Molecular Markers and Regulatory Networks in Solventogenic Clostridium Species: Metabolic Engineering Conundrum
Abstract
:1. Introduction
2. Sigma Factor 54 Directed Regulation
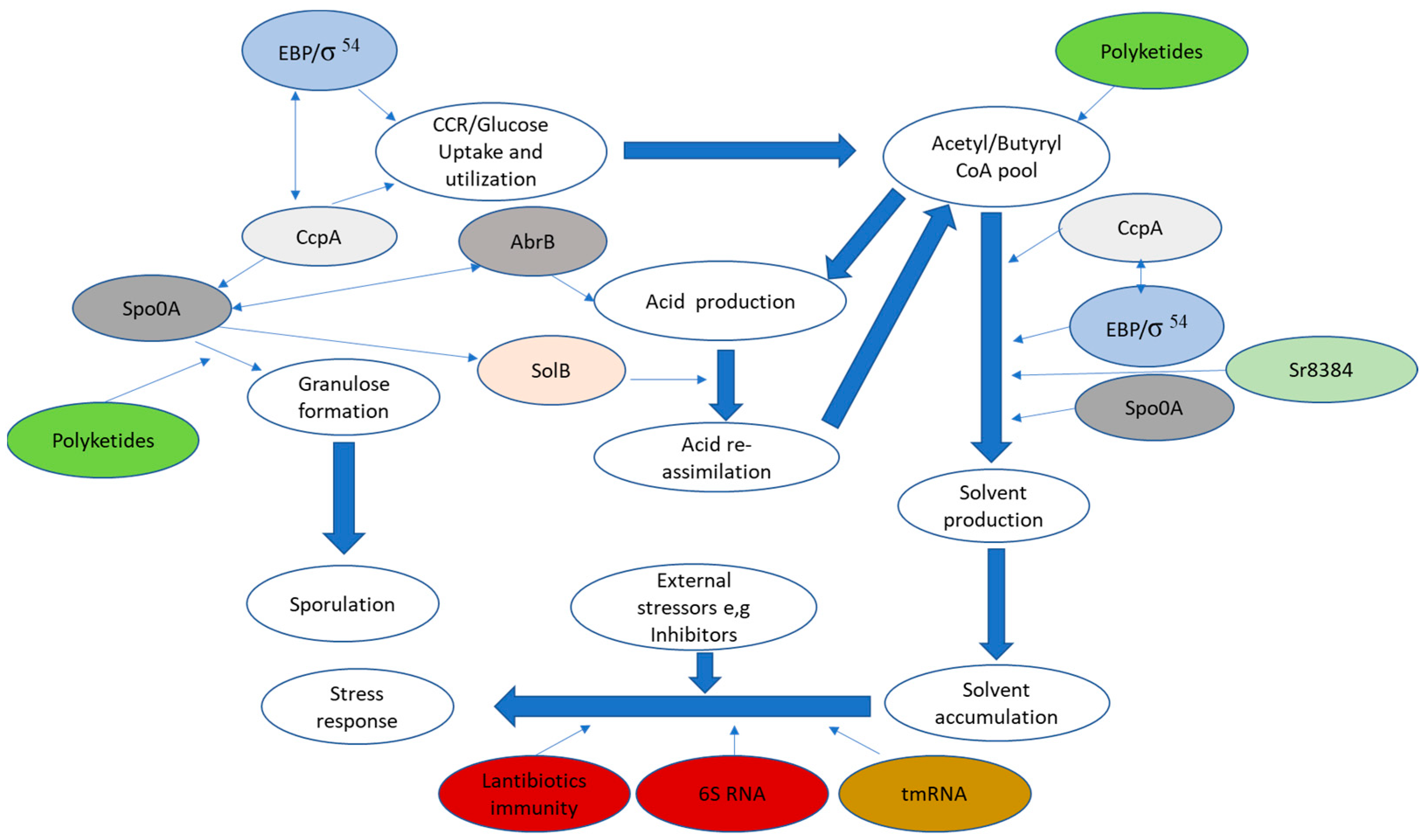
2.1. Interaction between σ54 Mediated Regulation and Carbon Catabolite Repression/Activation
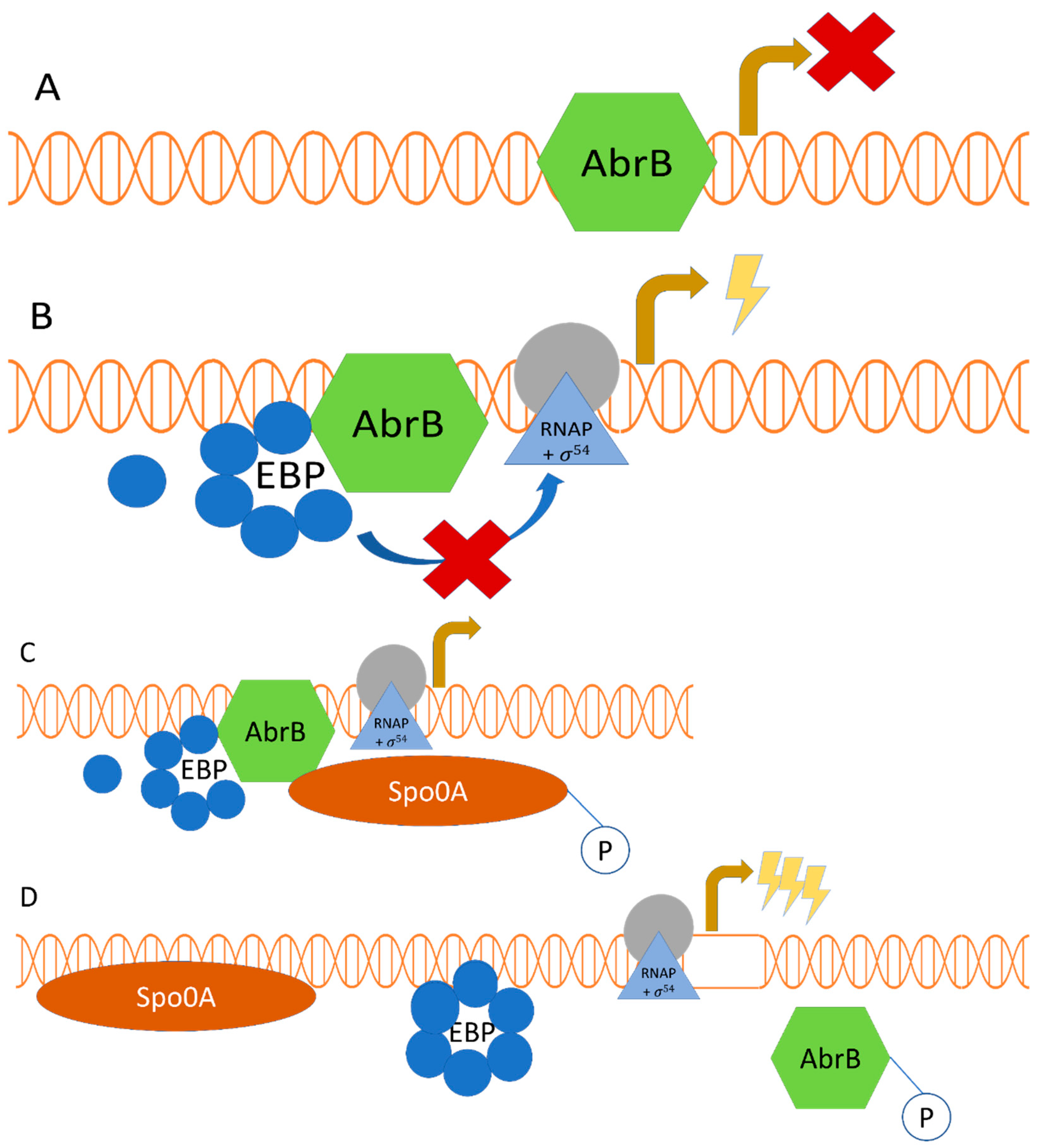
2.2. Regulatory Interplay between σ54-Mediated Regulation and the Transition Phase Regulator AbrB, along with the Sporulation Master Regulator Spo0A
3. Regulation by Small Non-Coding RNAs
3.1. SolB Mediated Regulation
3.2. The 6S RNA and tmRNA Mediated Regulation
3.3. Sr8384 Mediated Regulation of Growth and ABE Production
4. Bacteria Secondary Metabolites and Their Potential Role in Regulation
Non-Ribosomal Peptides (NRPs) Mediated Regulation
5. Potential Target Points for Metabolic Engineering
5.1. EBP/Sigma Factor 54 as Potential Targets for Metabolic Engineering
5.2. sRNAs as Potential Targets for Metabolic Engineering
5.3. Secondary Metabolites as Potential Targets for Metabolic Engineering
6. Conclusions
Funding
Acknowledgments
Conflicts of Interest
References
- Ezeji, T.C.; Atiyeh, H.; Mariano, A.P.; Rakshit, S.K. Innovative bioconversion of non-food substrates to fuels. Front. Bioeng. Biotechnol. 2023, 11, 1163513. [Google Scholar] [CrossRef] [PubMed]
- Panahi, S.H.K.; Dehhaghi, M.; Guillemin, G.J.; Okonkwo, C.C.; Kinder, J.E.; Ezeji, T.C. Biobutanol Production: Scope, Significance, and Applications. In Advances in Pollution Research, Advances and Developments in Biobutanol Production; Segovia-Hernandez, J.G., Behera, S., Sanchez-Ramirez, E., Eds.; Woodhead Publishing: Sawston, UK, 2023; pp. 1–45. ISBN 9780323911788. [Google Scholar] [CrossRef]
- Zhang, Y.; Ezeji, T.C. Transcriptional analysis of Clostridium beijerinckii NCIMB 8052 to elucidate role of furfural stress during acetone butanol ethanol fermentation. Biotechnol. Biofuels 2013, 6, 66. [Google Scholar] [CrossRef] [PubMed]
- Okonkwo, C.C.; Azam, M.M.; Ezeji, T.C.; Qureshi, N. Enhancing ethanol production from cellulosic sugars using Scheffersomyces (Pichia) stipitis. Bioprocess Biosyst. Eng. 2016, 39, 1023–1032. [Google Scholar] [CrossRef] [PubMed]
- Ezeji, T.; Qureshi, N.; Blaschek, H.P. Butanol production from agricultural residues: Impact of degradation products on Clostridium beijerinckii growth and butanol fermentation. Biotechnol. Bioeng. 2007, 97, 1460–1469. [Google Scholar] [CrossRef] [PubMed]
- Baral, N.R.; Shah, A. Microbial inhibitors: Formation and effects on acetone-butanol-ethanol fermentation of lignocellulosic biomass. Appl. Microbiol. Biotechnol. 2014, 98, 9151–9172. [Google Scholar] [CrossRef] [PubMed]
- Ujor, V.; Okonkwo, C.; Ezeji, T.C. Unorthodox methods for enhancing solvent production in solventogenic Clostridium species. Appl. Microbiol. Biotechnol. 2016, 100, 1089–1099. [Google Scholar] [CrossRef] [PubMed]
- Agu, C.V.; Ujor, V.; Ezeji, T.C. Metabolic engineering of Clostridium beijerinckii to improve glycerol metabolism and furfural tolerance. Biotechnol. Biofuels 2019, 12, 50. [Google Scholar] [CrossRef] [PubMed]
- Okonkwo, C.C.; Ujor, V.; Ezeji, T.C. Chromosomal integration of aldo-keto-reductase and short-chain dehydrogenase/reductase genes in Clostridium beijerinckii NCIMB 8052 enhanced tolerance to lignocellulose-derived microbial inhibitory compounds. Sci. Rep. 2019, 9, 7634. [Google Scholar] [CrossRef] [PubMed]
- Patil, R.C.; Suryawanshi, P.G.; Kataki, R.; Goud, V.V. Current challenges and advances in butanol production. In Sustainable Bioenergy; Elsevier: Amsterdam, The Netherlands, 2019; pp. 225–256. [Google Scholar] [CrossRef]
- Thunuguntla, R.; Atiyeh, H.K.; Huhnke, R.L.; Tanner, R.S. CO2-based production of C2–C6 acids and alcohols: The potential of novel Clostridia. Bioresour. Technol. Rep. 2024, 25, 101713. [Google Scholar] [CrossRef]
