Novel Lactic Acid Bacteria Strains from Regional Peppers with Health-Promoting Potential
Abstract
:1. Introduction
2. Materials and Methods
2.1. Isolation and LAB Identification
2.1.1. Source
2.1.2. Isolation of Peppers’ Bacterial Strains
2.1.3. Identification of LAB Isolates
2.1.4. Bacterial Strain Identification
2.2. Stress Tolerance
2.2.1. pH Resistance
2.2.2. Bile Tolerance
2.2.3. Resistance to Sequential Exposition of Simulated Gastric and Intestinal Juices
2.3. Adhesion Capacity
2.3.1. Surface Hydrophobicity
2.3.2. Auto-Aggregation
2.4. Safety Assessment
2.4.1. Haemolytic Activity
2.4.2. Antibiotic Sensitivity Test
2.5. Technological Properties
2.5.1. Compatibility of Strains
2.5.2. NaCl Resistance
2.5.3. Acidification Capacity
2.6. Biosurfactant Production
2.6.1. Surfactant Capacity
2.6.2. Emulsifying Capacity
2.7. Antipathogenic Activity
2.7.1. Inhibition of Bacterial Pathogenic Biofilm Adhesion
2.7.2. Co-Aggregation with Pathogens
2.8. Antimutagenic Activity
2.9. Statistical Analysis
3. Results
3.1. Isolation and Strains Identification
3.2. Bacterial Stress Tolerance
3.2.1. pH Resistance
3.2.2. Bile Tolerance
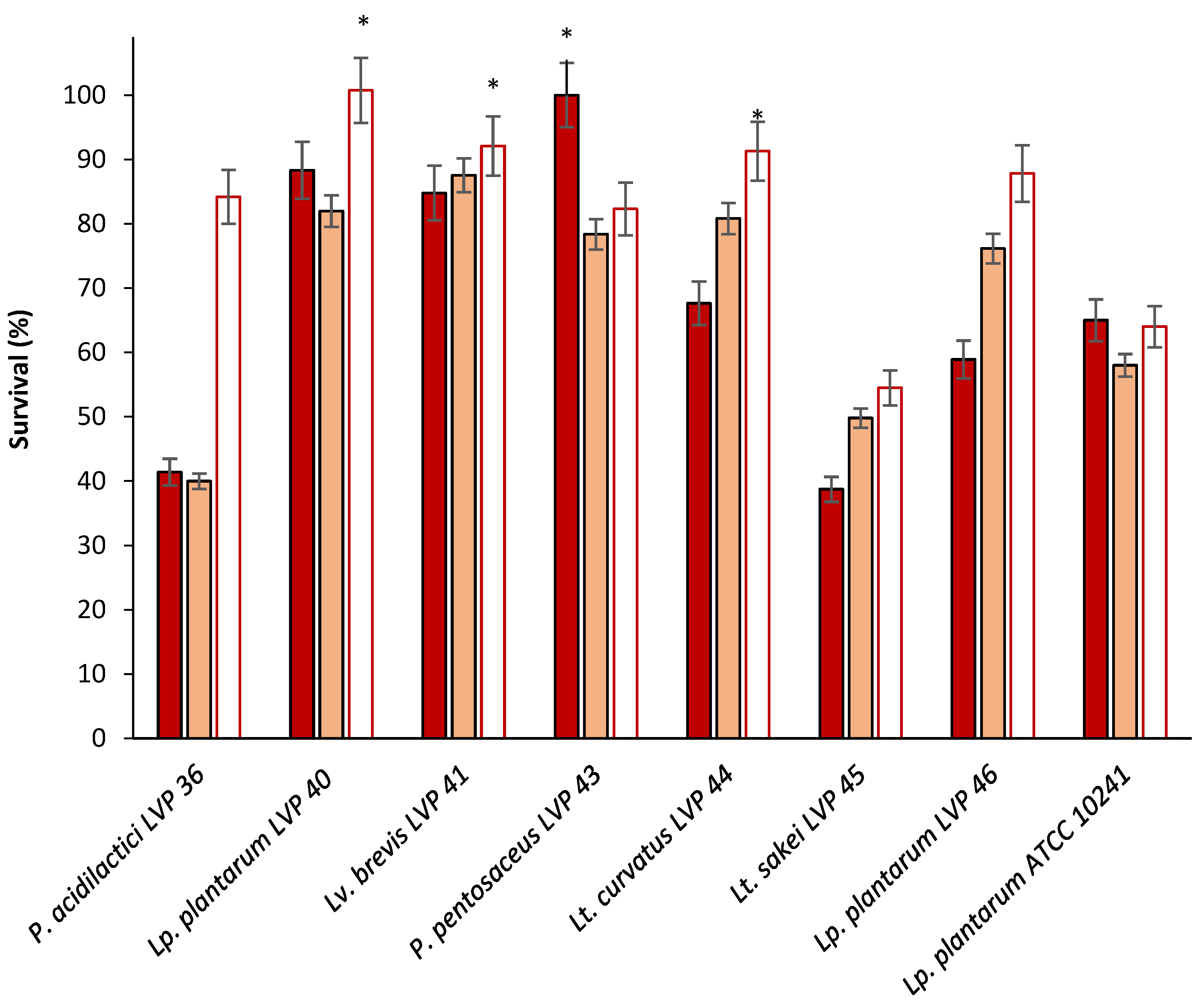
3.2.3. Resistance to Sequential Exposition to Stimulated Gastric and Intestinal Juice
3.3. Adhesion Capacity
3.3.1. Surface Hydrophobicity
3.3.2. Auto-Aggregation Ability
3.4. Safety Assessment
3.4.1. Haemolytic Activity
3.4.2. Antibiotic Sensitivity Test
3.5. Technological Properties
3.5.1. Compatibility of Strains
3.5.2. NaCl Resistance
3.5.3. Acidification Capacity
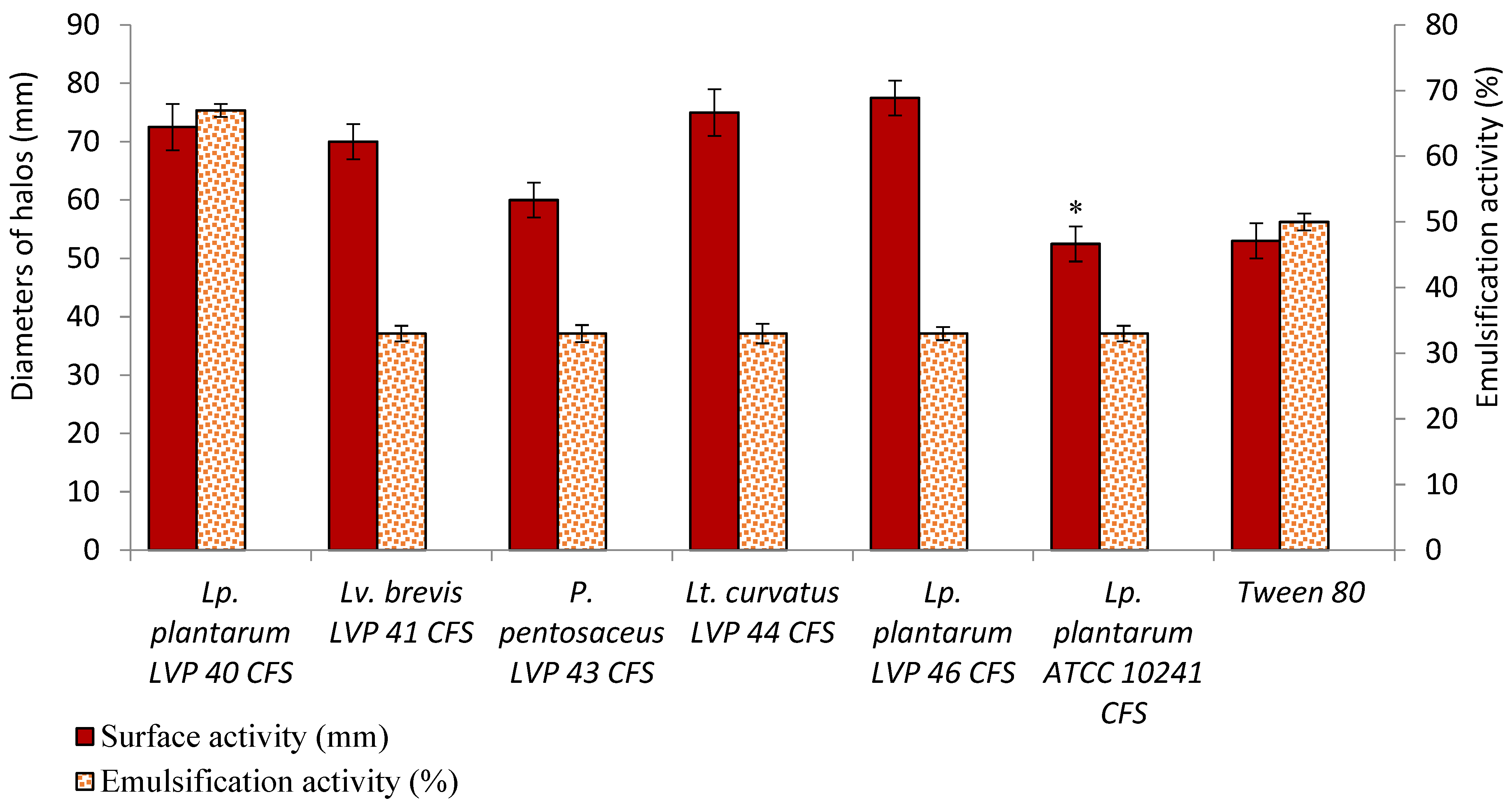
3.6. Surfactant and Emulsifying Properties
3.7. Antipathogenic Activity
3.7.1. Inhibition of Bacterial Pathogenic Biofilm Adhesion
3.7.2. Co-Aggregation with Pathogens
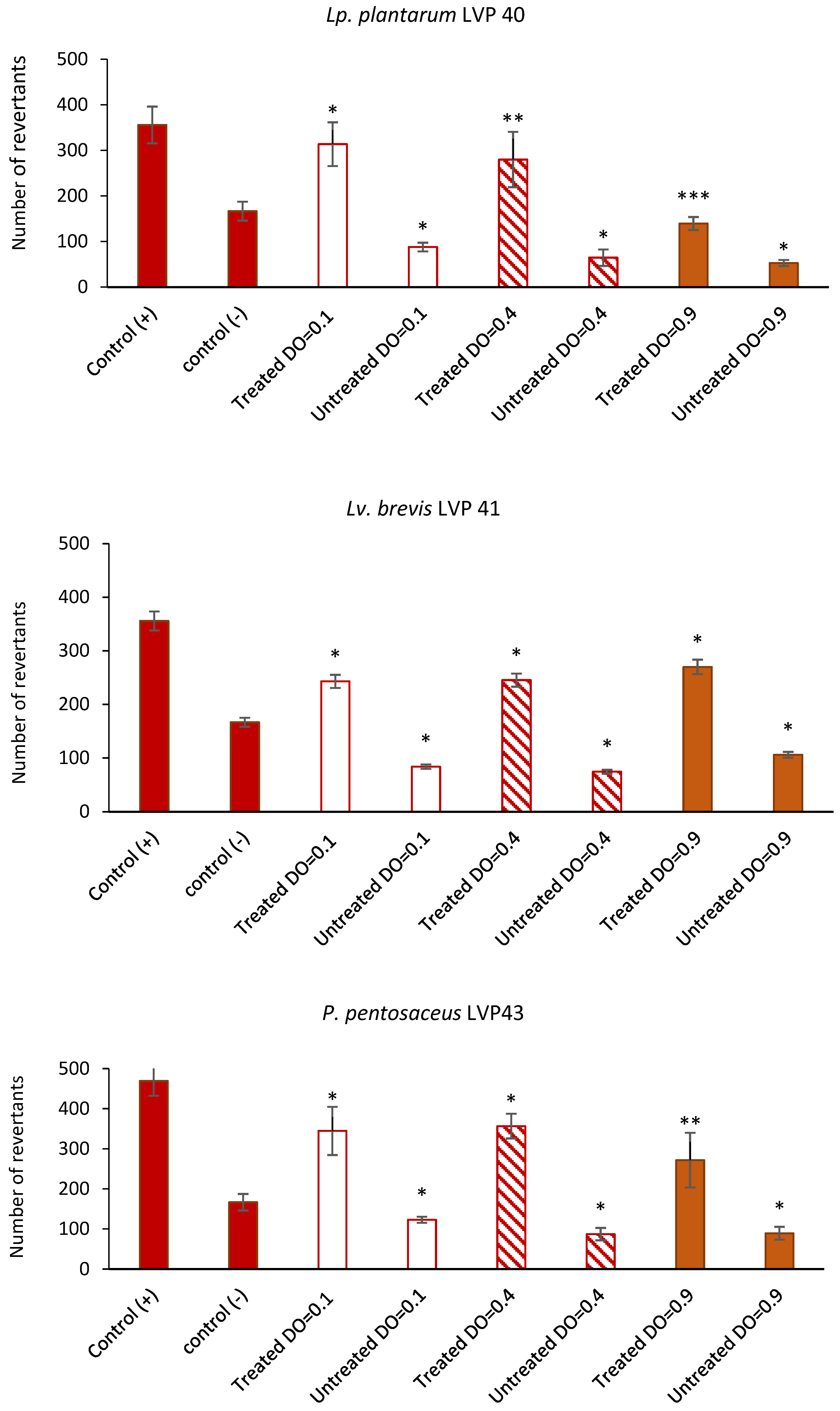
3.8. Antimutagenic Capacity of Lactic Acid Bacteria
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Chakraborty, R.; Roy, S. Exploration of the diversity and associated health benefits of traditional pickles from the Himalayan and adjacent hilly regions of Indian subcontinent. J. Food Sci. Technol. 2018, 55, 1599–1613. [Google Scholar] [CrossRef]
- Zapaśnik, A.; Sokołowska, B.; Bryła, M. Role of lactic acid bacteria in food preservation and safety. Foods 2022, 11, 1283. [Google Scholar] [CrossRef] [PubMed]
- Behera, S.S.; El Sheikha, A.F.; Hammami, R.; Kumar, A. Traditionally fermented pickles: How the microbial diversity associated with their nutritional and health benefits? J. Funct. Foods 2020, 70, 103971. [Google Scholar] [CrossRef]
- Casarotti, S.N.; Borgonovi, T.F.; Batista, C.L.; Penna, A.L.B. Guava, orange and passion fruit by-products: Characterization and its impacts on kinetics of acidification and properties of probiotic fermented products. LWT-Food Sci. Technol. 2018, 98, 69–76. [Google Scholar] [CrossRef]
- Apás, A.L.; Gonzalez, S.N.; Arena, M.E. Potential of goat probiotic to bind mutagens. Anaerobe 2014, 28, 8–12. [Google Scholar] [CrossRef] [PubMed]
- Lane, D.J. 16S/23S rRNA sequencing. In Nucleic Acid Techniques in Bacterial Systematics; Stackebrandt, E., Goodfellow, M., Eds.; Wiley: New York, NY, USA, 1991; pp. 115–175. [Google Scholar]
