Abstract
Fermented Chinese mustard greens are popular fermented vegetable foods in Guangdong Province, China. In this study, the quality characteristics and microbial composition of fermented Chinese mustard greens from different regions, including Shantou (ST), Meizhou (MZ), Yunfu (YF), and Guangzhou (GZ), were evaluated. The colour and texture of fermented Chinese mustard greens were significantly different from those of ST, MZ, YF, and GZ. L* values were 48.62, 42.30, 32.43, and 34.02 in the stem parts of ST, MZ, YF, and GZ, respectively. The chewiness value was greater in GZ (131.26 N) than in MZ (53.25 N), YF (39.99 N), and GZ (24.22 N) zones. The microbial community structure determined by high-throughput sequencing (HTS) demonstrated that Firmicutes, Proteobacteria, and Campilobacterota were the predominant phyla. Lactobacillus was the most predominant microorganism in the MZ and GZ samples and accounted for a greater proportion of the microorganisms in the ST and YF samples. In addition to Lactobacillus, the relative abundances of Cobetia and Weissella were greater in the ST group, while those of Halomonas and Pediococcus were greater in the YF group. There was a significant correlation between the microbial composition and quality indices (colour and texture) among the samples from the four regions. The quality of the fermented Chinese mustard greens in MZ and GZ was significantly different from that of other samples in ST and YF. The Lactobacillus genus (Lactobacillus plantarum and Lactobacillus selangorensis) in MZ and GZ contributed to changes in colour (b*, C*, L*, a*) and texture (firmness and chewiness). This study provided a comprehensive correlation between quality and microbial composition of fermented Chinese mustard greens from different regions in Guangdong Province. The evaluation and correlation between quality and microbiota are helpful for guiding future improvements in fermentation processes and manufacturing high-quality fermented Chinese mustard greens.
1. Introduction
Fermented vegetables likely originated more than 2000 years ago in Asia. This fermented material included fresh vegetables such as cabbages, turnips, radishes, carrots, and other indigenous varieties [1]. Fermented vegetables are recognized as foods with high nutritional value and good health benefits. They have the characteristics of simple processing equipment, a simple production process, and a low cost of raw materials. The planting area of vegetables exceeds 21 million hm2, and the production of vegetables is 769 million tons in China. In 2018, the total output of Sichuan kimchi alone exceeded 5 million tons, with a total output value of more than 42 billion yuan. In 2019, the output of fermented vegetables reached 70 billion yuan and increased annually in China [2]. Chinese mustard greens are the primary vegetable used in the preparation of fermented mustard in Guangdong Province. Interestingly, despite their close geographic proximity, the quality and flavour of fermented Chinese mustard greens exhibit variations influenced by the traditional lifestyle and preferences of the local populace. Mustard, scientifically known as Brassica juncea and classified within the Brassica genus of the Cruciferae family, is recognized for its distinct species groups: integrifolia, tsatsai, juncea, and napiformis [3,4]. Mustard greens are further categorized into four major groups, namely, root mustard, stem mustard, leaf mustard, and sedge mustard, encompassing a total of 16 varieties and several varietal types [5]. Notably, the predominant varieties used for salting, pickling, and fermentation include Brassica juncea var. tsatsai Mao, Brassica juncea var. tumida Tsen & Lee, and Brassica juncea var. multiceps [6,7,8,9,10,11]. Additionally, mustard greens have gained popularity in other Asian countries and regions due to their appealing characteristics of crispness and flavour [12]. Chinese mustard greens are rich in nutrients, including carbohydrates, dietary fibre, trace vitamins, essential minerals, and other beneficial nutrients. Furthermore, they contain a diverse array of bioactive components, such as thioglucosides and their degradation products, flavonoids, and phenolic compounds [13,14]. Moreover, research has demonstrated the positive health effects of fermented mustard extracts, including favourable antioxidant properties and potential anticancer effects [15,16,17]. However, it is important to note that the nutrient content of mustard changes during fermentation, particularly with respect to the geographic position, temperature, and time of fermentation.
Next-generation sequencing (NGS) is a main technology that has gained prominence during the rapid development of molecular biology. Nonculturable microorganisms can be obtained using NGS technology with high-throughput sequencing. Furthermore, NGS has contributed to evaluating the composition of microbial communities. In the context of fermented vegetables, high-throughput sequencing based on NGS technology has been employed to investigate the microbial community composition, taking into account factors such as geographic location and vegetable raw materials. Wang et al. conducted a comprehensive analysis of the bacterial community composition in traditional fermented sauerkraut from both northern and southern cities in China. Their findings revealed higher abundances of Lactobacillus spp., Bacillus acetobacter spp., Lauteromyces spp., and Weissella spp. in the fermented sauerkraut from the southern regions. Conversely, fermented sauerkraut from the northern regions exhibited increased abundances of Bacillus octococcus spp., Schizococcus spp., Morganella spp., Bacteroides oceanicus spp., Pseudomonas spp., and Providencia spp. [18]. Another study by Yang employed high-throughput sequencing to investigate the microbial composition and succession patterns in traditional sauerkraut fermentation among different farmers in Northeast China. The results indicated that the dominant genus in these sauerkraut samples was Weissella [19]. Furthermore, Peng et al. conducted an analysis of the bacterial flora composition in fermented vegetables made from Chinese cabbage, mung beans, bamboo shoots, and watermelon across different regions of Hainan (east, central, and west). Their study revealed that Lactobacillus spp., Chlorella spp., and Weissella spp. were the three dominant genera observed in the evolutionary relationships of the flora across different regions [20].
Traditional Chinese fermented mustard greens are distributed in several main regions, including Shantou (ST), Meizhou (MZ), Yunfu (YF), and Guangzhou (GZ), in Guangdong Province. It is necessary to research the differences in the quality and microbial community composition of Chinese fermented mustard greens. However, some studies have focused on the metabolites and flavours of traditional fermented vegetables [21]. There are still gaps in the knowledge on the colour and texture of fermented vegetables, particularly regarding the differences in the bacterial communities of fermented Chinese mustard greens from different regions. Additionally, the correlations between the quality and microbial communities of fermented Chinese mustard greens remain unknown. In this study, we used NGS technology to evaluate the microbial community composition of traditional fermented Chinese mustard greens from four regions of Guangdong Province, China. The relationships between quality (colour and texture) and microorganism diversity were analysed.
2. Materials and Methods
2.1. Sample Collection and Preparation
Chinese mustard greens should be exposed to sunlight for one day to increase their degree of drying. Salt was added to the surface, and the Chinese mustard greens were rubbed until they reached a wrinkled texture. The prepared mustard was placed in a glass container arranged in a spiral pattern, ensuring that the container was filled to approximately 70% of its capacity. A few Capsicum frutescens can be added for seasoning purposes, accompanied by a small amount of salt. The container was sealed to initiate the fermentation process, which required a minimum duration of 15 days. All samples were collected from four households in Guangdong Province, China: Shantou (ST) (116°43′ 36″ E, 23°17′9″ N), Meizhou (MZ) (116°7′1″ E, 24°18′36″ N), Yunfu (YF) (112°0′12″ E, 23°4′16″ N), and Guangzhou (GZ) (113°13′7″ E, 23°24′12″ N). Sampling was carried out according to previous methods [22]. The samples were collected from the pickle jar at the top, middle, and bottom. The samples were mixed evenly and packed in sterile self-sealing bags. Twenty-four samples from ST, MZ, YF, and GZ were obtained at the fermentation end of fermented Chinese mustard greens and then transported to the laboratory for further analysis.
2.2. Evaluation of Colour and Texture
The colours of the stem parts and leaf parts of the fermented Chinese mustard greens were measured. The colours, including L* (lightness), C* (chroma), a* (green chromaticity), and b* (yellow chromaticity), were determined using a CR400/CR410 colourimeter (Minolta, Tokyo, Japan) [23]. The firmness and chewiness of the samples were measured using a TA. XT texture analyser (Stable Micro Systems Ltd., Go-dalming, UK). The stem part of each leaf was used to evaluate the texture index. We measured the firmness and chewiness based on the force (N) using a P5 compression probe [24]. Each experiment was carried out three times.
2.3. DNA Extraction and PCR Amplification
The genomic DNA of microbes was extracted from different samples, including the ST, MZ, YF, and GZ regions, using a DNA kit (Omega Biotek, Norcross, GA, USA) according to the manufacturer’s instructions. The integrity, concentration, and purity of the genomic DNA were determined using 1% agarose gel electrophoresis and a NanoDrop 2000 UV–vis spectrophotometer (Thermo Scientific, Wilmington, DE, USA). The primer pairs 515F (5′-GTGYCAGCMGCCGCGGTAA-3′) and 806R (5′-GGACTACNVGGGTWTCTAA-3′) were used to amplify the hypervariable region V3-V4 of the bacterial 16S rRNA gene via an ABI GeneAmp® 9700 PCR thermocycler (ABI, Los Angeles, CA, USA) [25]. The PCR amplification conditions were 95 °C for 3 min; 27 cycles of 95 °C for 30 s, 55 °C for 30 s and 72 °C for 10 min; and 72 °C for 10 min. The mixtures included 4 μL of buffer (5 × TransStart FastPfu), 2 μL of dNTPs (2.5 mM), 0.8 μL of primer (5 μM), 0.4 μL of DNA polymerase (TransStart FastPfu), 10 ng of genomic DNA, and ddH2O (20 μL). The amplified DNA was detected by 2% agarose gel electrophoresis followed by purification with an AxyPrep DNA Gel Extraction Kit (Axygen Biosciences, Union City, CA, USA) according to the manufacturer’s instructions.
2.4. Illumina MiSeq Sequencing and Processing
The purified amplicons were pooled in equimolar amounts and paired-end sequenced (2 × 300) using Illumina MiSeq sequencing (Illumina, San Diego, CA, USA) at Majorbio Bio-Pharm Technology Co., Ltd. (Shanghai, China).
