scholar] [CrossRef]
- Joseph-Home, T.; Manning, N.J.; Hollomon, D.; Kelly, S.L. Defective Sterol Δ 5(6) Desaturase as a Cause of Azole Resistance in Ustilago maydis. FEMS Microbiol. Lett. 1995, 127, 29–34. [Google Scholar] [CrossRef]
- Bolton, M.D.; Ebert, M.K.; Faino, L.; Rivera-Varas, V.; de Jonge, R.; Van de Peer, Y.; Thomma, B.P.H.J.; Secor, G.A. RNA-Sequencing of Cercospora beticola DMI-Sensitive and -Resistant Isolates after Treatment with Tetraconazole Identifies Common and Contrasting Pathway Induction. Fungal Genet. Biol. 2016, 92, 1–13. [Google Scholar] [CrossRef] [PubMed]
- Cowen, L.E.; Lindquist, S. Hsp90 Potentiates the Rapid Evolution of New Traits: Drug Resistance in Diverse Fungi. Science 2005, 309, 2185–2189. [Google Scholar] [CrossRef] [PubMed]
- Mellado, E.; Diaz-Guerra, T.M.; Cuenca-Estrella, M.; Rodriguez-Tudela, J.L. Identification of Two Different 14-α Sterol Demethylase-Related Genes (cyp51A and cyp51B) in Aspergillus fumigatus and Other Aspergillus Species. J. Clin. Microbiol. 2001, 39, 2431–2438. [Google Scholar] [CrossRef] [PubMed]
- Jones, L.; Riaz, S.; Morales-Cruz, A.; Amrine, K.C.; McGuire, B.; Gubler, W.D.; Walker, M.A.; Cantu, D. Adaptive Genomic Structural Variation in the Grape Powdery Mildew Pathogen, Erysiphe necator. BMC Genom. 2014, 15, 1081. [Google Scholar] [CrossRef] [PubMed]
- Hamamoto, H.; Hasegawa, K.; Nakaune, R.; Lee, Y.J.; Makizumi, Y.; Akutsu, K.; Hibi, T. Tandem Repeat of a Transcriptional Enhancer Upstream of the Sterol 14α-Demethylase Gene (CYP51) in Penicillium digitatum. Appl. Environ. Microbiol. 2000, 66, 3421–3426. [Google Scholar] [CrossRef] [PubMed]
- Ma, Z.; Proffer, T.J.; Jacobs, J.L.; Sundin, G.W. Overexpression of the 14α-Demethylase Target Gene (CYP51) Mediates Fungicide Resistance in Blumeriella jaapii. Appl. Environ. Microbiol. 2006, 72, 2581–2585. [Google Scholar] [CrossRef]
- Nikou, D.; Malandrakis, A.; Konstantakaki, M.; Vontas, J.; Markoglou, A.; Ziogas, B. Molecular Characterization and Detection of Overexpressed C-14 Alpha-Demethylase-Based DMI Resistance in Cercospora beticola Field Isolates. Pestic. Biochem. Physiol. 2009, 95, 18–27. [Google Scholar] [CrossRef]
- Luo, C.-X.; Schnabel, G. The Cytochrome P450 Lanosterol 14α-Demethylase Gene Is a Demethylation Inhibitor Fungicide Resistance Determinant in Monilinia fructicola Field Isolates from Georgia. Appl. Environ. Microbiol. 2008, 74, 359–366. [Google Scholar] [CrossRef]
- Cools, H.J.; Bayon, C.; Atkins, S.; Lucas, J.A.; Fraaije, B.A. Overexpression of the Sterol 14α-Demethylase Gene (MgCYP51) in Mycosphaerella graminicola Isolates Confers a Novel Azole Fungicide Sensitivity Phenotype. Pest Manag. Sci. 2012, 68, 1034–1040. [Google Scholar] [CrossRef]
- Sun, X.; Xu, Q.; Ruan, R.; Zhang, T.; Zhu, C.; Li, H. PdMLE1, a Specific and Active Transposon Acts as a Promoter and Confers Penicillium digitatum with DMI Resistance. Environ. Microbiol. Rep. 2013, 5, 135–142. [Google Scholar] [CrossRef]