- Maldonado, N.C.; Aristimuño Ficoseco, C.; Mansilla, F.I.; Melián, C.; Hébert, E.M.; Vignolo, G.M.; Nader-Macías, M.E.F. Identification, characterization and selection of autochthonous lactic acid bacteria as probiotic for feedlot cattle. Livest. Sci. 2018, 212, 99–110. [Google Scholar] [CrossRef]
- Versalovic, J. Genomic fingerprinting of bacteria using repetitive sequence-based polymerase chain reaction. Methods Mol. Cell Biol. 1994, 5, 25–40. [Google Scholar]
- Heras, J.; Domínguez, C.; Mata, E.; Pascual, V.; Lozano, C.; Torres, C.; Zarazaga, M. GelJ–a tool for analyzing DNA fingerprint gel images. BMC Bioinform. 2015, 16, 270. [Google Scholar] [CrossRef] [PubMed]
- Ishii, S.; Sadowsky, M.J. Applications of the rep-PCR DNA fingerprinting technique to study microbial diversity, ecology and evolution. Environ. Microbiol. 2009, 11, 733–740. [Google Scholar] [CrossRef] [PubMed]
- Gilliland, S.E.; Staley, T.E.; Bush, L.J. Importance of bile tolerance of Lactobacillus acidophilus used as dietary adjunct. J. Dairy Sci. 2004, 67, 3045–3051. [Google Scholar] [CrossRef] [PubMed]
- Vera-Mejía, R.; Ormaza-Donoso, J.; Muñoz-Cedeño, J.; Arteaga-Chávez, F.; Sánchez-Miranda, L. Cepas de Lactobacillus plantarum con potencialidades probióticas aisladas de cerdos criollos. Rev. Salud Anim. 2018, 40, 1–12. [Google Scholar]
- Zárate, G.; Chaia, A.P.; González, S.; Oliver, G. Viability and β-galactosidase activity of dairy propionibacteria subjected to digestion by artificial gastric and intestinal fluids. J. Food Prot. 2000, 63, 1214–1221. [Google Scholar] [CrossRef]
- Hojjati, M.; Behabahani, B.A.; Falah, F. Aggregation, adherence, anti-adhesion and antagonistic activity properties relating to surface charge of probiotic Lactobacillus brevis gp104 against Staphylococcus aureus. Microb. Pathog. 2020, 147, 104420. [Google Scholar] [CrossRef] [PubMed]
- Tyfa, A.; Kunicka-Styczyńska, A.; Zabielska, J. Evaluation of hydrophobicity and quantitative analysis of biofilm formation by Alicyclobacillus sp. Acta Biochim. Pol. 2015, 62, 785–790. [Google Scholar] [CrossRef] [PubMed]
- Parente, E.; Brienza, C.; Moles, M.; Ricciardi, A. A comparison of methods for the measurement of bacteriocin activity. J. Microbiol. Methods 1995, 22, 95–108. [Google Scholar] [CrossRef]
- Cartagena, E.; Orphèe, C.H.; Verni, M.C.; Arena, M.E.; González, S.N.; Argañaraz, M.I.; Bardón, A. Medio de Cultivo Promotor y Bacterias No Patógenas Detoxificantes de Compuestos Mutagénicos/Carcinogénicos. Instituto Nacional de la Propiedad Industrial-INPI. Patent No. 20190102418, 25 March 2021. [Google Scholar]
- Walter, V.; Syldatk, C.; Hausmann, R. Screening Concepts for the Isolation of Biosurfactant Producing Microorganisms. In Biosurfactants. Advances in Experimental Medicine and Biology; Sen, R., Ed.; Springer: New York, NY, USA, 2010; pp. 1–13. [Google Scholar]
- Sambanthamoorthy, K.; Feng, X.; Patel, R.; Patel, S.; Paranavitana, C. Antimicrobial and antibiofilm potential of biosurfactants isolated from lactobacilli against multi-drug-resistant pathogens. BMC Microbiol. 2014, 14, 197. [Google Scholar] [CrossRef] [PubMed]