2.5. Statistical Analysis
All the experiments were conducted in triplicate as independent experiments. The data were analysed using SPSS software (version 14.0; SPSS, Chicago, IL, USA). The significance of differences between the variables was tested using one-way ANOVA. The means were compared using Duncan’s multiple range test. Statistical significance was determined at p < 0.05.
3. Results
3.1. Evaluation of Colour in Fermented Chinese Mustard Greens
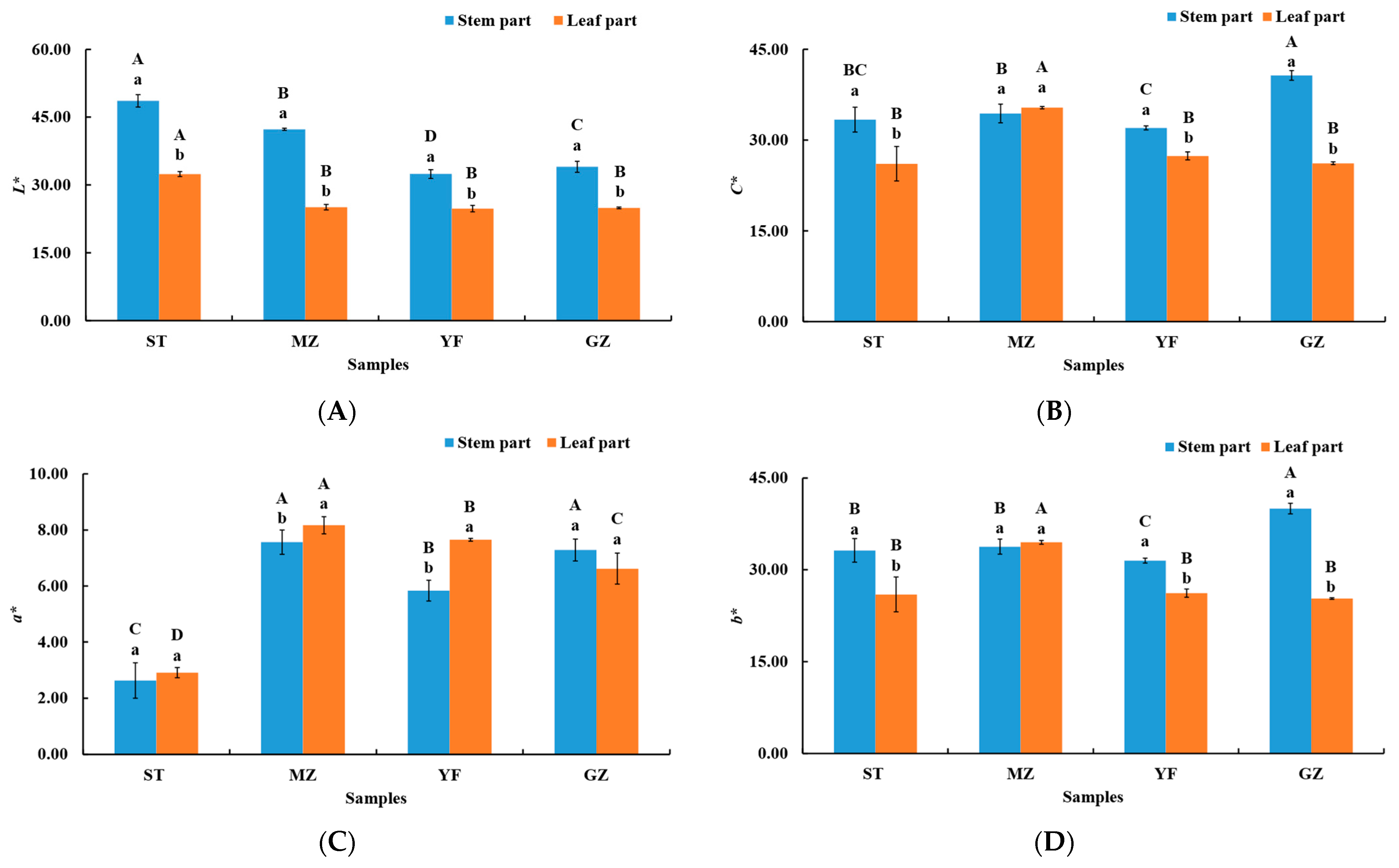
The colour of fermented Chinese mustard greens plays a significant role in determining its acceptability by consumers. The colour of the fermented Chinese mustard greens was represented with L*, C*, a*, and b* in different regions (ST, MZ, YF, and GZ) (Figure 1). The colours of the stem and leaf parts of the fermented mustard greens significantly differed. The L* values of the stem parts from the ST, MZ, YF, and GZ samples were greater than that of the leaf parts (p < 0.05) (Figure 1A). The L* values of the stem parts of the ST, MZ, GZ, and YF samples were 48.62, 42.30, 34.02, and 32.43, respectively. The L* of ST also showed the highest value (32.40) in the leaf part among all samples. The enzymatic browning reaction caused by polyphenol oxidase (PPO) and phenolic compounds happen during the fermentation period. This may be due to the higher PPO activity in leaf part, producing lower L* values. The C* and b* values of the stem part were greater than those of the leaf part in the ST, YF, and GZ samples (Figure 1B,D). The C* value (40.67) and b* value (40.02) of the stem part from the GZ sample were greater than those of the other samples (Figure 1B,D). The C* value (35.35), a* value (8.17), and b* value (34.48) of the leaf part from the MZ sample were greater than those of the other samples (Figure 1B–D). Interestingly, C* and b* values did not significantly differ between the stem and leaf parts in the MZ samples.
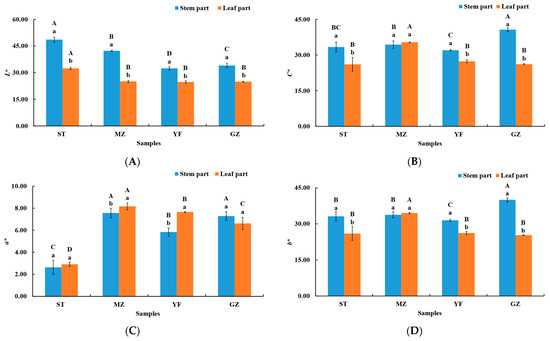
Figure 1.
The colour of the stem and leaf parts of fermented Chinese mustard greens. (A) L*, (B) C*, (C) a*, (D) b*. ST is Shantou, MZ is Meizhou, YF is Yunfu, and GZ is Guangzhou. Means designated by the same letters (uppercase, among different samples; lowercase, among stem parts and leaf parts) are significantly different according to Duncan’s test. Bars represent the means ± SD (n = 3, p < 0.05).
3.2. Analysis of Textures in Fermented Chinese Mustard Greens
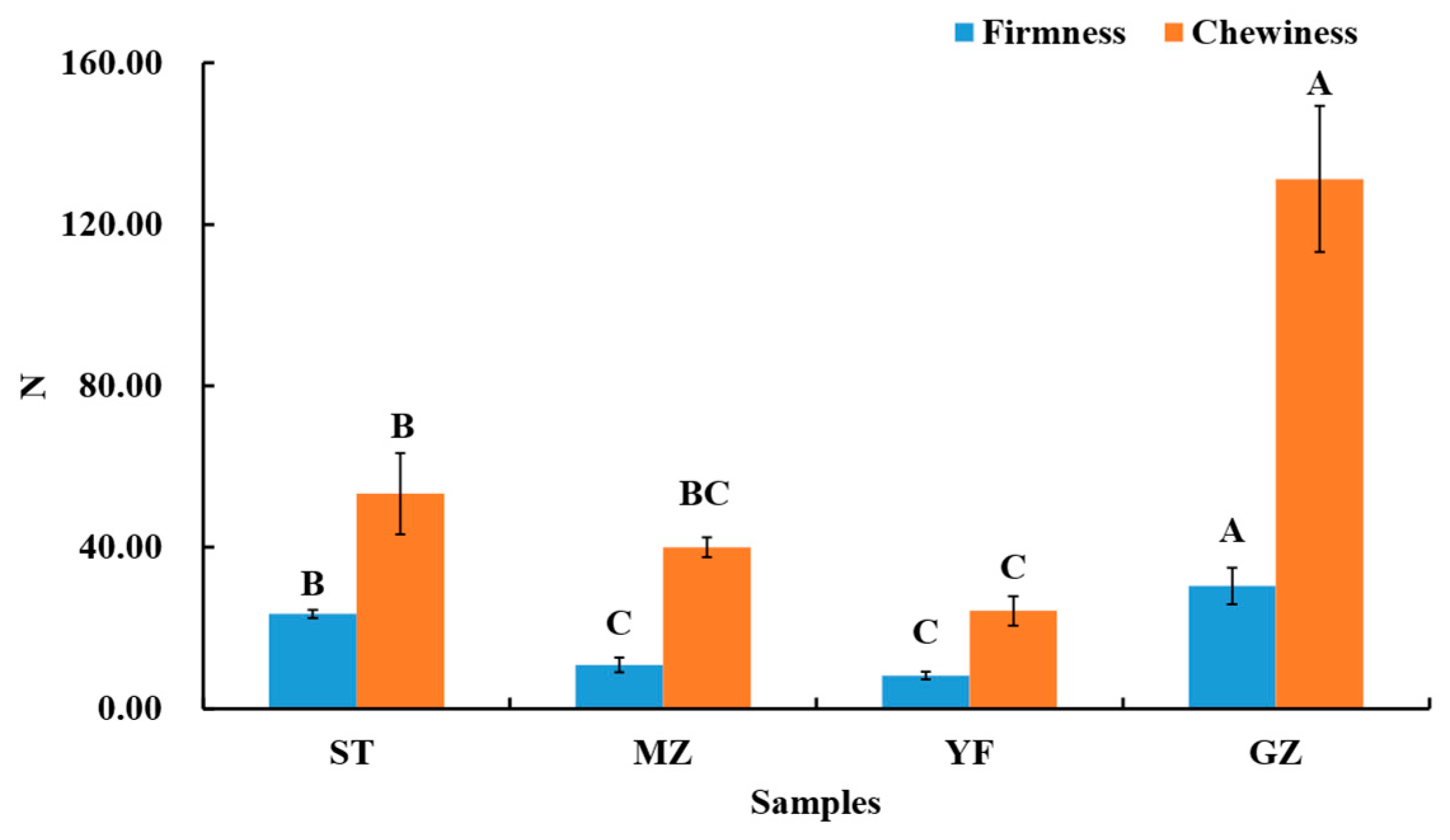
The composition and structure of the cell walls determine the firmness of Chinese mustard greens. Disassembly of the middle lamella and primary cell wall structures results in softening. The firmness and chewiness of the fermented Chinese mustard greens were evaluated, as shown in Figure 2. The firmness of the GZ samples was 30.37, which was the highest value, followed by those of the ST samples (23.49), MZ samples (10.80), and YF samples (8.20). The chewiness of the GZ samples is also significantly greater than that of the ST, MZ, and YF samples. This is due to the longer fermentation time of the GZ samples than that of others. Another study reported that the chewiness of fermented radish displayed a significant increase in different containers (glass jars, porcelain jars, and plastic jars) during the fermentation period [2]. The pectin in the cell wall of Chinese mustard greens gradually hydrolyses under acidic conditions. Some pectinases secreted by fermented microorganisms can also hydrolyse pectin. Therefore, differences in firmness and chewiness might largely depend on the microbial community.