- de Waard, M.A.; Andrade, A.C.; Hayashi, K.; Schoonbeek, H.; Stergiopoulos, I.; Zwiers, L.-H. Impact of Fungal Drug Transporters on Fungicide Sensitivity, Multidrug Resistance and Virulence. Pest Manag. Sci. 2006, 62, 195–207. [Google Scholar] [CrossRef] [PubMed]
- Hayashi, K.; Schoonbeek, H.; De Waard, M.A. Expression of the ABC Transporter BcatrD from Botrytis cinerea Reduces Sensitivity to Sterol Demethylation Inhibitor Fungicides. Pestic. Biochem. Physiol. 2002, 73, 110–121. [Google Scholar] [CrossRef]
- Hulvey, J.; Popko, J.T.; Sang, H.; Berg, A.; Jung, G. Overexpression of ShCYP51B and ShatrD in Sclerotinia homoeocarpa Isolates Exhibiting Practical Field Resistance to a Demethylation Inhibitor Fungicide. Appl. Environ. Microbiol. 2012, 78, 6674–6682. [Google Scholar] [CrossRef] [PubMed]
- Stergiopoulos, I.; Zwiers, L.-H.; De Waard, M.A. Secretion of Natural and Synthetic Toxic Compounds from Filamentous Fungi by Membrane Transporters of the ATP-Binding Cassette and Major Facilitator Superfamily. Eur. J. Plant Pathol. 2002, 108, 719–734. [Google Scholar] [CrossRef]
- Cannon, R.D.; Lamping, E.; Holmes, A.R.; Niimi, K.; Baret, P.V.; Keniya, M.V.; Tanabe, K.; Niimi, M.; Goffeau, A.; Monk, B.C. Efflux-Mediated Antifungal Drug Resistance. Clin. Microbiol. Rev. 2009, 22, 291–321. [Google Scholar] [CrossRef] [PubMed]
- Kretschmer, M.; Leroch, M.; Mosbach, A.; Walker, A.-S.; Fillinger, S.; Mernke, D.; Schoonbeek, H.-J.; Pradier, J.-M.; Leroux, P.; De Waard, M.A.; et al. Fungicide-Driven Evolution and Molecular Basis of Multidrug Resistance in Field Populations of the Grey Mould Fungus Botrytis cinerea. PLoS Pathog. 2009, 5, e1000696. [Google Scholar] [CrossRef] [PubMed]
- Leroch, M.; Plesken, C.; Weber, R.W.S.; Kauff, F.; Scalliet, G.; Hahn, M. Gray Mold Populations in German Strawberry Fields Are Resistant to Multiple Fungicides and Dominated by a Novel Clade Closely Related to Botrytis cinerea. Appl. Environ. Microbiol. 2013, 79, 159–167. [Google Scholar] [CrossRef]
- Grabke, A.; Stammler, G. A Botrytis cinerea Population from a Single Strawberry Field in Germany Has a Complex Fungicide Resistance Pattern. Plant Dis. 2015, 99, 1078–1086. [Google Scholar] [CrossRef] [PubMed]
- Omrane, S.; Sghyer, H.; Audéon, C.; Lanen, C.; Duplaix, C.; Walker, A.-S.; Fillinger, S. Fungicide Efflux and the MgMFS1 Transporter Contribute to the Multidrug Resistance Phenotype in Zymoseptoria tritici Field Isolates: Fungicide Efflux & MgMFS 1 Contribute to MDR in Z. tritici. Environ. Microbiol. 2015, 17, 2805–2823. [Google Scholar] [CrossRef]
- Holmes, A.R.; Cardno, T.S.; Strouse, J.J.; Ivnitski-Steele, I.; Keniya, M.V.; Lackovic, K.; Monk, B.C.; Sklar, L.A.; Cannon, R.D. Targeting Efflux Pumps to Overcome Antifungal Drug Resistance. Future Med. Chem. 2016, 8, 1485–1501. [Google Scholar] [CrossRef]
- Palani, P.V.; Lalithakumari, D. Resistance of Venturia inaequalis to the Sterol Biosynthesis-Inhibiting Fungicide, Penconazole [1-(2-(2,4-Dichlorophenyl) Pentyl)-1H-1,2,4-Triazole]. Mycol. Res. 1999, 103, 1157–1164. [Google Scholar] [CrossRef]