- Verni, M.C.; Orphèe, C.H.; González, S.N.; Bardón, A.; Arena, M.E.; Cartagena, E. Flourensia fiebrigii SF Blake in combination with Lactobacillus paracasei subsp. paracasei CE75. A novel antipathogenic and detoxifying strategy. LWT 2022, 156, 113023. [Google Scholar]
- Tateda, K.; Comte, R.; Pechere, J.C.; K¨ohler, T.; Yamaguchi, K.; Van Delden, C. Azithromycin inhibits quorum sensing in Pseudomonas aeruginosa. Antimicrob. Agents Chemother. 2001, 45, 1930–1933. [Google Scholar] [CrossRef]
- Sandasi, M.; Leonard, C.M.; Van Vuuren, S.F.; Viljoen, A.M. Peppermint (Mentha piperita) inhibits microbial biofilms in vitro. S. Afr. J. Bot. 2011, 77, 80–85. [Google Scholar] [CrossRef]
- Díaz, M.A.; Alberto, M.R.; Vega-Hissi, E.G.; Gonzalez, S.N.; Arena, M.E. Interference in Staphylococcus aureus. Biofilm and virulence factors production by human probiotic bacteria with antimutagenic activity. Arab. J. Sci. Eng. 2022, 47, 241–253. [Google Scholar] [CrossRef]
- Maron, D.M.; Ames, B.N. Revised methods for the Salmonella mutagenicity test. Mutat. Res. Sect. Environ. Mutagen. Relat. Subj. 1983, 113, 173–215. [Google Scholar] [CrossRef] [PubMed]
- Şanlier, N.; Gökcen, B.B.; Sezgin, A.C. Health benefits of fermented foods. Crit. Rev. Food Sci. Nutr. 2019, 59, 506–527. [Google Scholar] [CrossRef]
- García-Burgos, M.; Moreno-Fernández, J.; Alférez, M.J.; Díaz-Castro, J.; López-Aliaga, I. New perspectives in fermented dairy products and their health relevance. J. Funct. Foods 2020, 72, 104059. [Google Scholar] [CrossRef]
- Sionek, B.; Szydłowska, A.; Kucukgoz, K.; Kołożyn-Krajewska, D. Traditional and new microorganisms in lactic acid fermentation of food. Fermentation 2023, 9, 1019. [Google Scholar] [CrossRef]
- Thierry, A.; Baty, C.; Marché, L.; Chuat, V.; Picard, O.; Lortal, S.; Valence, F. Lactofermentation of vegetables: An ancient method of preservation matching new trends. Trends Food Sci. Technol. 2023, 139, 104112. [Google Scholar] [CrossRef]
- Khalil, E.S.; Abd Manap, M.Y.; Mustafa, S.; Alhelli, A.M.; Shokryazdan, P. Probiotic properties of exopolysaccharide-producing Lactobacillus strains isolated from tempoyak. Molecules 2018, 23, 398. [Google Scholar] [CrossRef] [PubMed]
- Ali, M.S.; Lee, E.B.; Lim, S.K.; Suk, K.; Park, S.C. Isolation and identification of Limosilactobacillus reuteri PSC102 and evaluation of its potential probiotic, antioxidant and antibacterial properties. Antioxidants 2023, 12, 238. [Google Scholar] [CrossRef] [PubMed]
- Hyrslova, I.; Drab, V.; Cihlar, J.; Krausova, G.; Mrvikova, I.; Kana, A.; Stetina, J.; Musilova, S. Functional and probiotic characterization of newly isolated strains from infant feces and breast milk. Fermentation 2023, 9, 960. [Google Scholar] [CrossRef]
- Han, K.; Park, S.; Sathiyaseelan, A.; Wang, M.H. Isolation and characterization of Enterococcus faecium from fermented Korean soybean paste with antibacterial effects. Fermentation 2023, 9, 760. [Google Scholar] [CrossRef]
- de Souza, B.M.S.; Borgonovi, T.F.; Casarotti, S.N.; Todorov, S.D.; Penna, A.L.B. Lactobacillus casei and Lactobacillus fermentum strains isolated from mozzarella cheese: Probiotic potential, safety, acidifying kinetic parameters and viability under gastrointestinal tract condition. Probiotics Antimicrob. Proteins 2019, 11, 382–396. [Google Scholar] [CrossRef] [PubMed]