- Ezeji, T.; Milne, C.; Price, N.D.; Blaschek, H.P. Achievements and perspectives to overcome the poor solvent resistance in acetone and butanol-producing microorganisms. Appl. Microbiol. Biotechnol. 2010, 85, 1697–1712. [Google Scholar] [CrossRef]
- Li, C.J.; Zhang, Z.; Zhan, P.C.; Lv, A.P.; Li, P.P.; Liu, L.; Li, W.J.; Yang, L.L.; Zhi, X.Y. Comparative Genomic Analysis and Proposal of Clostridium yunnanense sp. nov., Clostridium rhizosphaerae sp. nov., and Clostridium paridis sp. nov., Three Novel Clostridium sensu stricto Endophytes with Diverse Capabilities of Acetic Acid and Ethanol production. Anaerobe 2023, 79, 102686. [Google Scholar] [PubMed]
- Mitra, R.; Raja, V.B.K.K.; Panneerselvam, E. Comparison of Wrinkle Patterns Generated by Intradermal and Intramuscular Botulinum Toxin Injections by Clinical Evaluation. J. Maxillofac. Oral Surg. 2024. [Google Scholar] [CrossRef]
- Staedtke, V.; Roberts, N.J.; Bai, R.Y.; Zhou, S. Clostridium novyi-NT in cancer therapy. Genes Dis. 2016, 3, 144–152. [Google Scholar] [CrossRef] [PubMed]
- Dang, L.H.; Bettegowda, C.; Huso, D.L.; Kinzler, K.W.; Vogelstein, B. Combination bacteriolytic therapy for the treatment of experimental tumors. Proc. Natl. Acad. Sci. USA 2001, 98, 15155–15160. [Google Scholar] [CrossRef] [PubMed]
- Patyar, S.; Joshi, R.; Byrav, D.P.; Prakash, A.; Medhi, B.; Das, B. Bacteria in cancer therapy: A novel experimental strategy. J. Biomed. Sci. 2010, 17, 21. [Google Scholar] [CrossRef] [PubMed]
- Nölling, J.; Breton, G.; Omelchenko, M.V.; Makarova, K.S.; Zeng, Q.; Gibson, R.; Lee, H.M.; Dubois, J.; Qiu, D.; Hitti, J.; et al. Genome Sequence and Comparative Analysis of the Solvent-Producing Bacterium Clostridium acetobutylicum. J. Bacteriol. 2001, 183, 4823–4838. [Google Scholar] [CrossRef] [PubMed]
- Lin, Z.; Cong, W.; Zhang, J. Biobutanol Production from Acetone–Butanol–Ethanol Fermentation: Developments and Prospects. Fermentation 2023, 9, 847. [Google Scholar] [CrossRef]
- Choua, K.J.; Croft, T.; Hebdon, S.D.; Magnusson, L.R.; Xionga, W.; Reyes, L.H.; Chen, X.; Miller, E.J.; Riley, D.M.; Dupuis, S.; et al. Engineering the cellulolytic bacterium, Clostridium thermocellum, to co-utilize hemicellulose. Metab. Eng. 2024, 83, 193–205. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.L.; Sun, Y.Q.; Pan, D.T.; Xiu, Z.-L. Kinetics-based development of two-stage continuous fermentation of 1,3-propanediol from crude glycerol by Clostridium butyricum. Biotechnol. Biofuels Bioprod. 2024, 17, 38. [Google Scholar] [CrossRef] [PubMed]
- Jones, D.T.; Schulz, F.; Roux, S.; Brown, S.D. Solvent-Producing Clostridia Revisited. Microorganisms 2023, 11, 2253. [Google Scholar] [CrossRef]
- Höfele, F.; Schoch, T.; Oberlies, C.; Dürre, P. Heterologous Production of Isopropanol Using Metabolically Engineered Acetobacterium woodii Strains. Bioengineering 2023, 10, 1381. [Google Scholar] [CrossRef] [PubMed]
- Schulz, S.; Molitor, B.; Angenent, L.T. Acetate augmentation boosts the ethanol production rate and specificity by Clostridium ljungdahlii during gas fermentation with pure carbon monoxide. Bioresour. Technol. 2023, 369, 128387. [Google Scholar] [CrossRef] [PubMed]
- Gedam, P.S.; Raut, A.N.; Dhamole, P.B.; Gole, V.L. Challenges in Biobutanol Fermentation and Separation. In Biorefinery: A Sustainable Approach for the Production of Biomaterials, Biochemicals and Biofuels; Pathak, P.D., Mandavgane, S.A., Eds.; Springer: Singapore, 2023. [Google Scholar] [CrossRef]
- Thang Ma, Y.; Guo, N.; Wang, S.; Wang, Y.; Jiang, Z.; Guo, L.; Wei Luo, W.; Wang, Y. Metabolically engineer Clostridium saccharoperbutylacetonicum for comprehensive conversion of acid whey into valuable biofuels and biochemicals. Bioresour. Technol. 2024, 400, 130640. [Google Scholar] [CrossRef] [PubMed]