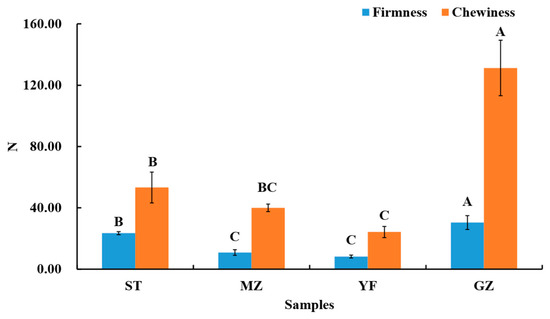
Figure 2.
The firmness and chewiness of fermented Chinese mustard greens. ST is Shantou, MZ is Meizhou, YF is Yunfu, and GZ is Guangzhou. Means designated by the same letters (uppercase, among different samples) are significantly different according to Duncan’s test. Bars represent means ± SDs (n = 3, p < 0.05).
3.3. Comparison of the Diversity Indices of Fermented Chinese Mustard Greens
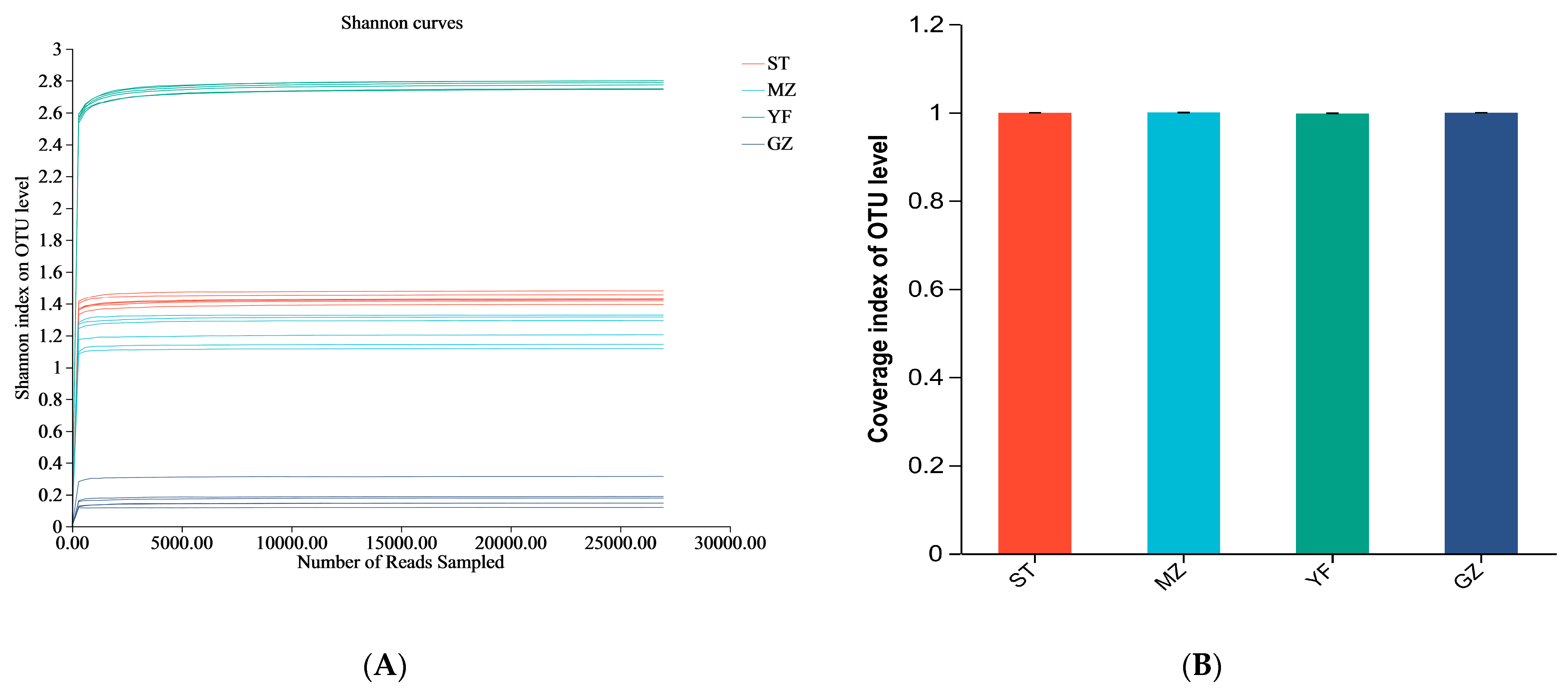
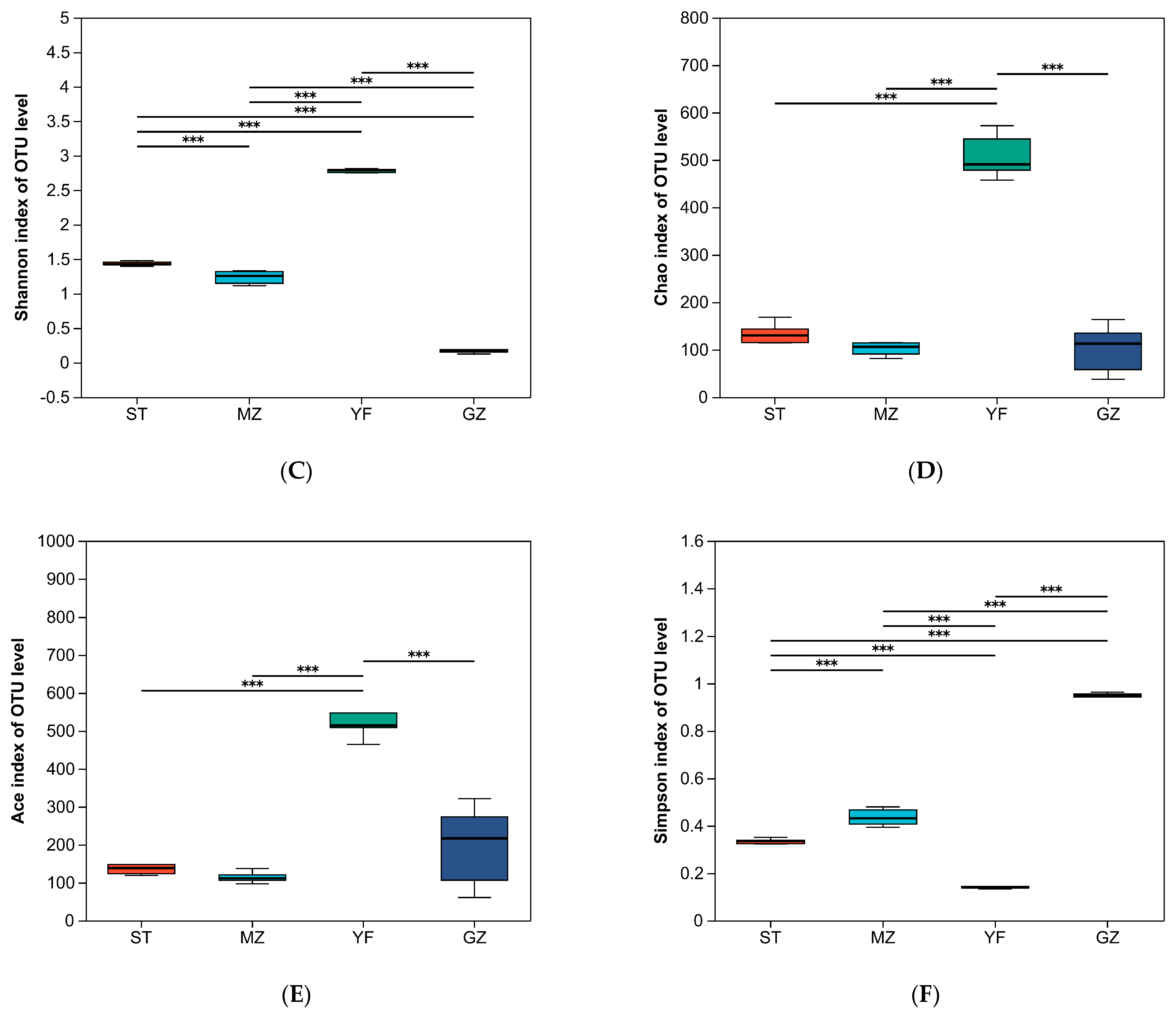
ST, MZ, YF, and GZ are traditional fermented Chinese mustard greens from four different regions in Guangdong Province. We compared the differences in the diversity indices of fermented Chinese mustard greens from different regions. The indices of alpha diversity, including the sequence data and OTU numbers, are shown in Table 1. The mean OTUs of the ST, MZ, YF, and GZ samples were 105, 101, 449, and 65, respectively. The convergence of rarefaction curves among the 24 samples continued to stabilize (Figure 3A), and the coverage indices ranged from 0.996 to 1 (Figure 3B), indicating that the depth of sequencing was sufficient to analyse the microbial community among all samples. The Shannon, Chao, Ace, and Simpson indices differed among the fermented Chinese mustard greens from the four regions. The YF samples had significantly greater Shannon, Chao, and Ace index values than the ST, MZ, and GZ samples did (Figure 3C–E). In the GZ sample, the Shannon index was the lowest, and the Simpson index was the highest (Figure 3C,F). In contrast to the other three groups, the YF sample showed significantly greater bacterial richness and bacterial community diversity.