- Damaceno, Q.S.; Gallotti, B.; Reis, I.M.; Totte, Y.C.; Assis, G.B.; Figueiredo, H.C.; Silva, T.F.; Azevedo, V.; Nicoli, J.R.; Martins, F.S. Isolation and identification of potential probiotic bacteria from human milk. Probiotics Antimicrob. Proteins 2021, 15, 491–501. [Google Scholar] [CrossRef] [PubMed]
- Rodrigues da Cunha, L.; Ferreira, C.L.F.; Durmaz, E.; Goh, Y.J.; Sanozky-Dawes, R.; Klaenhammer, T. Characterization of Lactobacillus gasseri isolates from a breastfed infant. Gut Microbes 2012, 3, 15–24. [Google Scholar] [CrossRef] [PubMed]
- Georgieva, R.; Yocheva, L.; Tserovska, L.; Zhelezova, G.; Stefanova, N.; Atanasova, A.; Danguleva, A.; Ivanova, G.; Karapetkov, N.; Rumyan, N.; et al. Antimicrobial activity and antibiotic susceptibility of Lactobacillus and Bifidobacterium spp. intended for use as starter and probiotic cultures. Biotechnol. Biotechnol. Equip. 2015, 29, 84–91. [Google Scholar] [CrossRef] [PubMed]
- Campedelli, I.; Mathur, H.; Salvetti, E.; Clarke, S.; Rea, M.C.; Torriani, S.; Ross, R.P.; Hill, C.; O’Tool, P.W. Genus-wide assessment of antibiotic resistance in Lactobacillus spp. Appl. Environ. Microbiol. 2019, 85, e01738-18. [Google Scholar] [CrossRef] [PubMed]
- Jeyagowri, N.; Ranadheera, C.S.; Manap, M.Y.; Gamage, A.; Merah, O.; Madhujith, T. Phenotypic characterisation and molecular identification of potentially probiotic Lactobacillus sp. isolated from fermented rice. Fermentation 2023, 9, 807. [Google Scholar] [CrossRef]
- Yao, A.A.; Dortu, C.; Egounlety, M.; Pinto, C.; Vinodh, A.E.; Huch, M.; Franz, C.M.A.P.; Holzapfel, W.; Mbugua, S.; Mengu, M.; et al. Production of freeze-dried lactic acid bacteria starter culture for cassava fermentation into gari. Afr. J. Biotechnol. 2009, 8, 4996–5004. [Google Scholar]
- Markande, A.R.; Patel, D.; Varjani, S. A review on biosurfactants: Properties, applications and current developments. Bioresour. Technol. 2021, 330, 124963. [Google Scholar] [CrossRef]
- Vecino, X.; Rodríguez-López, L.; Ferreira, D.; Cruz, J.M.; Moldes, A.B.; Rodrigues, L.R. Bioactivity of glycolipopeptide cell-bound biosurfactants against skin pathogens. Int. J. Biol. Macromol. 2018, 109, 971–979. [Google Scholar] [CrossRef]
- Yan, X.; Gu, S.; Cui, X.; Shi, Y.; Wen, S.; Chen, H.; Ge, J. Antimicrobial, anti-adhesive and anti-biofilm potential of biosurfactants isolated from Pediococcus acidilactici and Lactobacillus plantarum against Staphylococcus aureus CMCC26003. Microb. Pathog. 2019, 127, 12–20. [Google Scholar] [CrossRef]
- Verni, M.C.; Garay, J.A.; Mendoza, L.; Bardón, A.; Borkosky, S.; Arena, M.E.; Cartagena, E. Lipophilic 9, 10-dehydrofukinone action on pathogenic and non-pathogenic bacterial biofilms. Why is this main volatile metabolite in Senecio? Chem. Biodivers. 2020, 17, e1900507. [Google Scholar] [CrossRef]
- Goŀek, P.; Bednarski, W.; Brzozowski, B.; Dziuba, B. The obtaining and properties of biosurfactants synthesized by bacteria of the genus Lactobacillus. Ann. Microbiol. 2009, 59, 119–126. [Google Scholar] [CrossRef]
- Fracchia, L.; Cavallo, M.; Allegrone, G.; Martinotti, M.G.A. Lactobacillus-derived biosurfactant inhibits biofilm formation of human pathogenic Candida albicans biofilm producers. Appl. Microbiol. Biotechnol. 2010, 2, 827–837. [Google Scholar]