- Koopman, T.A.; Meitz-Hopkins, J.C.; Tobutt, K.R.; Bester, C.; Lennox, C.L. Pathogenicity and Virulence of South African Isolates of Venturia inaequalis. Eur. J. Plant Pathol. 2022, 164, 45–58. [Google Scholar] [CrossRef]
- Corkley, I.; Fraaije, B.; Hawkins, N. Fungicide Resistance Management: Maximizing the Effective Life of Plant Protection Products. Plant Pathol. 2022, 71, 150–169. [Google Scholar] [CrossRef]
- Van Den Bosch, F.; Paveley, N.; Van Den Berg, F.; Hobbelen, P.; Oliver, R. Mixtures as a Fungicide Resistance Management Tactic. Phytopathology 2014, 104, 1264–1273. [Google Scholar] [CrossRef] [PubMed]
- Chen, J.; Wei, J.; Fu, L.; Wang, S.; Liu, J.; Guo, Q.; Jiang, J.; Tian, Y.; Che, Z.; Chen, G.; et al. Tebuconazole Resistance of Fusarium graminearum Field Populations from Wheat in Henan Province. J. Phytopathol. 2021, 169, 525–532. [Google Scholar] [CrossRef]
- Keinath, A.P.; Rennberger, G.; Wechter, P. Widespread Resistance to Tebuconazole and Cross-Resistance to Other DMI Fungicides in Stagonosporopsis Citrulli Isolated from Watermelon in South Carolina. Plant Dis. 2023. PDIS-03-23-0478-RE. [Google Scholar] [CrossRef] [PubMed]
- Beckerman, J.L.; Sundin, G.W.; Rosenberger, D.A. Do Some IPM Concepts Contribute to the Development of Fungicide Resistance? Lessons Learned from the Apple Scab Pathosystem in the United States. Pest Manag. Sci. 2015, 71, 331–342. [Google Scholar] [CrossRef] [PubMed]
- Franco Ortega, S.; Prencipe, S.; Gullino, M.L.; Spadaro, D. New Molecular Tool for a Quick and Easy Detection of Apple Scab in the Field. Agronomy 2020, 10, 581. [Google Scholar] [CrossRef]
- Gadoury, D.M.; Seem, R.C.; MacHardy, W.E.; Wilcox, W.F.; Rosenberger, D.A.; Stensvand, A. A Comparison of Methods Used to Estimate the Maturity and Release of Ascospores of Venturia inaequalis. Plant Dis. 2004, 88, 869–874. [Google Scholar] [CrossRef]
- Meitz-Hopkins, J.C.; Von Diest, S.G.; Koopman, T.A.; Bahramisharif, A.; Lennox, C.L. A Method to Monitor Airborne Venturia inaequalis Ascospores Using Volumetric Spore Traps and Quantitative PCR. Eur. J. Plant Pathol. 2014, 140, 527–541. [Google Scholar] [CrossRef]
- Daniëls, B.; De Landtsheer, A.; Dreesen, R.; Davey, M.W.; Keulemans, J. Real-Time PCR as a Promising Tool to Monitor Growth of Venturia spp. in Scab-Susceptible and -Resistant Apple Leaves. Eur. J. Plant Pathol. 2012, 134, 821–833. [Google Scholar] [CrossRef]
- Prencipe, S.; Sillo, F.; Garibaldi, A.; Gullino, M.L.; Spadaro, D. Development of a Sensitive TaqMan qPCR Assay for Detection and Quantification of Venturia inaequalis in Apple Leaves and Fruit and in Air Samples. Plant Dis. 2020, 104, 2851–2859. [Google Scholar] [CrossRef] [PubMed]
- Goddard, M.R.; Godfray, H.C.J.; Burt, A. Sex Increases the Efficacy of Natural Selection in Experimental Yeast Populations. Nature 2005, 434, 636–640. [Google Scholar] [CrossRef] [PubMed]
- Ehrlich, P. Address in Pathology, ON CHEMIOTHERAPY: Delivered before the Seventeenth International Congress of Medicine. BMJ 1913, 2, 353–359. [Google Scholar] [CrossRef] [PubMed]