Table 1.
Diversity indices of fermented Chinese mustard green from four regions in Guangdong Province.
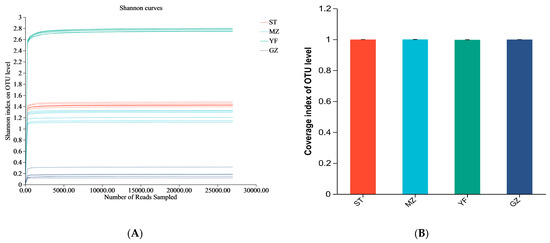
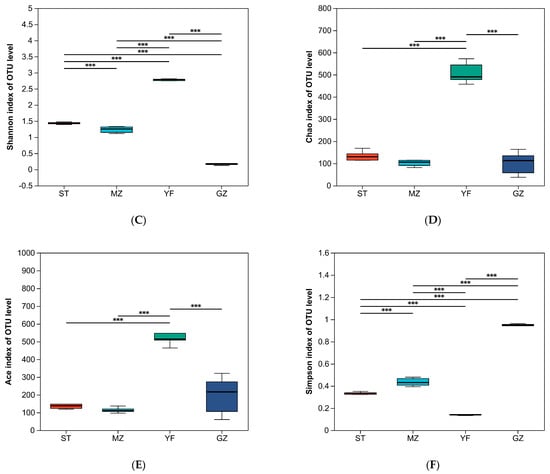
Figure 3.
Alpha diversity of the bacterial community of fermented Chinese mustard greens. (A) Shannon–non curve for each sample. (B) Coverage index. (C) Shannon index. (D) Chao1 values. (E) Ace index. (F) Simpson index. *** p < 0.051. ST is Shantou, MZ is Meizhou, YF is Yunfu, and GZ is Guangzhou.
3.4. Microbial Community Comparisons
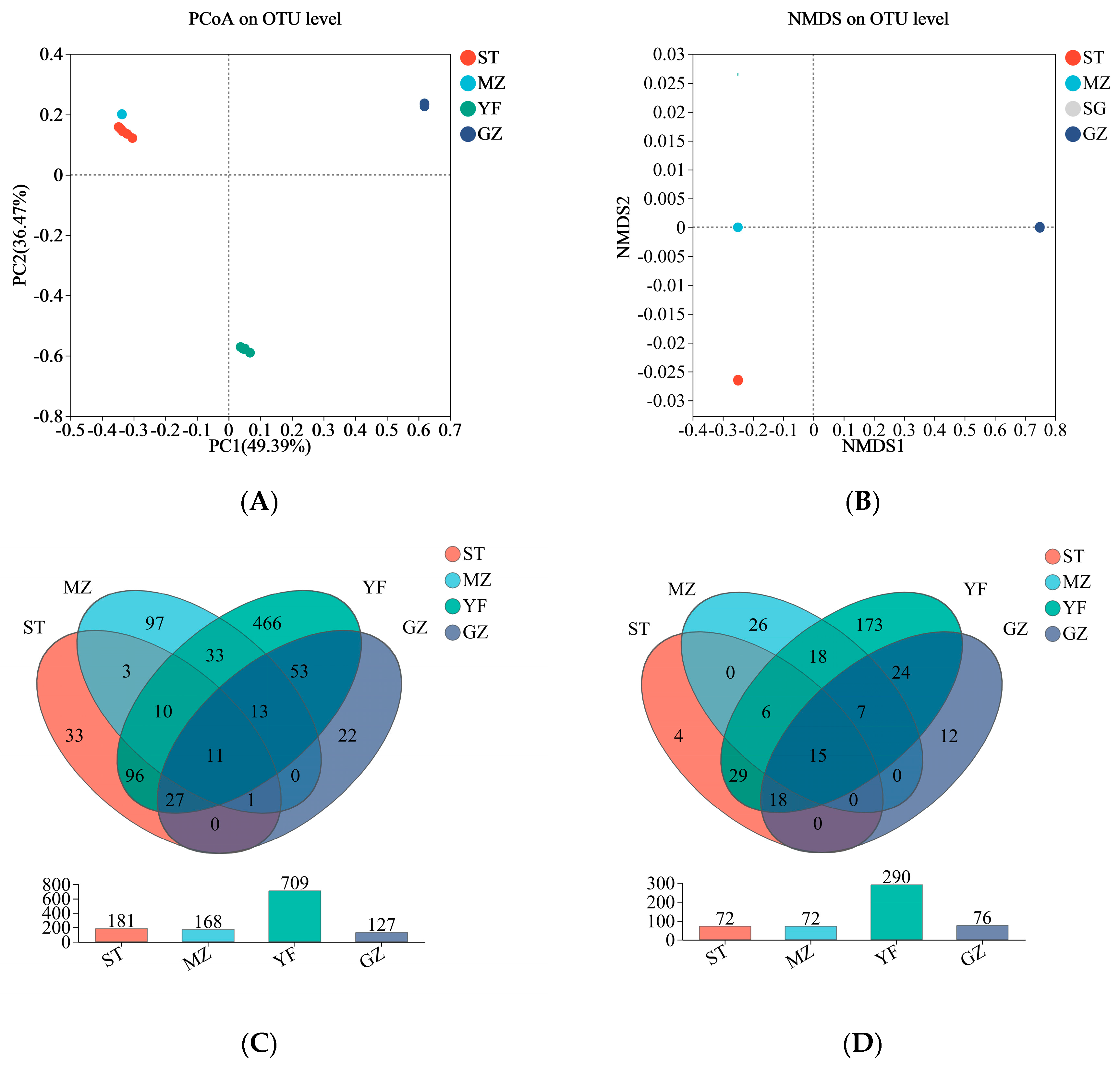
The microbial composition of the fermented Chinese mustard greens was compared via principal coordinate analysis (PCoA) and nonmetric multidimensional scaling (NMDS). The microbial communities in both the YF and GZ samples were significantly different from those in the other regions according to the PCoA results (Figure 4A). The NMDS analysis was consistent with the PCoA results. There was an apparent separation between the YF sample and the other samples and between the GZ sample and the other samples. There was obvious overlap between the ST samples and MZ samples. The bacterial composition of the YF and GZ samples differed significantly from that of the other two regions (Figure 4B). A Venn diagram of the OTUs and genera from the four regions of the samples is shown in Figure 4C,D. The intersection region among the four regions of samples showed 11 OTUs at the OUT level. The 466 OTUs from the YF samples differed from those from the other three regions (Figure 4C). At the genus level, the YF sample showed that 173 genera were different from those in the other region samples (Figure 4D), implying that the microbial community of the YF sample was most different from that of the other region samples.
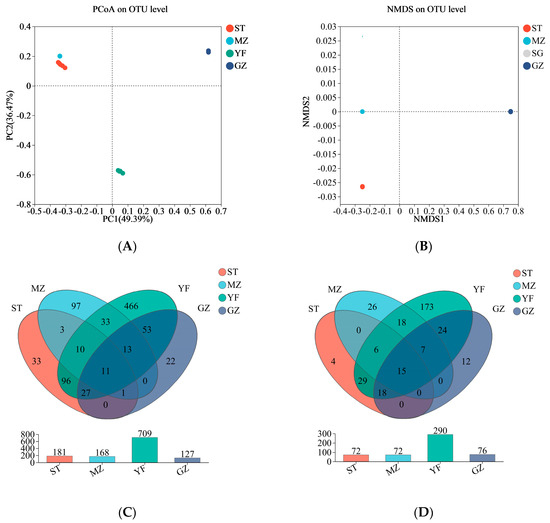
Figure 4.
Beta diversity and Venn diagrams of the bacterial community of fermented Chinese mustard greens. (A) Principal coordinate analysis (PCoA). (B) Nonmetric multidimensional scaling (NMDS). Venn diagrams at the OTU (C) and genus (D) levels according to bacterial biodiversity. ST is Shantou, MZ is Meizhou, YF is Yunfu, and GZ is Guangzhou.