- Ismaeel, M.C.; Ibrahim, K.M.; Al-Malikey, M.K. The effect of surlactin produced by Lactobacillus acidophilus on eye infectious bacteria in rabbits. Baghdad Sci. J. 2013, 10, 133–143. [Google Scholar] [CrossRef]
- De Gregorio, P.R.; Parolin, C.; Abruzzo, A.; Luppi, B.; Protti, M.; Mercolini, L.; Silva, J.A.; Giordani, B.; Marangoni, A.; Nader-Macías, M.E.F.; et al. Biosurfactant from vaginal Lactobacillus crispatus BC1 as a promising agent to interfere with Candida adhesion. Microb. Cell Factories 2020, 19, 133. [Google Scholar] [CrossRef]
- Bader, D.; Lee, D. Structure-properties of Soft Tissues Articular Cartilage. In Structural Biological Materials: Design and Structure Property Relationships; Elices, M., Ed.; Pergamon: Amsterdam, The Netherlands, 2000; pp. 73–104. [Google Scholar]
- Carniello, V.; Peterson, B.W.; Van der Mei, H.C.; Busscher, H.J. Physico-chemistry from initial bacterial adhesion to surface-programmed biofilm growth. Adv. Colloid Interface Sci. 2018, 261, 1–14. [Google Scholar] [CrossRef] [PubMed]
- Gudiña, E.J.; Rocha, V.; Teixeira, J.A.; Rodrigues, L.R. Antimicrobial and antiadhesive properties of a biosurfactant isolated from Lactobacillus paracasei ssp. paracasei A20. Lett. Appl. Microbiol. 2010, 50, 419–424. [Google Scholar] [CrossRef] [PubMed]
- Batoni, G.; Kaya, E.; Catelli, E.; Quinti, S.; Botti, M.; De Carli, A.; Bianchi, M.; Maisetta, G.; Esin, S. Lactobacillus probiotic strains differ in their ability to adhere to human lung epithelial cells and to prevent adhesion of clinical isolates of Pseudomonas aeruginosa from cystic fibrosis lung. Microorganisms 2023, 11, 1707. [Google Scholar] [CrossRef] [PubMed]
- Ramos, A.N.; Sesto Cabral, M.E.; Arena, M.E.; Arrighi, C.F.; Arroyo Aguilar, A.A.; Valdez, J.C. Compounds from Lactobacillus plantarum culture supernatants with potential pro-healing and antipathogenic properties in skin chronic wounds. Pharm. Biol. 2015, 53, 350–358. [Google Scholar] [CrossRef] [PubMed]
- Valdéz, J.C.; Peral, M.C.; Rachid, M.; Santana, M.; Perdigón, G. Interference of Lactobacillus plantarum with Pseudomonas aeruginosa in vitro and in infected burns: The potential use of probiotics in wound treatment. Clin. Microbiol. Infect. 2005, 11, 472–479. [Google Scholar] [CrossRef]
- Díaz, M.A.; González, S.N.; Alberto, M.R.; Arena, M.E. Human probiotic bacteria attenuate Pseudomonas aeruginosa biofilm and virulence by quorum-sensing inhibition. Biofouling 2020, 36, 597–609. [Google Scholar] [CrossRef] [PubMed]
- Davares, A.K.L.; Arsene, M.M.J.; Viktorovna, P.I.; Vyacheslavovna, Y.N.; Vladimirovna, Z.A.; Aleksandrovna, V.E.; Nikolayevich, S.A.; Nadezhda, S.; Anatolievna, G.O.; Nikolaevna, S.I.; et al. Quorum-sensing inhibitors from probiotics as a strategy to combat bacterial cell-to-cell communication involved in food spoilage and food safety. Fermentation 2022, 8, 711. [Google Scholar] [CrossRef]
- Kampouris, I.D.; Karayannakidis, P.D.; Banti, D.C.; Sakoula, D.; Konstantinidis, D.; Yiangou, M.; Samaras, P.E. Evaluation of a novel quorum quenching strain for MBR biofouling mitigation. Water Res. 2018, 143, 56–65. [Google Scholar] [CrossRef] [PubMed]