- Han, Y.F.; Xie, B.T.; Wu, G.X.; Guo, Y.Q.; Li, D.M.; Huang, Z.Y. Combination of trace metal to improve solventogenesis of Clostridium carboxidivorans P7 in syngas fermentation. Front. Microbiol. 2020, 11, 577266. [Google Scholar] [CrossRef] [PubMed]
- Buckel, W.; Barker, H.A. Two Pathways of Glutamate Fermentation by Anaerobic Bacteria. J. Bacteriol. 1974, 117, 1248–1260. [Google Scholar] [CrossRef] [PubMed]
- Aristilde, L.; Lewis, I.A.; Park, J.O.; Rabinowitz, J.D. Hierarchy in Pentose Sugar Metabolism in Clostridium acetobutylicum. Appl. Environ. Microbiol. 2015, 81, 1452–1462. [Google Scholar] [CrossRef] [PubMed]
- Gottumukkala, L.D.; Haigh, K.; Görgens, J. Trends and advances in conversion of lignocellulosic biomass to biobutanol: Microbes, bioprocesses and industrial viability. Renew. Sustain. Energy Rev. 2017, 76, 963–973. [Google Scholar] [CrossRef]
- Han, B.; Ujor, V.; Lai, L.B.; Gopalan, V.; Ezeji, T.C. Use of proteomic analysis to elucidate the role of calcium on acetone-butanol-ethanol (ABE) fermentation in Clostridium beijerinckii NCIMB 8052. Appl. Environ. Microbiol. 2013, 79, 282–293. [Google Scholar] [CrossRef] [PubMed]
- Xue, C.; Zhao, J.; Chen, L.; Yang, S.-T.; Bai, F. Recent advances and state-of-the-art strategies in strain and process engineering for biobutanol production by Clostridium acetobutylicum. Biotechnol. Adv. 2017, 35, 310–322. [Google Scholar] [CrossRef] [PubMed]
- Yang, Y.; Nie, X.; Jiang, Y.; Yang, C.; Gu, Y.; Jiang, W. Metabolic regulation in solventogenic Clostridia: Regulators, mechanisms and engineering. Biotechnol. Adv. 2018, 36, 905–914. [Google Scholar] [CrossRef]
- Smith, I. Regulatory Proteins That Control Late-Growth Development. In Bacillus subtilis and Other Gram-Positive Bacteria; John Wiley & Sons, Ltd.: Hoboken, NJ, USA, 1993; pp. 785–800. [Google Scholar] [CrossRef]
- Tangney, M.; Galinier, A.; Deutscher, J.; Mitchell, W. Analysis of the Elements of Catabolite Repression in Clostridium acetobutylicum ATCC 824. J. Mol. Microbiol. Biotechnol. 2003, 6, 6–11. [Google Scholar] [CrossRef]
- Scotcher, M.C.; Rudolph, F.B.; Bennett, G.N. Expression of abrB310 and sinR, and Effects of Decreased abrB310 Expression on the Transition from Acidogenesis to Solventogenesis, in Clostridium acetobutylicum ATCC 824. Appl. Environ. Microbiol. 2005, 71, 1987–1995. [Google Scholar] [CrossRef]
- Ren, C.; Gu, Y.; Hu, S.; Wu, Y.; Wang, P.; Yang, Y.; Yang, C.; Yang, S.; Jiang, W. Identification and inactivation of pleiotropic regulator CcpA to eliminate glucose repression of xylose utilization in Clostridium acetobutylicum. Metab. Eng. 2010, 12, 446–454. [Google Scholar] [CrossRef]
- Noguchi, T.; Tashiro, Y.; Yoshida, T.; Zheng, J.; Sakai, K.; Sonomoto, K. Efficient butanol production without carbon catabolite repression from mixed sugars with Clostridium saccharoperbutylacetonicum N1-4. J. Biosci. Bioeng. 2013, 116, 716–721. [Google Scholar] [CrossRef]
- Wietzke, M.; Bahl, H. The redox-sensing protein Rex, a transcriptional regulator of solventogenesis in Clostridium acetobutylicum. Appl. Microbiol. Biotechnol. 2012, 96, 749–761. [Google Scholar] [CrossRef]
- Tan, Y.; Liu, Z.-Y.; Liu, Z.; Zheng, H.-J.; Li, F.-L. Comparative transcriptome analysis between csrA-disruption Clostridium acetobutylicum and its parent strain. Mol. BioSystems 2015, 11, 1434–1442. [Google Scholar] [CrossRef]
- Qureshi, N.; Lin, X.; Liu, S.; Saha, B.C.; Mariano, A.P.; Polaina, J.; Ezeji, T.C.; Friedl, A.; Maddox, I.S.; Klasson, K.T.; et al. Global View of Biofuel Butanol and Economics of Its Production by Fermentation from Sweet Sorghum Bagasse, Food Waste, and Yellow Top Presscake: Application of Novel Technologies. Fermentation 2020, 6, 58. [Google Scholar] [CrossRef]