- Brent, K.J.; Hollomon, D.W. Fungicide Resistance in Crop Pathogens: How Can It Be Managed? In FRAC Monograph, 2nd ed.; Fungicide Resistance Action Committee: Brussels, Belgium, 2007; ISBN 90-72398-07-6. [Google Scholar]
- Van Den Bosch, F.; Paveley, N.; Shaw, M.; Hobbelen, P.; Oliver, R. The Dose Rate Debate: Does the Risk of Fungicide Resistance Increase or Decrease with Dose? The Dose Rate Debate. Plant Pathol. 2011, 60, 597–606. [Google Scholar] [CrossRef]
- Koh, H.S.; Sohn, S.H.; Lee, Y.S.; Koh, Y.J.; Song, J.H.; Jung, J.S. Specific and Sensitive Detection of Venturia nashicola, the Scab Fungus of Asian Pears, by Nested PCR. Plant Pathol. J. 2013, 29, 357–363. [Google Scholar] [CrossRef]
- Gutierrez Vazquez, Y.; Adams, I.P.; McGreig, S.; Walshaw, J.; van den Berg, F.; Sanderson, R.; Pufal, H.; Conyers, C.; Langton, D.; Broadhead, R.; et al. Profiling Azole Resistant Haplotypes within Zymoseptoria tritici Populations Using Nanopore Sequencing. Front. Agron. 2022, 4, 943440. [Google Scholar] [CrossRef]
- Giolai, M.; Verweij, W.; Pearson, N.; Nicholson, P.; Leggett, R.M.; Clark, M.D. Air-Seq: Measuring Air Metagenomic Diversity in an Agricultural Ecosystem. bioRxiv 2022. [Google Scholar] [CrossRef]
- van den Bosch, F.; Fraaije, B.; Oliver, R.; van den Berg, F.; Paveley, N. The Use of Mathematical Models to Guide Fungicide Resistance Management Decisions. In Fungicide Resistance in Plant Pathogens; Ishii, H., Hollomon, D.W., Eds.; Springer: Tokyo, Japan, 2015; pp. 49–62. ISBN 978-4-431-55641-1. [Google Scholar]


| Linkage Group | Size (bp) | SNPs | Informative SNPs |
|---|---|---|---|
| 1 | 17,896,836 | 12,003 | 241 |
| 2 | 15,525,007 | 10,572 | 201 |
| 3 | 6,231,899 | 5506 | 84 |
| 4 | 3,556,600 | 2427 | 53 |
| 5 | 2,751,175 | 2122 | 33 |
| 6 | 9,505,949 | 7500 | 147 |
| 7 | 4,036,205 | 3407 | 51 |
| 8 | 2,557,858 | 2250 | 33 |
| 9a | 1,643,167 | 588 | 11 |
| 9b | 322 | 6 | |
| 10 | 612,523 | 30 | 1 |
| Dose | LG1 QTL | LG7 QTL | LG1 QTL:LG7 QTL | Variability Explained (%) |
|---|---|---|---|---|
| ED10 | 5.56 × 10−5 | 5.41 × 10−4 | 0.19 | 45.42 |
| ED50 | 5.09 × 10−3 | 9.09 × 10−6 | 0.69 | 44.31 |
| ED90 | 0.54 | 1.18 × 10−3 | 0.66 | 22.73 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Heaven, T.; Armitage, A.D.; Xu, X.; Goddard, M.R.; Cockerton, H.M. Dose-Dependent Genetic Resistance to Azole Fungicides Found in the Apple Scab Pathogen. J. Fungi 2023, 9, 1136. https://doi.org/10.3390/jof9121136
Heaven T, Armitage AD, Xu X, Goddard MR, Cockerton HM. Dose-Dependent Genetic Resistance to Azole Fungicides Found in the Apple Scab Pathogen. Journal of Fungi. 2023; 9(12):1136. https://doi.org/10.3390/jof9121136
Chicago/Turabian StyleHeaven, Thomas, Andrew D. Armitage, Xiangming Xu, Matthew R. Goddard, and Helen M. Cockerton. 2023. "Dose-Dependent Genetic Resistance to Azole Fungicides Found in the Apple Scab Pathogen" Journal of Fungi 9, no. 12: 1136. https://doi.org/10.3390/jof9121136