- Liang, Y.; Pan, Y.; Li, Q.; Wu, B.; Hu, M. RNA-seq-based transcriptomic analysis of AHL-induced biofilm and pyocyanin inhibition in Pseudomonas aeruginosa by Lactobacillus brevis. Int. Microbiol. 2022, 25, 447–456. [Google Scholar] [CrossRef] [PubMed]
- Collado, M.C.; Meriluot, J.; Salminen, S. Measurement of aggregation properties between probiotics and pathogens: In vitro evaluation of different methods. J. Microbiol. Methods 2007, 71, 71–74. [Google Scholar] [CrossRef] [PubMed]
- Kumar, M.; Kumar, A.; Nagpal, R.; Mohania, D.; Behare, P.; Verma, V.; Kumar, P.; Poddar, D.; Aggarwal, P.K.; Henry, C.J.K.; et al. Cancer-preventing attributes of probiotics: An update. Int. J. Food Sci. Nutr. 2010, 61, 473–496. [Google Scholar] [CrossRef] [PubMed]



| Bacteria | Access Number GenBank | BLAST | Peppers | Origin |
|---|---|---|---|---|
| Lactilactobacillus curvatus LVP 32 | SUB5345111 LVP32 MK659876 | 100% | Red/Green | DG |
| Lactiplantibacillus plantarum LVP 33 | SUB5659046 LVP33 MK965193 | 100% | Green | M |
| Lactilactobacillus curvatus LVP 34 | SUB5345224 LVP34 MK659883 | 99.86% | Leaf | DG |
| Enterococcus casseliflavus LVP 35 | SUB5345348 LVP35 MK659877 | 99.93% | Green | M |
| Pediococcus acidilactici LVP 36 | SUB5659133 LVP36 MK965101 | 99.93% | Red | M |
| Leuconostoc mesenteroides LVP 37 | SUB5345695 LVP37 MK659879 | 97.38% | Red | DG |
| Lactilactobacillus curvatus LVP 38 | SUB5345760 LVP38 MK659880 | 99.93% | Green | DG |
| Lactilactobacillus curvatus LVP 39 | SUB5349021 LVP39 MK676004 | 100% | Green | DG |
| Lactiplantibacillus plantarum LVP 40 | SUB5349099 LVP40 MK676008 | 100% | Green | M |
| Levilactobacillus brevis LVP 41 | SUB5349336 LVP41 MK676009 | 99.16% | Green | M |
| Weisella cibaria LVP 42 | SUB5515270 LVP42 MK825577 | 100% | Green | M |
| Pediococcus pentosaceus LVP 43 | SUB5349351 LVP43 MK676007 | 100% | Red | DG |
| Lactilactobacillus curvatus LVP 44 | SUB5349355 LVP44 MK676006 | 99.79% | Green | DG |
| Lactilactobacillus sakei LVP 45 | SUB5349726 LVP45 MK676005 | 99.93% | Green | DG |
| Lactiplantibacillus plantarum LVP 46 | SUB5515733 LVP46 MK825575 | 100% | Green | M |
| Bacteria | pH 3 | pH 4 | pH 7 |
|---|---|---|---|
| Enterococcus casseliflavus LVP 35 | 3.20 | 0.07 | 0.18 |
| Pediococcus acidilactici LVP 36 | 2.97 | 0.14 | 0.04 |
| Leuconostoc mesenteroides LVP 37 | 3.85 | 0.58 | 0.007 |
| Lactilactobacillus curvatus LVP 38 | 4.11 | 0.13 | 0.11 |
| Lactiplantibacillus plantarum LVP 40 | 2.33 | 0 | 0.52 |
| Levilactobacillus brevis LVP 41 | 1.46 | 0.26 | 0.07 |
| Weisella cibaria LVP 42 | 7.08 | 1.23 | 0.30 |
| Pediococcus pentosaceus LVP 43 | 1.57 | 0.11 | 0.11 |
| Lactilactobacillus curvatus LVP 44 | 0.78 | 0.003 | 0 |
| Lactilactobacillus sakei LVP 45 | 2.74 | 0.39 | 0.25 |
| Lactiplantibacillus plantarum LVP 46 | 2.67 | 0 | 0.41 |
| Lactiplantibacillus plantarum ATCC 10241 | 6.07 | 0.98 | 0.85 |
| Bacteria | Time (h) to Reach OD = 0.3 | Δ Time (h) | Δ Time (min) | Growth Retardation Criteria | |
|---|---|---|---|---|---|
| Without Bile (Control) | With Bile (Treated) | ||||
| P. acidilactici LVP 36 | 4.6 | 6.2 | 1.4 | 81 | Not tolerant |
| Lp. plantarum LVP 40 | 3.6 | 4 | 0.4 | 24 | Tolerant |