- Ujor, V.C.; Lai, L.B.; Okonkwo, C.C.; Gopalan, V.; Ezeji, T.C. Ribozyme-Mediated Downregulation Uncovers DNA Integrity Scanning Protein A (DisA) as a Solventogenesis Determinant in Clostridium beijerinckii. Front. Bioeng. Biotechnol. 2021, 9, 669462. [Google Scholar] [CrossRef]
- Olorunsogbon, T.; Adesanya, Y.; Atiyeh, H.K.; Okonkwo, C.C.; Ujor, V.C.; Ezeji, T.C. Effects of Clostridium beijerinckii and Medium Modifications on Acetone–Butanol–Ethanol Production From Switchgrass. Front. Bioeng. Biotechnol. 2022, 10, 942701. [Google Scholar] [CrossRef]
- Davis, M.C.; Kesthely, C.A.; Franklin, E.A.; MacLellan, S.R. The essential activities of the bacterial sigma factor. Can. J. Microbiol. 2017, 63, 89–99. [Google Scholar] [CrossRef]
- Buck, M.; Cannon, W. Activator-independent formation of a closed complex between σ54-holoenzyme and nifH and nifU promoters of Klebsiella pneumoniae. Mol. Microbiol. 1992, 6, 1625–1630. [Google Scholar] [CrossRef]
- Shen, C.-H. Gene expression: Transcription of the genetic code. In Diagnostic Molecular Biology; Academic Press: Cambridge, MA, USA, 2019; pp. 59–86. [Google Scholar] [CrossRef]
- Sauer, U.; Santangelo, J.D.; Treuner, A.; Buchholz, M.; Dürre, P. Sigma factor and sporulation genes in Clostridium. FEMS Microbiol. Rev. 1995, 17, 331–340. [Google Scholar] [CrossRef]
- Nie, X.; Yang, B.; Zhang, L.; Gu, Y.; Yang, S.; Jiang, W.; Yang, C. PTS regulation domain-containing transcriptional activator CelR and sigma factor σ54 control cellobiose utilization in Clostridium acetobutylicum. Mol. Microbiol. 2016, 100, 289–302. [Google Scholar] [CrossRef]
- Hocq, R.; Bouilloux-Lafont, M.; Lopes Ferreira, N.; Wasels, F. Σ54 (σL) plays a central role in carbon metabolism in the industrially relevant Clostridium beijerinckii. Sci. Rep. 2019, 9, 7228. [Google Scholar] [CrossRef]
- Yang, B.; Nie, X.; Gu, Y.; Jiang, W.; Yang, C. Control of solvent production by sigma-54 factor and the transcriptional activator AdhR in Clostridium beijerinckii. Microb. Biotechnol. 2020, 13, 328–338. [Google Scholar] [CrossRef]
- Nie, X.; Dong, W.; Yang, C. Genomic reconstruction of σ54 regulons in Clostridiales. BMC Genom. 2019, 20, 565. [Google Scholar] [CrossRef]
- Morett, E.; Buck, M. In Viva Studies on the Interaction of RNA Polymerase-a54 with the Klebsiella pneumoniae and Rhizobium meliloti nzfw Promoters. J. Mol. Biol. 1989, 210, 65–77. [Google Scholar] [CrossRef]
- Danson, A.E.; Jovanovic, M.; Buck, M.; Zhang, X. Mechanisms of σ54-dependent transcription initiation and regulation. J. Mol. Biol. 2019, 431, 3960–3974. [Google Scholar] [CrossRef]
- Jones, S. Sigma 54 minds the gap. Nat. Rev. Microbiol. 2009, 7, 3. [Google Scholar] [CrossRef]
- Bush, M.; Dixon, R. The Role of Bacterial Enhancer Binding Proteins as Specialized Activators of 54-Dependent Transcription. Microbiol. Mol. Biol. Rev. 2012, 76, 497–529. [Google Scholar] [CrossRef]
- Deutscher, J.; Aké, F.M.D.; Derkaoui, M.; Zébré, A.C.; Cao, T.N.; Bouraoui, H.; Kentache, T.; Mokhtari, A.; Milohanic, E.; Joyet, P. The Bacterial Phosphoenolpyruvate: Carbohydrate Phosphotransferase System: Regulation by Protein Phosphorylation and Phosphorylation-Dependent Protein-Protein Interactions. Microbiol. Mol. Biol. Rev. 2014, 78, 231–256. [Google Scholar] [CrossRef] [PubMed]
- Ren, C.; Gu, Y.; Wu, Y.; Zhang, W.; Yang, C.; Yang, S.; Jiang, W. Pleiotropic functions of catabolite control protein CcpA in Butanol-producing Clostridium acetobutylicum. BMC Genom. 2012, 13, 349. [Google Scholar] [CrossRef] [PubMed]
- Yang, Y.; Zhang, L.; Huang, H.; Yang, C.; Yang, S.; Gu, Y.; Jiang, W. A Flexible Binding Site Architecture Provides New Insights into CcpA Global Regulation in Gram-Positive Bacteria. MBio 2017, 8, e02004-16. [Google Scholar] [CrossRef] [PubMed]
- Deutscher, J.; Francke, C.; Postma, P.W. How Phosphotransferase System-Related Protein Phosphorylation Regulates Carbohydrate Metabolism in Bacteria. Microbiol. Mol. Biol. Rev. 2006, 70, 939–1031. [Google Scholar] [CrossRef] [PubMed]