3.5. Bacterial Profiles of Fermented Chinese Mustard Greens
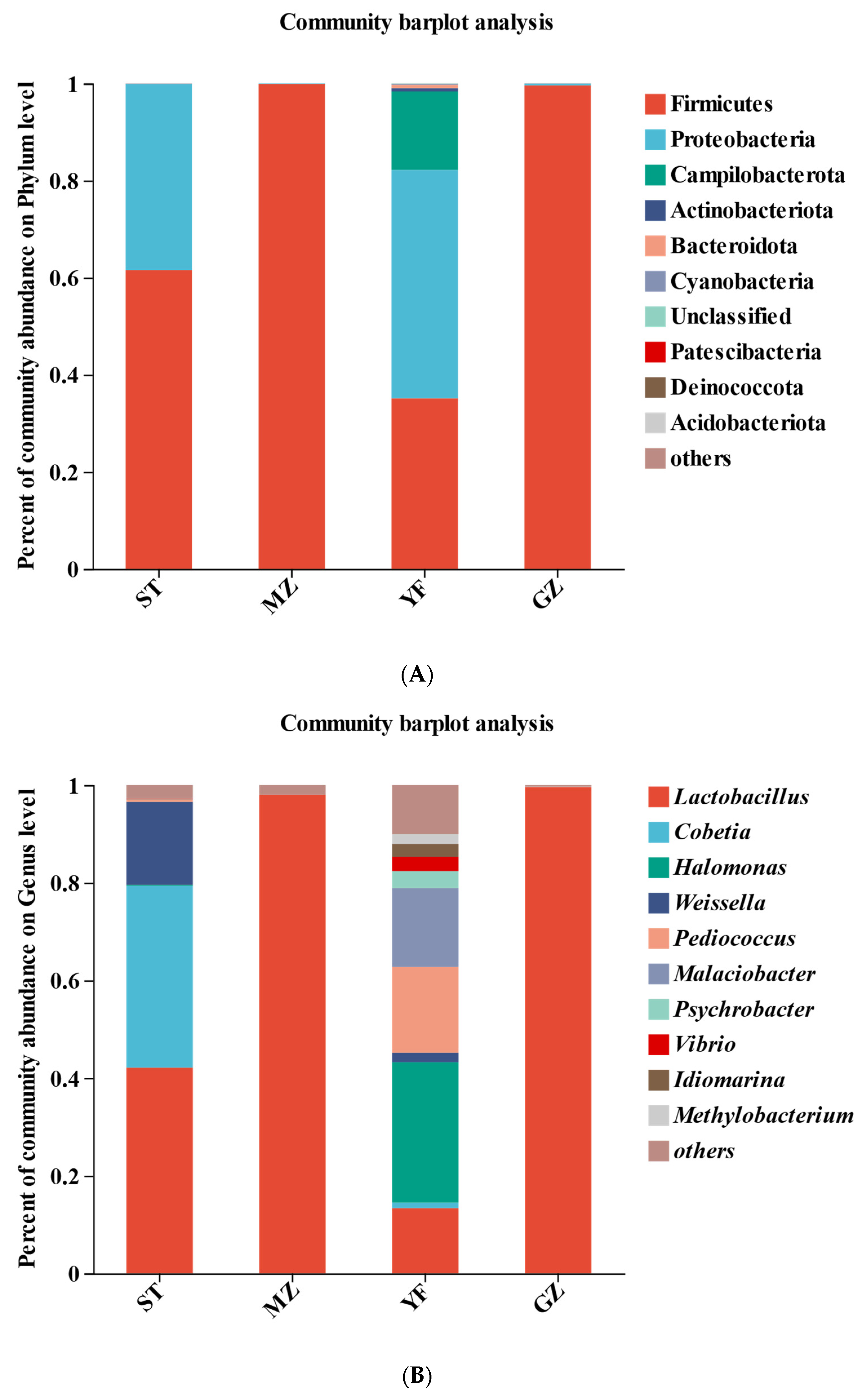
The bacterial community compositions of the samples from the four regions are shown in Figure 5. Firmicutes and Proteobacteria were the main phyla in the ST samples; Firmicutes were the main phyla in the MZ and GZ samples; and Firmicutes, Proteobacteria, and Campilobacterota were the main phyla in the YF samples, accounting for more than 90% of the annotated reads at the phylum level (Figure 5A). The abundance of Firmicutes was relatively greater in the MZ (99.86%) and GZ (99.6%) zones, followed by the ST (61.54%) and YF (35.14%) zones. Proteobacteria, the second most predominant phylum, accounted for 38.31% and 47.06% of the total bacteria in the ST and YF samples, respectively. Campilobacterota was observed only in the YF samples, accounting for 16.16%.
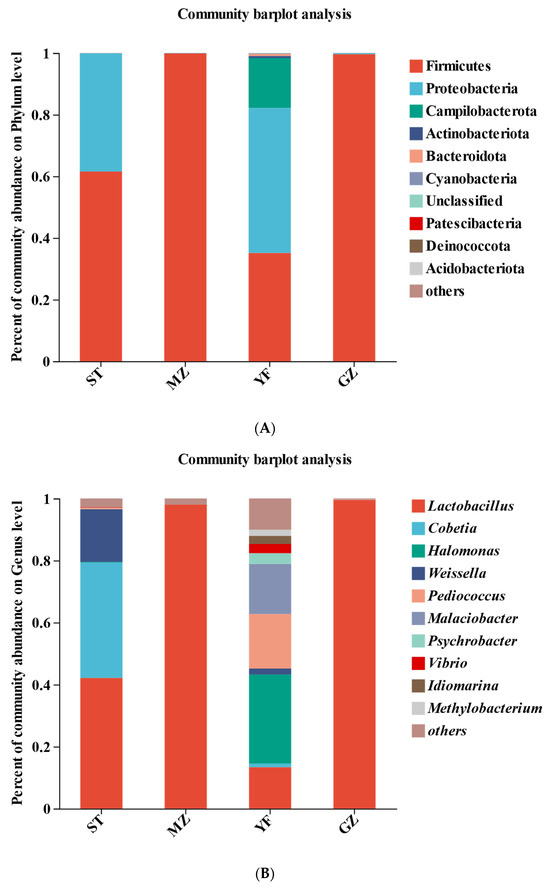
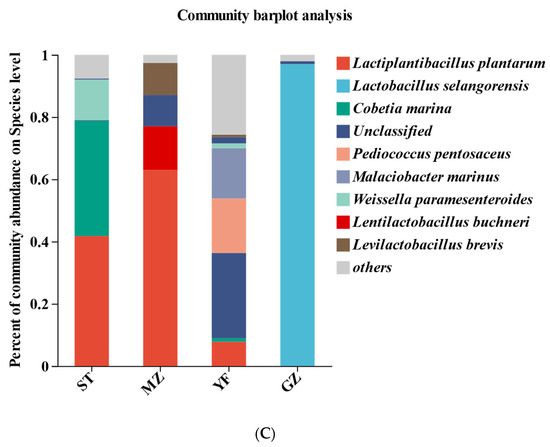
Figure 5.
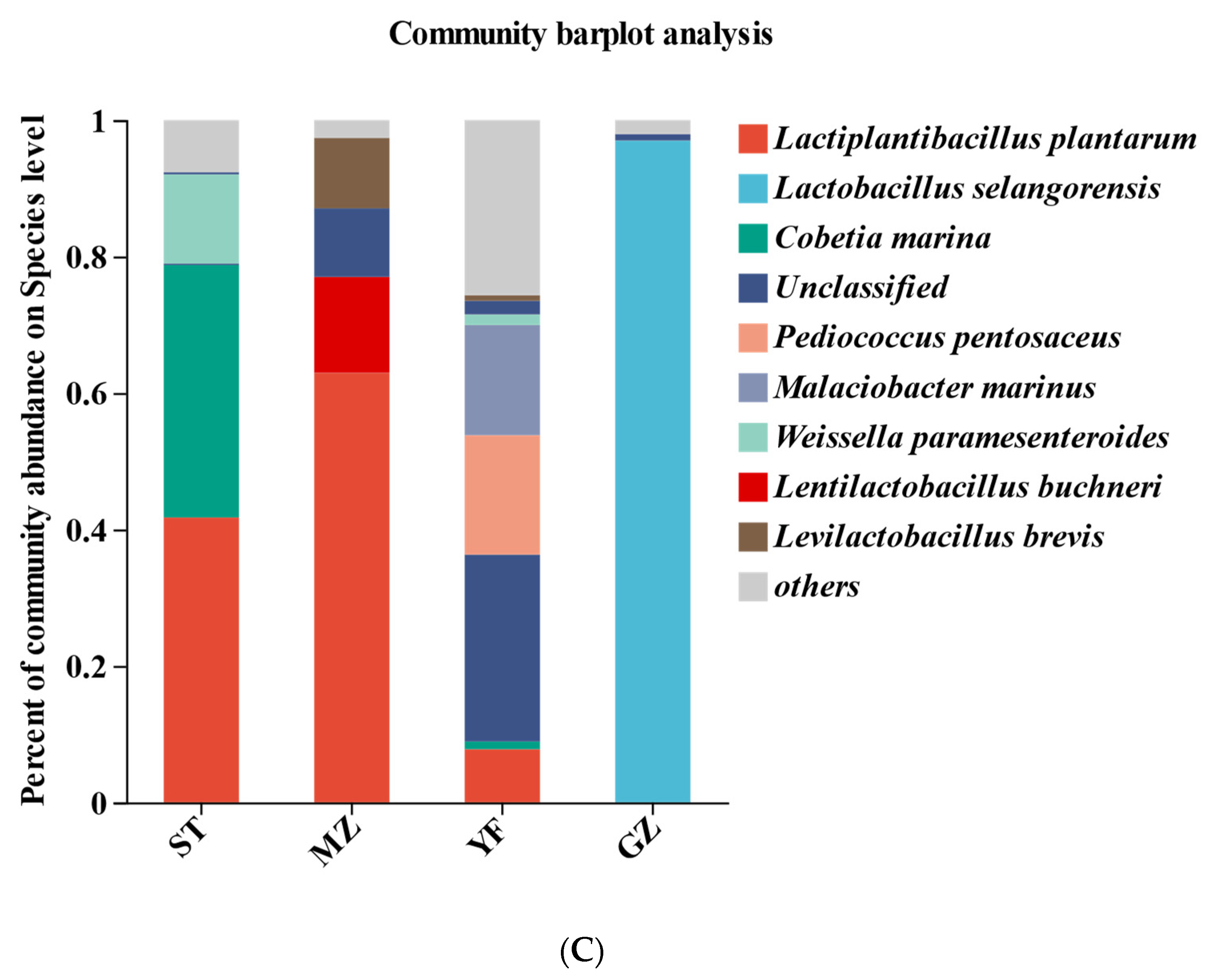
Composition of bacterial communities in fermented Chinese mustard greens at the phylum (A), genus (B), and species (C) levels. ST is Shantou, MZ is Meizhou, YF is Yunfu, and GZ is Guangzhou.
Lactobacillus, Cobetia, Halomonas, Weissella, Pediococcus, Malaciobacter, and Psychrobacter were the most represented genera in all the samples (Figure 5B). Lactobacillus is a key and dominant bacterial genus in fermented vegetables. Notably, Lactobacillus was the dominant genus in the MZ (98.02%) and GZ (99.56%) zones. Lactobacillus (42.14%), Cobetia (37.22%), and Weissella (16.98%) were highly abundant in the ST samples. However, Lactobacillus accounted for 13.37% of the YF samples, while Halomonas (28.71%), Pediococcus (17.55%), Malaciobacter (16.16%), Psychrobacter (3.47%), Vibrio (2.96), Idiomarina (2.62%), Methylobacterium (2.0%), Weissella (1.96%), and Cobetia (1.14%) were observed in the YF samples.