| Lv. brevis LVP 41 | 3.2 | 3.3 | 0.1 | 6 | Resistant |
| P. pentosaceus LVP 43 | 4.4 | 4.9 | 0.5 | 30 | Tolerant |
| Lt. curvatus LVP 44 | 2.1 | 2.6 | 0.5 | 30 | Tolerant |
| Lt. sakei LVP 45 | 2.8 | 4.4 | 1.6 | 96 | Not tolerant |
| Lp. plantarum LVP 46 | 2.6 | 3.2 | 0.6 | 36 | Tolerant |
| Lp. plantarum ATCC 10241 | 3.8 | 4.8 | 1.0 | 60 | Little tolerant |
| Bacteria | Log CFU/mL | |||
|---|---|---|---|---|
| Initial | After Juice Exposition: | |||
| Gastric | Intestinal | |||
| Lp. plantarum LVP 40 | pH 3 | 9.21 | 8.23 | 7.06 |
| pH 4 | 9.40 | 8.29 | 7.83 | |
| Lv. brevis LVP 41 | pH 3 | 9.80 | 7.88 | 7.09 |
| pH 4 | 9.81 | 7.99 | 7.38 | |
| P. pentosaceus LVP 43 | pH 3 | 9.75 | 7.79 | 7.14 |
| pH 4 | 9.88 | 7.92 | 7.17 | |
| Lt. curvatus LVP 44 | pH 3 | 9.96 | 8.04 | 7.18 |
| pH 4 | 9.89 | 8.10 | 7.30 | |
| Lp. plantarum LVP 46 | pH 3 | 9.14 | 8.30 | 8.00 |
| pH 4 | 9.93 | 8.30 | 8.20 | |
| Lp. plantarum ATCC 10241 | pH 3 | 9.20 | 6.40 | 6.03 |
| pH 4 | 9.84 | 7.80 | 7.01 | |
| Bacteria | Strains | Hydrophobicity (%) | ||
|---|---|---|---|---|
| Xylene | Chloroform | Ethyl Acetate | ||
| Lp. plantarum | LVP 40 | 3.88 ± 0.02 a | 9.50 ± 0.00 c | 11.61 ± 0.09 a |
| Lv. brevis | LVP 41 | 84.45 ± 0.19 c | 98.92 ± 0.13 e | 96.92 ± 0.15 e |
| P. pentosaceus | LVP 43 | – | 9.21 ± 0.09 c | 17.16 ± 0.12 b |
| Lt. curvatus | LVP 44 | – | 5.61 ± 0.08 b | 24.55 ± 0.24 c |
| Lp. plantarum | LVP 46 | 11.35 ± 0.09 b | 1.78 ± 0.02 a | 18.06 ± 0.05 b |
| Lp. plantarum | ATCC 10241 | 87.26 ± 0.45 c | 71.70 ± 0.77 d | 74.57 ± 0.73 d |
| Bacteria | Strains | Auto-Aggregation (%) | ||||
|---|---|---|---|---|---|---|
| 1 h | 2 h | 3 h | 4 h | 24 h | ||
| Lp. plantarum | LVP 40 | 11.09 ± 1.50 b | 14.30 ± 0.64 b | 15.28 ± 1.16 b | 18.61 ± 0.04 c | 72.84 ± 7.54 b |
| Lv. brevis | LVP 41 | 4.85 ± 1.54 a | 6.29 ± 0.91 a | 10.61 ± 0.07 a | 10.46 ± 0.61 a | 30.83 ± 2.06 a |
| P. pentosaceus | LVP 43 | 14.57 ± 0.27 b | 21.78 ± 0.53 c | 28.98 ± 1.59 d | 33.67 ± 1.10 d | 72.36 ± 1.59 b |
| Lt. curvatus | LVP 44 | 14.11 ± 1.90 b | 14.09 ± 2.19 b | 23.45 ± 2.00 c | 30.57 ± 4.67 d | 74.27 ± 0.47 b |
| Lp. plantarum | LVP 46 | 10.71 ± 0.93 b | 12.01 ± 0.33 b | 18.09 ± 0.84 b | 19.83 ± 1.64 c | 75.55 ± 1.59 b |
| Lp. plantarum | ATCC 10241 | 4.35 ± 1.92 a | 8.68 ± 0.46 a | 15.27 ± 2.05 b | 16.17 ± 0.35 b | 72.61 ± 3.91 b |
| Groups | ATB | Antimicrobial Susceptibility | |||||
|---|---|---|---|---|---|---|---|
| Lp. plantarum LVP 40 | Lv. brevis LVP 41 | P. pentosaceus LVP 43 | Lt. curvatus LVP 44 | Lp. plantarum LVP 46 | Lp. plantarum ATCC 10241 | ||
| Group 1 inhibitors of cell wall synthesis | Ampicillin | S | S | S | S | S | S |
| Cephalothin | S | S | S | S | S | S | |
| Vancomycin | R | R | R | R | R | R | |
| Group 2 inhibitors of protein synthesis | Chloramphenicol | S | S | S | S | S | S |
| Clindamycin | S | S | S | S | S | S | |
| Erythromycin | S | S | S | S | S | S | |
| Gentamicin | S | S | S | S | S | S | |
| Group 3 inhibitors of nucleic acid synthesis | Ciprofloxacin | R | R | R | R | R | R |
| Norfloxacin | R | R | R | R | R | R | |
| Rifampicin | S | S | S | S | S | S | |
| Lactic Acid Bacteria | Pathogenic Bacteria | % Co-aggregation | |||
|---|---|---|---|---|---|