- Alsaker, K.V.; Spitzer, T.R.; Papoutsakis, E.T. Transcriptional Analysis of spo0A Overexpression in Clostridium acetobutylicum and Its Effect on the Cell’s Response to Butanol Stress. J. Bacteriol. 2004, 186, 1959–1971. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Li, X.; Mao, Y.; Blaschek, H.P. Genome-wide dynamic transcriptional profiling in Clostridium beijerinckii NCIMB 8052 using single-nucleotide resolution RNA-Seq. BMC Genom. 2012, 13, 102. [Google Scholar] [CrossRef] [PubMed]
- Xue, Q.; Yang, Y.; Chen, J.; Chen, L.; Yang, S.; Jiang, W.; Gu, Y. Roles of three AbrBs in regulating two-phase Clostridium acetobutylicum fermentation. Appl. Microbiol. Biotechnol. 2016, 100, 9081–9089. [Google Scholar] [CrossRef]
- Ravagnani, A.; Jennert, K.C.; Steiner, E.; Grünberg, R.; Jefferies, J.R.; Wilkinson, S.R.; Young, D.I.; Tidwell, E.C.; Brown, D.P.; Youngman, P.; et al. Spo0A directly controls the switch from acid to solvent production in solvent-forming clostridia. Mol. Microbiol. 2000, 37, 1172–1185. [Google Scholar] [CrossRef]
- Thormann, K.; Feustel, L.; Lorenz, K.; Nakotte, S.; Dürre, P. Control of butanol formation in Clostridium acetobutylicum by transcriptional activation. J. Bacteriol. 2002, 184, 1966–1973. [Google Scholar] [CrossRef]
- Dürre, P.; Hollergschwandner, C. Initiation of endospore formation in Clostridium acetobutylicum. Anaerobe 2004, 10, 69–74. [Google Scholar] [CrossRef]
- Bengelsdorf, F.R.; Poehlein, A.; Flitsch, S.K.; Linder, S.; Schiel-Bengelsdorf, B.; Stegmann, B.A.; Krabben, P.; Green, E.; Zhang, Y.; Minton, N. Host organisms: Clostridium acetobutylicum/Clostridium beijerinckii and related organisms. Ind. Biotechnol. Microorg. 2017, 1, 327–364. [Google Scholar]
- Harris, L.M.; Welker, N.E.; Papoutsakis, E.T. Northern, Morphological, and Fermentation Analysis of spo0A Inactivation and Overexpression in Clostridium acetobutylicum ATCC 824. J. Bacteriol. 2002, 184, 3586–3597. [Google Scholar] [CrossRef] [PubMed]
- Sullivan, L.; Bennett, G.N. Proteome analysis and comparison of lostridium acetobutylicum ATCC 824 and Spo0A strain variants. J. Ind. Microbiol. Biotechnol. 2006, 33, 298–308. [Google Scholar] [CrossRef] [PubMed]
- Jiang, Y.; Xu, C.; Dong, F.; Yang, Y.; Jiang, W.; Yang, S. Disruption of the acetoacetate decarboxylase gene in solvent-producing Clostridium acetobutylicum increases the butanol ratio. Metab. Eng. 2009, 11, 284–291. [Google Scholar] [CrossRef] [PubMed]
- Han, B.; Gopalan, V.; Ezeji, T.C. Acetone production in solventogenic Clostridium species: New insights from non-enzymatic decarboxylation of acetoacetate. Appl. Microbiol. Biotechnol. 2011, 91, 565–576. [Google Scholar] [CrossRef] [PubMed]
- Chen, G.; Kumar, A.; Wyman, T.H.; Moran, C.P., Jr. Spo0A-dependent activation of an extended− 10 region promoter in Bacillus subtilis. J. Bacteriol. 2006, 188, 1411–1418. [Google Scholar] [CrossRef] [PubMed]
- Vogel, J.; Sharma, C.M. How to find small non-coding RNAs in bacteria. Biol. Chem. 2005, 386, 1219–1238. [Google Scholar] [CrossRef]
- Brosse, A.; Guillier, M. Bacterial Small RNAs in Mixed Regulatory Networks. In Regulating with RNA in Bacteria and Archaea; John Wiley & Sons, Ltd.: Hoboken, NJ, USA, 2018; pp. 453–469. [Google Scholar] [CrossRef]
- Lalaouna, D.; Simoneau-Roy, M.; Lafontaine, D.; Massé, E. Regulatory RNAs and target mRNA decay in prokaryotes. Biochim. Biophys. Acta (BBA)-Gene Regul. Mech. 2013, 1829, 742–747. [Google Scholar] [CrossRef]
- Argaman, L.; Hershberg, R.; Vogel, J.; Bejerano, G.; Wagner, E.G.H.; Margalit, H.; Altuvia, S. Novel small RNA-encoding genes in the intergenic regions of Escherichia coli. Curr. Biol. 2001, 11, 941–950. [Google Scholar]