Lactiplantibacillus plantarum, Lactobacillus selangorensis, Cobetia marina, Pediococcus pentosaceus, Malaciobacter marinus, Weissella paramesenteroides, Lentilactobacillus buchneri, and Levilactobacillus brevis were the most representative species in all the samples at the species level (Figure 5C). Lactobacillus plantarum (41.75%), Cobetia marina (37%), and Weissella paramesenteroides (13.08%) were abundant in the ST samples. Lactobacillus plantarum (62.98%) was most abundant in the MZ samples, followed by Lactobacillus buchneri (14.03%) and Lactobacillus brevis (10.31%). Notably, Lactobacillus selangorensis (96.97%) was dominant in the GZ samples. However, Lactiplantibacillus plantarum accounted for only 7.79% of the YF samples, while Pediococcus pentosaceus (17.5%), Malaciobacter marinus (16.14%), Weissella paramesenteroides (1.55%), and Cobetia marina (1.13%) were abundant in the YF samples.
3.6. Differential Bacteria in the Fermented Chinese Mustard Greens
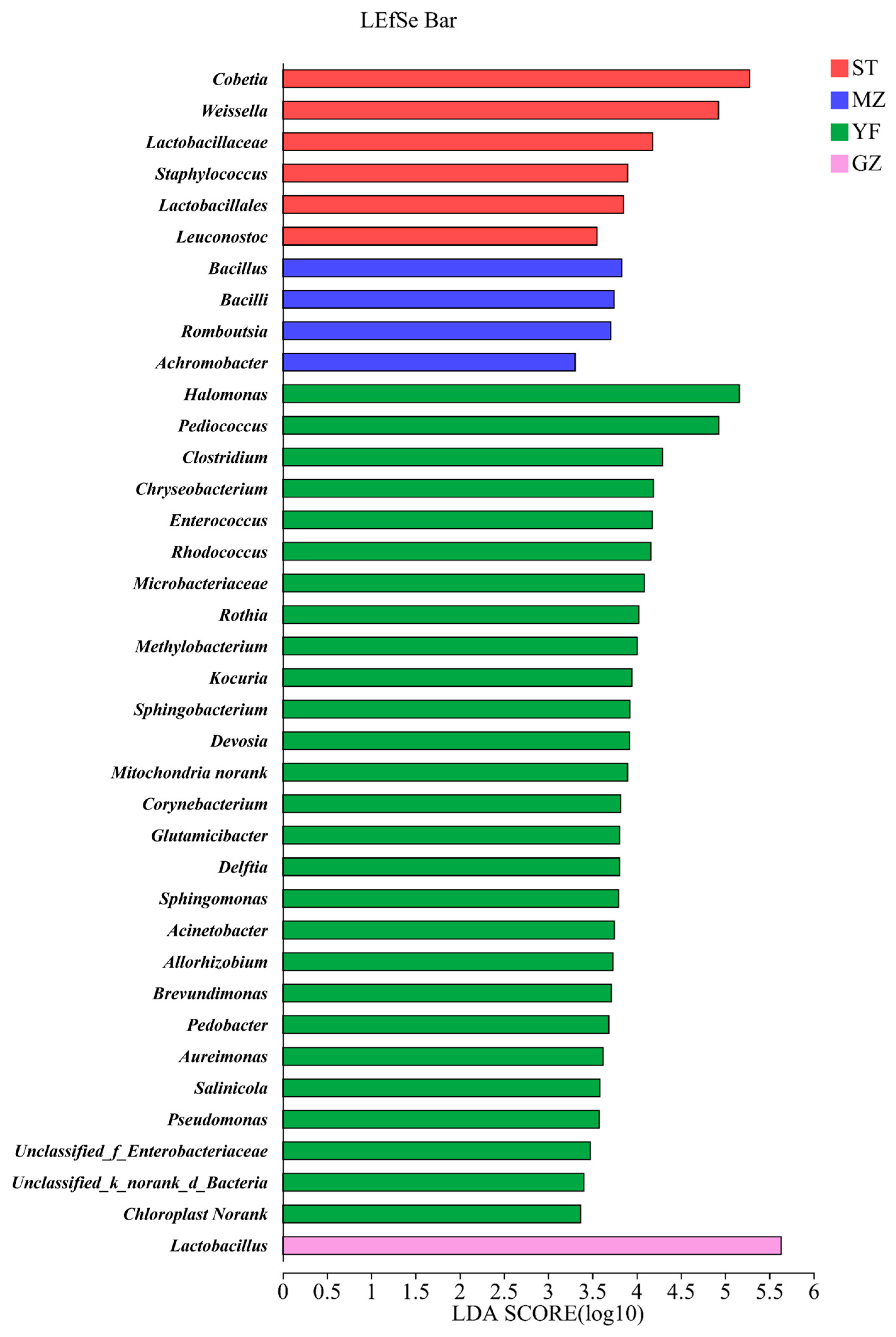
The microbial communities in different groups were analysed according to the relative abundance of the microbial composition via linear discriminant analysis effect size (LEfSe) (Figure 6). Cobetia, Weissella, Lactobacillaceae, Staphylococcus, Lactobacillales, and Leuconostoc were enriched in the ST samples. Bacillus, Bacilli, Romboutsia, and Achromobacter were enriched in the MZ samples. Twenty-seven genera were enriched in the YF samples. Only Lactobacillus was enriched in the GZ samples.
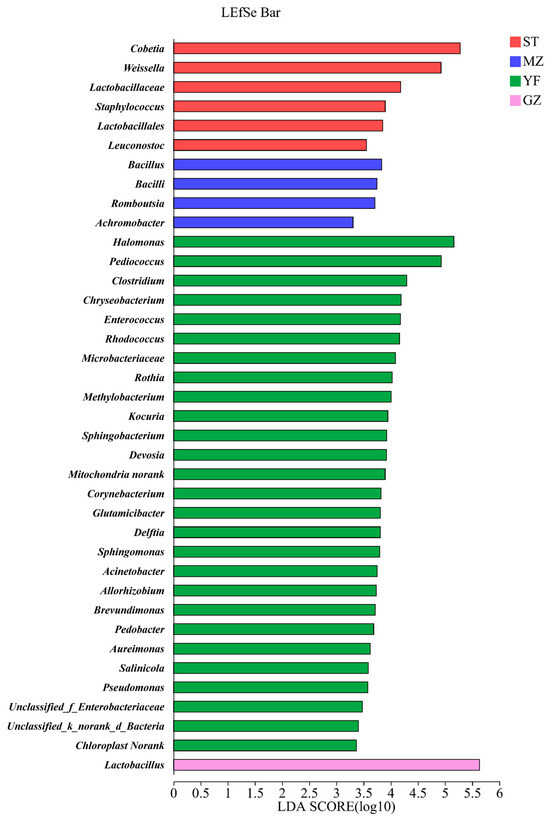
Figure 6.
LEfSe comparison of bacterial communities among fermented Chinese mustard greens. Histogram of the results of the microbiota with a threshold value of 2. ST is Shantou, MZ is Meizhou, YF is Yunfu, and GZ is Guangzhou.
3.7. Correlation of Bacterial Communities and the Quality Indices of Fermented Chinese Mustard Greens
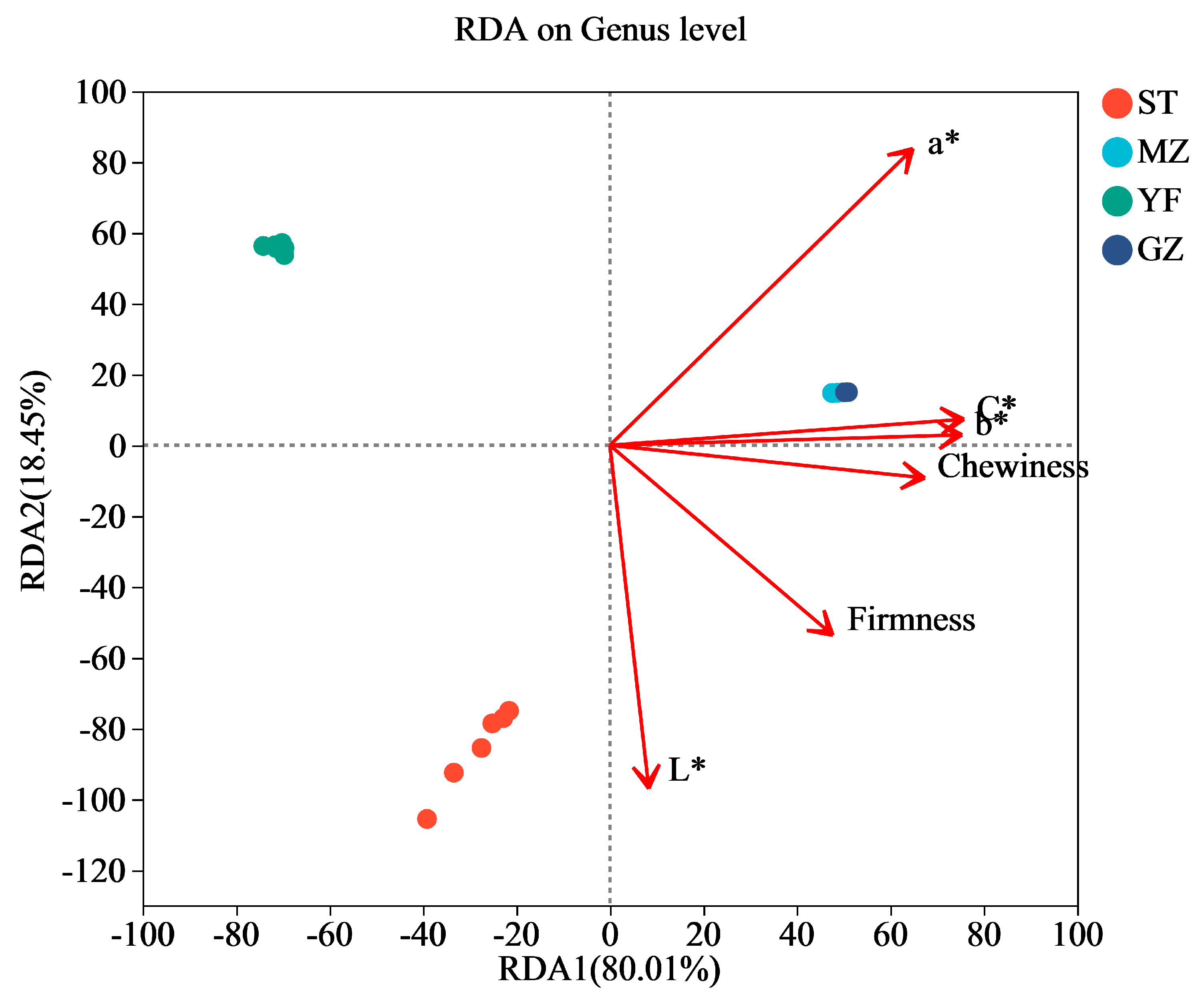
The results of the redundancy analysis (RDA) suggested that there was a strong correlation between quality (colour and texture) and the microbial communities (Figure 7). The horizontal axis was positively related to the contents of chewiness, rirmness, L*, a*, b*, and C*. The a* values of the colour of the Chinese mustard greens had the greatest impact on the bacterial communities, followed by the L*, C*, b* values, firmness, and chewiness. According to the Mantel test analysis, a* (r = 0.9291, p = 0.001) had the strongest correlation with microbial abundance, followed by L* (r = 0.7725, p = 0.001), C* (r = 0.4813, p = 0.001), b* (r = 0.4719, p = 0.001), firmness (r = 0.4199, p = 0.006), and chewiness (r = 0.382, p = 0.008). The colour indices (C* and b*) and texture (chewiness) were more strongly correlated with the MZ and GZ samples. L* values are more strongly correlated with the ST samples. For the YF samples, the correlation between the quality indices (colour and texture) and YF samples was not greater than that for the other samples. Furthermore, both RDA1 and RDA2 had an explanatory power of 98.46%, but further research is needed to investigate the relationships between species and quality indicators.