| 1 h | 4 h | 24 h | |||
| Lp. plantarum LVP40 | P. aeruginosa | ATCC 27853 | 7.83 ± 1.59 c | 25.06 ± 2.83 c | 51.20 ± 3.33 d |
| PAO1 | 9.84 ± 1.64 c | 24.97 ± 1.04 c | 48.14 ± 2.70 d | ||
| S. aureus | ATCC 6538 | 4.31 ± 3.04 B | 4.31 ± 3.04 A | 4.31 ± 3.04 A | |
| HT1 | 5.11 ± 0.74 B | 18.07 ± 2.12 C | 18.07 ± 2.12 B | ||
| Lv. brevis LVP41 | P. aeruginosa | ATCC 27853 | 19.43 ± 4.60 e | 67.46 ± 4.83 d | 100 ± 0.12 e |
| PAO1 | 21.46 ± 1.69 e | 89.16 ± 0.74 d | 100 ± 1.14 e | ||
| S. aureus | ATCC 6538 | 1.04 ± 0.74 A | 1.04 ± 0.74 A | 1.04 ± 0.74 A | |
| HT1 | 0.72 ± 0.51 A | 3.73 ± 1.49 A | 29.37 ± 6.67 C | ||
| P. pentosaceus LVP43 | P. aeruginosa | ATCC 27853 | 6.33 ± 0.11 c | 13.29 ± 3.05 a | 13.29 ± 3.05 a |
| PAO1 | 7.93 ± 1.84 c | 19.08 ± 4.78 b | 20.47 ± 0.99 b | ||
| S. aureus | ATCC 6538 | 9.00 ± 6.36 B,C,D | 15.42 ± 6.36 B,C | 36.99 ± 0.07 C | |
| HT1 | 1.50 ± 0.70 A | 8.03 ± 2.56 B | 37.75 ±18.66 C | ||
| Lt. curvatus LVP44 | P. aeruginosa | ATCC 27853 | 11.18 ± 4.66 d | 26.35 ± 1.74 c | 38.94 ± 0.46 c |
| PAO1 | 4.37 ± 0.27 b | 20.20 ± 1.00 b | 24.35 ± 2.94 b,c | ||
| S. aureus | ATCC 6538 | 7.47 ± 6.67 A,B,C | 68.29 ± 0.11 E | 76.89 ± 0.86 E | |
| HT1 | - | 11.39 ± 1.51 B,C | 36.29 ± 7.61 C | ||
| Lp. plantarum LVP46 | P. aeruginosa | ATCC 27853 | 7.64 ± 0.15 c | 19.52 ± 0.40 b | 34.32 ± 10.47 c |
| PAO1 | 12.39 ± 0.57 d | 23.55 ± 3.13 b,c | 30.22 ± 2.69 c | ||
| S. aureus | ATCC 6538 | - | - | 34.32 ± 10.47 C | |
| HT1 | 2.46 ± 0.50 A,B | 7.43 ± 1.64 B | 18.79 ± 14.39 A,B,C | ||
| Lp. plantarum ATCC10241 | P. aeruginosa | ATCC 27853 | 3.65 ± 0.86 b | 21.84 ± 5.55 b | 40.84 ± 24.42 b,c,d |
| PAO1 | 0.35 ± 0.24 a | 10.00 ± 0.00 a | 48.29 ± 5.03 d | ||
| S. aureus | ATCC 6538 | 14.26 ± 7.95 C,D | 14.26 ± 0.00 C | 14.26 ± 0.00 B | |
| HT1 | 7.46 ± 3.79 B,C | 27.87 ± 6.45 D | 44.82 ± 5.42 C,D | ||
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Nuñez, I.M.; Verni, M.C.; Argañaraz Martinez, F.E.; Babot, J.D.; Terán, V.; Danilovich, M.E.; Cartagena, E.; Alberto, M.R.; Arena, M.E. Novel Lactic Acid Bacteria Strains from Regional Peppers with Health-Promoting Potential. Fermentation 2024, 10, 209. https://doi.org/10.3390/fermentation10040209
Nuñez IM, Verni MC, Argañaraz Martinez FE, Babot JD, Terán V, Danilovich ME, Cartagena E, Alberto MR, Arena ME. Novel Lactic Acid Bacteria Strains from Regional Peppers with Health-Promoting Potential. Fermentation. 2024; 10(4):209. https://doi.org/10.3390/fermentation10040209
Chicago/Turabian StyleNuñez, Ivana Micaela, María Cecilia Verni, Fernando Eloy Argañaraz Martinez, Jaime Daniel Babot, Victoria Terán, Mariana Elizabeth Danilovich, Elena Cartagena, María Rosa Alberto, and Mario Eduardo Arena. 2024. "Novel Lactic Acid Bacteria Strains from Regional Peppers with Health-Promoting Potential" Fermentation 10, no. 4: 209. https://doi.org/10.3390/fermentation10040209
APA StyleNuñez, I. M., Verni, M. C., Argañaraz Martinez, F. E., Babot, J. D., Terán, V., Danilovich, M. E., Cartagena, E., Alberto, M. R., & Arena, M. E. (2024). Novel Lactic Acid Bacteria Strains from Regional Peppers with Health-Promoting Potential. Fermentation, 10(4), 209. https://doi.org/10.3390/fermentation10040209