- Richards, G.R.; Vanderpool, C.K. Molecular call and response: The physiology of bacterial small RNAs. Biochim. Biophys. Acta (BBA) Gene Regul. Mech. 2011, 1809, 525–531. [Google Scholar] [CrossRef]
- Storz, G.; Vogel, J.; Wassarman, K.M. Regulation by Small RNAs in Bacteria: Expanding Frontiers. Mol. Cell 2011, 43, 880–891. [Google Scholar] [CrossRef] [PubMed]
- Thomason, M.K.; Storz, G. Bacterial Antisense RNAs: How Many Are There, and What Are They Doing? Annu. Rev. Genet. 2010, 44, 167–188. [Google Scholar] [CrossRef] [PubMed]
- Georg, J.; Hess, W.R. Cis-Antisense RNA, Another Level of Gene Regulation in Bacteria. Microbiol. Mol. Biol. Rev. 2011, 75, 286–300. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.; Indurthi, D.C.; Jones, S.W.; Papoutsakis, E.T. Small RNAs in the Genus Clostridium. MBio 2011, 2, 10–1128. [Google Scholar] [CrossRef] [PubMed]
- Venkataramanan, K.P.; Jones, S.W.; McCormick, K.P.; Kunjeti, S.G.; Ralston, M.T.; Meyers, B.C.; Papoutsakis, E.T. The Clostridium small RNome that responds to stress: The paradigm and importance of toxic metabolite stress in C. acetobutylicum. BMC Genom. 2013, 14, 1–16. [Google Scholar] [CrossRef] [PubMed]
- Keiler, K.C. Biology of trans-Translation. Annu. Rev. Microbiol. 2008, 62, 133–151. [Google Scholar] [CrossRef] [PubMed]
- Craig, N.L.; Cohen-Fix, O.; Storz, G. Molecular Biology: Principles of Genome Function; Oxford University Press: Oxford, UK, 2014. [Google Scholar]
- Wach, W.; Harms, K.; Klingeberg, M.; Duerre, P.; Nold, N.; Schiel, B. Production of Acid and Solvent in Microorganisms. United. States Patent No. US8679799B2, 25 March 2014. [Google Scholar]
- Wassarman, K.M. 6S RNA: A regulator of transcription. Mol. Microbiol. 2007, 65, 1425–1431. [Google Scholar] [CrossRef] [PubMed]
- Zimmermann, T. Untersuchungen zur Butanolbildung von Hyperthermus butylicus und Clostridium acetobutylicum. Ph.D. Dissertation, Universität Ulm, Ulm, Germany, 2013. [Google Scholar] [CrossRef]
- Wang, Q.; Venkataramanan, K.; Huang, H.; Papoutsakis, E.T.; Wu, C.H. Transcription factors and genetic circuits orchestrating the complex, multilayered response of Clostridium acetobutylicum to butanol and butyrate stress. BMC Syst. Biol. 2013, 7, 120. [Google Scholar] [CrossRef] [PubMed]
- Jones, A.J.; Fast, A.G.; Clupper, M.; Papoutsakis, E.T. Small and Low but Potent: The Complex Regulatory Role of the Small RNA SolB in Solventogenesis in Clostridium acetobutylicum. Appl. Environ. Microbiol. 2018, 84, e00597-18. [Google Scholar] [CrossRef]
- André, G.; Even, S.; Putzer, H.; Burguière, P.; Croux, C.; Danchin, A.; Martin-Verstraete, I.; Soutourina, O. S-box and T-box riboswitches and antisense RNA control a sulfur metabolic operon of Clostridium acetobutylicum. Nucleic Acids Res. 2008, 36, 5955–5969. [Google Scholar] [CrossRef]
- Barrick, J.E.; Sudarsan, N.; Weinberg, Z.; Ruzzo, W.L.; Breaker, R.R. 6S RNA is a widespread regulator of eubacterial RNA polymerase that resembles an open promoter. RNA 2005, 11, 774–784. [Google Scholar] [CrossRef] [PubMed]
- Gildehaus, N.; Neußer, T.; Wurm, R.; Wagner, R. Studies on the function of the riboregulator 6S RNA from E. coli: RNA polymerase binding, inhibition of in vitro transcription and synthesis of RNA-directed de novo transcripts. Nucleic Acids Res. 2007, 35, 1885–1896. [Google Scholar] [CrossRef] [PubMed]
- Mahendran, G.; Jayasinghe, O.T.; Thavakumaran, D.; Arachchilage, G.M.; Silva, G.N. Key players in regulatory RNA realm of bacteria. Biochem. Biophys. Rep. 2022, 30, 101276. [Google Scholar] [CrossRef] [PubMed]
- Trotochaud, A.E.; Wassarman, K.M. 6S RNA Regulation of pspF Transcription Leads to Altered Cell Survival at High pH. J. Bacteriol. 2006, 188, 3936–3943. [Google Scholar] [CrossRef] [PubMed]
- Shin, J.-H.; Price, C.W. The SsrA-SmpB Ribosome Rescue System Is Important for Growth of Bacillus subtilis at Low and High Temperatures. J. Bacteriol. 2007, 189, 3729–3737. [Google Scholar] [CrossRef] [PubMed]