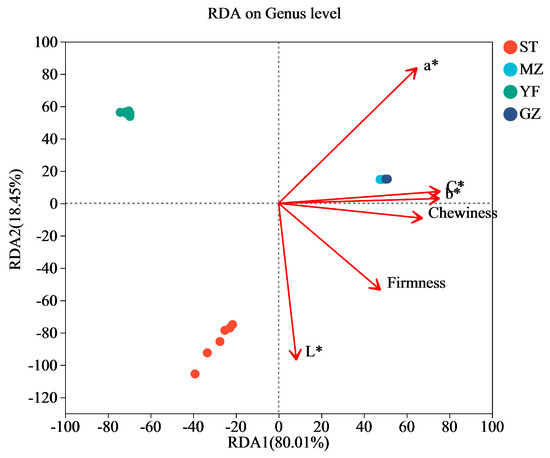
Figure 7.
Redundancy analysis (RDA) of bacterial communities among fermented Chinese mustard greens. ST is Shantou, MZ is Meizhou, YF is Yunfu, and GZ is Guangzhou.
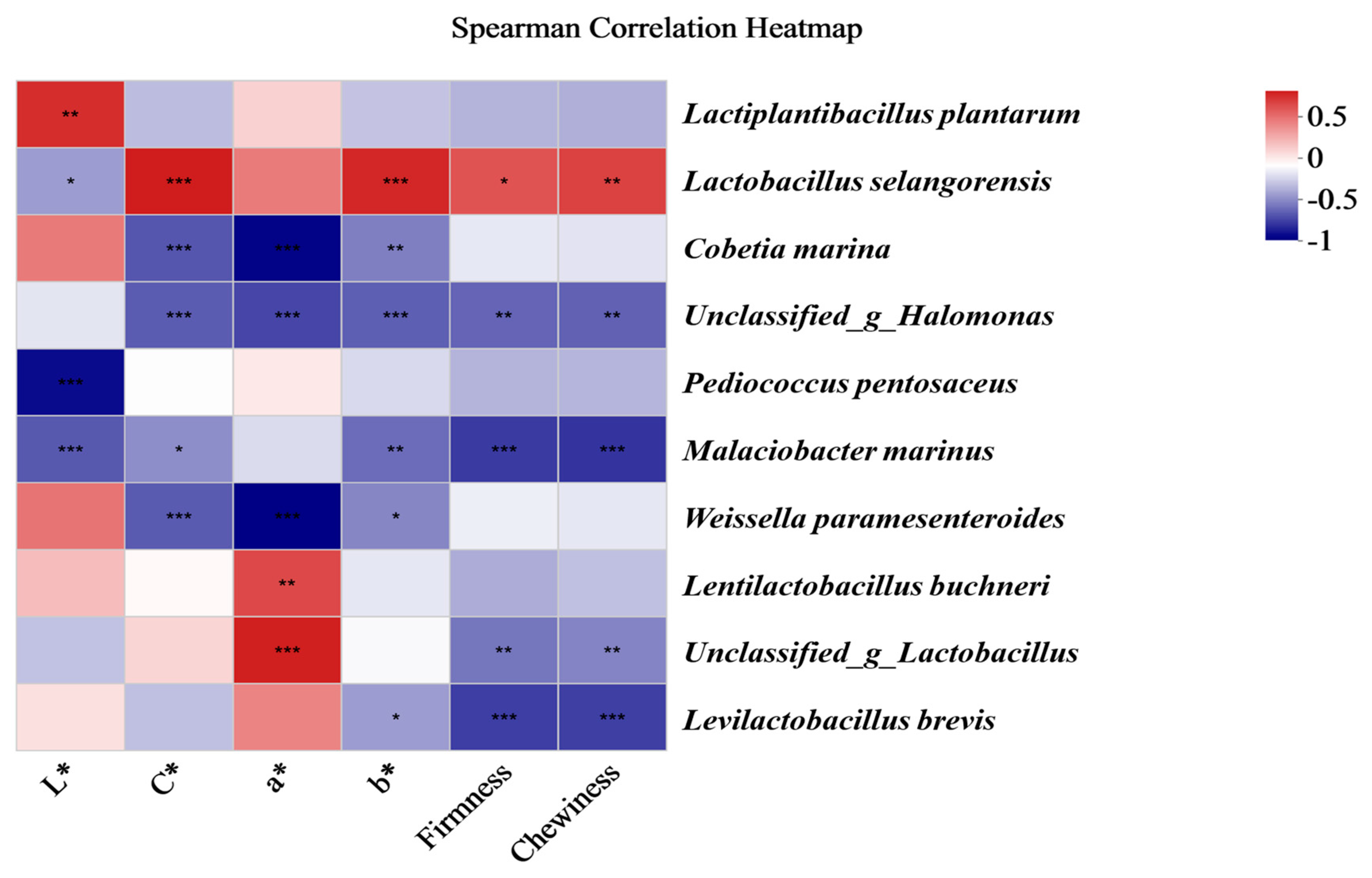
As shown in Figure 8, the colour quality indices (L*, C*, a*, and b*) and texture quality indices (firmness and chewiness) were correlated with the main bacterial community of the traditional Chinese mustard greens. L. plantarum was significantly positively correlated with L*. L. selangorensis was positively correlated with C*, b*, firmness, and chewiness. Lactobacillus buchneri was significantly positively correlated with a*. Cobetia marina was negatively significantly correlated with C*, a*, and b*, and Pediococcus pentosaceus was negatively significantly correlated with L*. Malacibacter marinus was significantly correlated with L*, C*, b*, firmness, and chewiness. Weissella paramesenteroides was significantly negatively correlated with C*, a*, and b*. Lactobacillus brecis was significantly negatively correlated with b*, firmness, and chewiness.
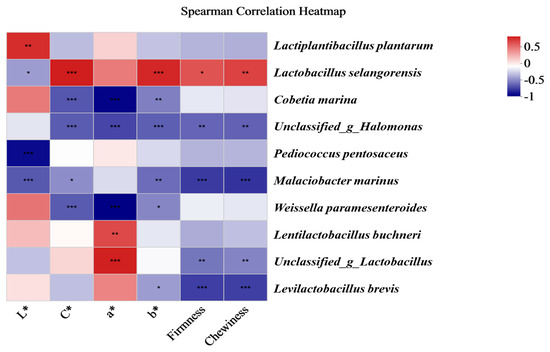
Figure 8.
Spearman correlation analysis of the quality indices and main species of bacterial communities among fermented Chinese mustard greens. The Spearman correlation coefficient r ranges from −0.5 to 0.5; r < 0 indicates a negative correlation, and r > 0 indicates a positive correlation. * p < 0.05, ** p < 0.01, *** p < 0.001. ST is Shantou, MZ is Meizhou, YF is Yunfu, and GZ is Guangzhou.