- Withey, J.H.; Friedman, D.I. A salvage pathway for protein synthesis: tmRNA and trans-translation. Annu. Rev. Microbiol. 2003, 57, 101–123. [Google Scholar] [CrossRef] [PubMed]
- Jones, A.J.; Venkataramanan, K.P.; Papoutsakis, T. Overexpression of two stress-responsive, small, non-coding RNAs, 6S and tmRNA, imparts butanol tolerance in Clostridium acetobutylicum. FEMS Microbiol. Lett. 2016, 363, fnw063. [Google Scholar] [CrossRef] [PubMed]
- Sharma, U.K.; Chatterji, D. Transcriptional switching in Escherichia coli during stress and starvation by modulation of σ70 activity. FEMS Microbiol. Rev. 2010, 34, 646–657. [Google Scholar] [CrossRef]
- Venkataramanan, K.P.; Min, L.; Hou, S.; Jones, S.W.; Ralston, M.T.; Lee, K.H.; Papoutsakis, E.T. Complex and extensive post-transcriptional regulation revealed by integrative proteomic and transcriptomic analysis of metabolite stress response in Clostridium acetobutylicum. Biotechnol. Biofuels 2015, 8, 81. [Google Scholar] [CrossRef]
- Mehdizadeh, G.I.; Edwards, A.N.; McBride, S.M.; McClane, B.A. The impact of orphan histidine kinases and phosphotransfer proteins on the regulation of clostridial sporulation initiation. mBio 2024, 15, e02248-23. [Google Scholar] [CrossRef]
- Furuya, K.; Kiyoshi, K.; Punjuy, C.; Yoshida, N.; Maruyama, R.; Yasuda, T.; Watanabe, K.; Kadokura, T.; Nakayama, S. Effect of spo0A, sigE, sigG, and sigK disruption on butanol production and spore formation in Clostridium saccharoperbutylacetonicum strain N1–4 (ATCC13564). J. Biosci. Bioeng. 2023, 136, 198–204. [Google Scholar] [CrossRef] [PubMed]
- Rhodius, V.A.; Suh, W.C.; Nonaka, G.; West, J.; Gross, C.A. Conserved and Variable Functions of the σE Stress Response in Related Genomes. PLoS Biol. 2005, 4, e2. [Google Scholar] [CrossRef] [PubMed]
- Konovalova, A.; Grabowicz, M.; Balibar, C.J.; Malinverni, J.C.; Painter, R.E.; Riley, D.; Mann, P.A.; Wang, H.; Garlisi, C.G.; Sherborne, B.; et al. Inhibitor of intramembrane protease RseP blocks the σ E response causing lethal accumulation of unfolded outer membrane proteins. Proc. Natl. Acad. Sci. USA 2018, 115, E6614–E6621. [Google Scholar] [CrossRef] [PubMed]
- Iqbal, S.; Begum, F.; Rabaan, A.A.; Aljeldah, M.; Al Shammari, B.R.; Alawfi, A.; Alshengeti, A.; Sulaiman, T.; Khan, A. Classification and Multifaceted Potential of Secondary Metabolites Produced by Bacillus subtilis Group: A Comprehensive Review. Molecules 2023, 28, 927. [Google Scholar] [CrossRef] [PubMed]
- Pahalagedara, A.S.N.W.; Flint, S.; Palmer, J.; Brightwell, G.; Gupta, T.B. Antimicrobial production by strictly anaerobic Clostridium spp. Int. J. Antimicrob. Agents 2020, 55, 105910. [Google Scholar] [CrossRef] [PubMed]
- Yang, M.; Li, J.; Li, A. Total synthesis of clostrubin. Nat. Commun. 2015, 6, 6445. [Google Scholar] [CrossRef] [PubMed]
- Li, J.S.; Barber, C.; Zhang, W. Natural Products from Anaerobes. J. Ind. Microbiol. Biotechnol. 2019, 46, 375–383. [Google Scholar] [CrossRef] [PubMed]
- Herman, N.A.; Kim, S.J.; Li, J.S.; Cai, W.; Koshino, H.; Zhang, W. The industrial anaerobe Clostridium acetobutylicum uses polyketides to regulate cellular differentiation. Nat. Commun. 2017, 8, 1514. [Google Scholar] [CrossRef]
- Li, J.; Barber, C.; Herman, N.; Cai, W.; Zafrir, E.; Du, Y.; Zhu, X.; Skyrud, W.; Zhang, W. Investigation of secondary metabolism in the industrial butanol hyper-producer Clostridium saccharoperbutylacetonicum N1-4. J. Ind. Microbiol. Biotechnol. 2020, 47, 319–328. [Google Scholar] [CrossRef]
- Pahalagedara, A.S.; Jauregui, R.; Maclean, P.; Altermann, E.; Flint, S.; Palmer, J.; Brightwell, G.; Gupta, T.B. Culture and genome-based analysis of four soil Clostridium isolates reveal their potential for antimicrobial production. BMC Genom. 2021, 22, 686. [Google Scholar] [CrossRef]
- Lincke, T.; Behnken, S.; Ishida, K.; Roth, M.; Hertweck, C. Closthioamide: An unprecedented polythioamide antibiotic from the strictly anaerobic bacterium Clostridium cellulolyticum. Angew. Chem. 2010, 122, 2055–2057. [Google Scholar] [CrossRef]
| Clostridium Species | Substrates | Products | References |
|---|---|---|---|
| C. acetobutylicum | Glucose, xylose, arabinose, cellobiose | Acetone, butanol, and ethanol | [19] |
| C. beijerinckii | Glucose, xylose, arabinose, cellobiose | Acetone, butanol, and ethanol | [19] |
| C. thermocellum | Lignocellulose, Cellulose | Ethanol | [20] |
| C. butyricum | Glycerol | 1,3-propanediol | [21] |
| C. Pasteurianum | Glycerol | 1,3-propanediol, butanol and ethanol | [22] |
| C. carboxidivorans | Carbon monoxide, hydrogen, and carbon dioxide | Ethanol, butanol and hexanol | [11] |
| C. aurantibutyricum | Glucose, lactose, maltose, galactose, xylose, starch | Acetone, isopropanol, and butanol | [23] |
| C. ljundahlii | Carbon monoxide and hydrogen | Ethanol | [24] |
| C. tetanomorphum | Glucose and maltose, glutamate | Butanol and ethanol | [25] |
| C. saccharoperbutylacetonicum | Glucose | Acetone, ethanol, and butanol | [26] |
| Organism | Number of EBPs Predicted to be Present in Genome | Type of EBP Regulatory Domains Identified | Number of σ54-Regulated Operons |
|---|---|---|---|
| C. acetobutylicum ATCC 824 | 3 | PRD-EIIA-PRD (1), PrpR-N-PAS (1), PTS_Hpr-PAS-PAS (1) | 2 |
| C. autoethanogenum DSM 10061 | 12 | PAS-PAS (1), GAF-PAS (6), PAS (3) 0 (2) | - |
| C. beijerinckii NCIMB 8052 | 19 | PRD-EIIA-PRD (12), CBS-PAS (1), PTS_Hpr-PAS- PAS (1) PAS-PAS (2). GAF-PAS (2), RR (1) | 26 |
| C. carboxydivorans P7 | 11 | PRD-EIIA-PRD (1), PTS_Hpr-PAS-PAS (1), PAS-PAS (1) GAF-PAS (2), PAS (4), 0 (1), PrpR-N-PAS (1) | - |
| C. ljungdahlii | 19 | PrpR-N-PAS (1), PAS-PAS (1), GAF-PAS (10), PAS (5), 0 (2) | - |
| C. saccharobutylicum DSM 13864 | 5 | PRD-EIIA-PRD (3), GAF-PAS (2) | - |
| C. saccharoperbutylacetonicum N1-4 (HMT) | 14 | PRD-EIIA-PRD (5), GAF-PAS (3), PAS-PAS (1), PAS (2), PrpR-N-PAS (2), V4R (1) | - |
| Clostridium Species | Number of sRNA Identified in Genome | Genome Size (mb) | GC Content (%) | Metabolism Type |
|---|---|---|---|---|
| Clostridium acetobutylicum ATCC 824 | 113 | 4.13 | 30.90 | Saccharolytic |
| C. beijerinckii NCIMB 8052 | 366 | 6.00 | 29.90 | Saccharolytic |
| C. cellulolyticum H10 | 45 | 4.07 | 37.40 | Cellulolytic |
| C. kluyveri DSM 555 | 126 | 4.02 | 32.02 | - |
| C. kluyveri NBRC 12016 | 136 | 3.96 | 32.02 | - |
| C. phytofermentans ISDg | 42 | 4.85 | 35.30 | Cellulolytic |
| C. thermocellum | 15 | 3.84 | 39.00 | Cellulolytic |
| C. carboxidivorans p7 | NA | 5.75 | 29.89 | - |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Olorunsogbon, T.; Okonkwo, C.C.; Ezeji, T.C. Molecular Markers and Regulatory Networks in Solventogenic Clostridium Species: Metabolic Engineering Conundrum. Fermentation 2024, 10, 297. https://doi.org/10.3390/fermentation10060297
Olorunsogbon T, Okonkwo CC, Ezeji TC. Molecular Markers and Regulatory Networks in Solventogenic Clostridium Species: Metabolic Engineering Conundrum. Fermentation. 2024; 10(6):297. https://doi.org/10.3390/fermentation10060297
Chicago/Turabian StyleOlorunsogbon, Tinuola, Christopher Chukwudi Okonkwo, and Thaddeus Chukwuemeka Ezeji. 2024. "Molecular Markers and Regulatory Networks in Solventogenic Clostridium Species: Metabolic Engineering Conundrum" Fermentation 10, no. 6: 297. https://doi.org/10.3390/fermentation10060297
APA StyleOlorunsogbon, T., Okonkwo, C. C., & Ezeji, T. C. (2024). Molecular Markers and Regulatory Networks in Solventogenic Clostridium Species: Metabolic Engineering Conundrum. Fermentation, 10(6), 297. https://doi.org/10.3390/fermentation10060297