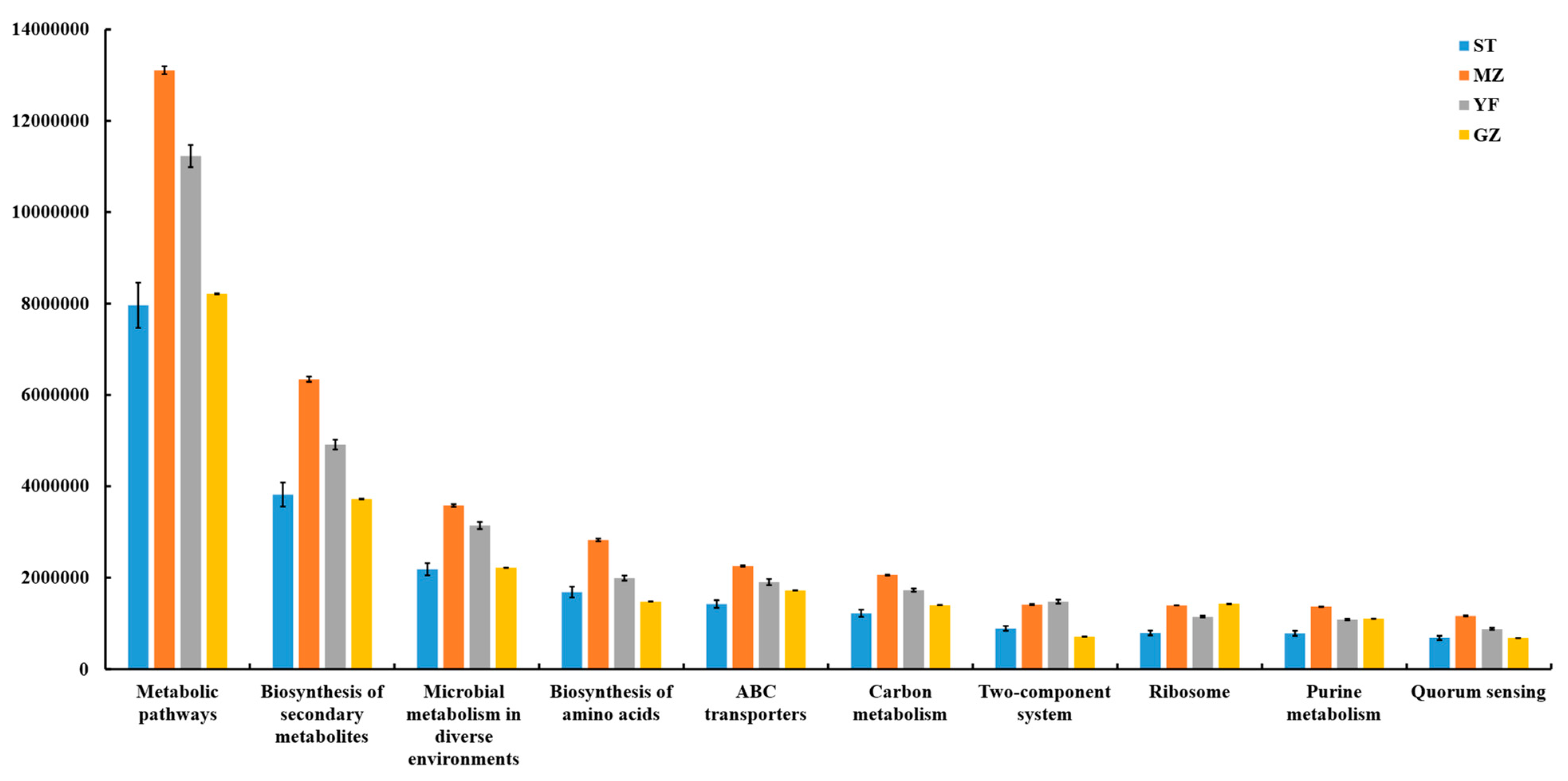
3.8. Predicted Functions of Microbial Communities
A PICRUSt model was applied to four regions of fermented Chinese mustard greens to predict their microbial community functions (Figure 9). Bacterial-related genes involved in metabolism, genetic information processing, environmental information processing, human diseases, cellular processes, and organism systems were detected in all the groups. Microbial communities are characterized by metabolism-related genes, which suggests that metabolism plays a major role. In all groups, the metabolic pathway with the greatest enrichment was at level 3, followed by biosynthesis of secondary metabolites, microbial metabolism in diverse environments, biosynthesis of amino acids, ABC transporters, carbon metabolism, two-component system, ribosome, purine metabolism, and quorum sensing, which showed significant differences among all groups. The ability of the MZ samples to predict metabolic pathways was the highest among all the samples.
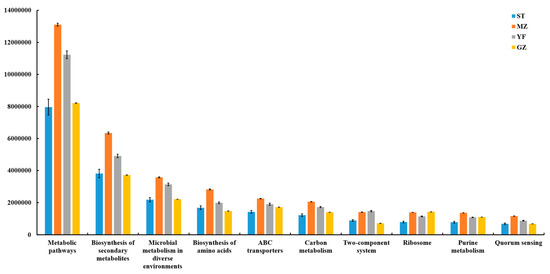
Figure 9.
The predictive functions of the bacterial community in fermented Chinese mustard greens (Pathway Level 3). ST is Shantou, MZ is Meizhou, YF is Yunfu, and GZ is Guangzhou.
4. Discussion
Our study employed high-throughput sequencing technology to investigate and compare the quality and bacterial communities of the ST, MZ, YF, and GZ samples. The relationship between quality (sensory and texture) and bacterial profiles was analysed among fermented Chinese mustard greens from different regions. There may be substantial differences among the microbial communities in the ST, MZ, YF, and GZ samples due to the different regions and fermentation processes. Lactobacillus was the most dominant genus in the MZ and GZ samples. Some studies have also shown that Lactobacillus is the main genus in fermented vegetables [26,27,28,29,30]. However, Lactobacillus, Cobetia, and Weissella were the predominant genera in the ST samples. This difference might be due to various influencing factors, including the process, temperature and time, equipment, and geography [31]. Some studies also show that Weissella was also highly abundant in fermented vegetables [32,33]. Another study reported that Cobetia was also the most abundant genus in fermented radish brine [34]. In addition to Lactobacillus, Halomonas, Prediococcus, Malaciobacter, and other genera were observed to be prevalent in the YF samples. These results for fermented vegetables are quite different from those of previous studies. The study reported that Halomonas has been isolated from seafood in Japan but is rarely isolated from fermented vegetables [35]. YF samples were found to contain high levels of Malaciobacter. These results also indicate that the microbial bacterial communities in the YF samples are distinct from those in the other regions. Malaciobacter can be isolated from fermented cheese in other studies [36]. These differences in the bacterial community might result from region, temperature, humidity, and fermentation processing. The salinity of fermented processing also significantly affects microorganism diversity and the final quality of fermented vegetables [37].
Furthermore, the species compositions of the four different regions were also significantly different. Lactiplantibacillus plantarum was the predominant bacteria in the ST and MZ samples. The abundance of Lentilacillus buchneri was also high in the MZ samples. The relative abundance of Lactobacillus selangorensis was much greater in the GZ samples than in the other four Chinese mustard greens from different regions. Another study showed that L. plantarum was the most abundant species in pickled vegetables [38]. Cobetia marina and W. paramesenteroides were significantly abundant in the ST samples, and C. marina was also found in the meat source food [39]. Nevertheless, the Lactobacillus species was relatively less abundant in the YF samples, and P. pentosaceus, M. marinus, and other species were more abundant. Therefore, these bacterial species may play an important role in the quality of YF samples. The samples were collected from different regions, including the ST, MZ, YF, and GZ samples in this study. Therefore, fermentation processing and region might be the reasons for the differences in the bacterial composition of these four fermented vegetables.
Colour and texture are important quality factors that influence consumer choice. There was a positive correlation between quality and the bacterial community based on the RDA results. This is the first study to report that the structure of bacterial communities affects colour and texture in traditional Chinese mustard greens from different regions. The results suggested that the colour indices (a*, b*, and c*) and chewiness were positively correlated with the bacterial communities of the MZ and GZ samples. This might be because Lactobacillus accounted for the highest proportion of the bacterial communities in the MZ and GZ samples. The L* values of the colour index and firmness index are also positively correlated with those of the ST samples. Further studies conclusively showed that Lactobacillus is also the main microorganism in ST samples. In addition, Lactobacillus accounted for a lower proportion of the bacteria in the YF samples, which was less strongly correlated with colour and texture. Therefore, among all microorganism genera, Lactobacillus might play a key role in colour and texture quality. Some studies have reported that the textural properties of fermented cheese vary considerably depending on the strain of Lactobacillus lactis used in its manufacture. This might be due to the increased protein hydration of fermented cheese [40]. Furthermore, other studies report that the seaweed species and the amount of kelp can affect the colour of the seaweed sauerkraut. An increase in the level of species results in a lower L* value and a higher a* value. A lighter colour is considered desirable in conventional seaweed sauerkraut [41]. Texture is a major index for evaluating sensory properties of fermented onion samples (white onion, red onion, and scallion) [42].
5. Conclusions
This study presented a detailed analysis and comparison of the quality and microbial community in traditional fermented Chinese mustard greens from four regions (ST, MZ, YF, and GZ) in Guangdong, China. The colour and texture of fermented Chinese mustard greens showed significant differences in different regions. The quality of fermented Chinese mustard greens in the GZ region was significantly greater than that in other regions according to the C*, b*, firmness, and chewiness values. Microbial community including Lactobacillus, Cobetia, Halomonas, Weissella, Pediococcus, Malaciobacter, and Psychrobacter were the more represented genera in some samples. Among those, Lactobacillus was the most predominate microorganism in the MZ and GZ regions and accounted for a greater proportion of the microorganisms in the ST and YF regions. Furthermore, there was a significant correlation between the microbial communities and quality indices (colour and texture) among the samples from the four regions. The Lactobacillus genus (L. plantarum, L. selangorensis) contributed to changes in colour (b*, C*, L*, a*) and texture (firmness and chewiness). There was also a microbial safety risk in traditional fermented Chinese mustard greens. Thus, future studies should focus on screening strains and optimising the fermentation process.
Author Contributions
Conceptualization, K.F. and W.H.; methodology, Y.K., Y.D., H.X. and X.T.; software, S. and K.F.; validation, W.H.; investigation, K.F.; resources, W.H.; data curation, K.F.; writing—original draft preparation, S.; writing—review and editing, K.F. and W.H.; project administration, K.F. and W.H.; funding acquisition, S. and K.F. All authors have read and agreed to the published version of the manuscript.
Funding
This study was supported by the Guangdong Basic and Applied Basic Research Foundation (Grant No. 2023A1515011473), Guangdong Province University Feature Innovation Project (2023KTSCX214), Zhuhai College of Science and Technology “Three Levels” Talent Construction Project, and Faculty Research Grants of Macau University of Science and Technology (FRG-24-024-FMD).
Data Availability Statement
The data used to support the findings of this study are available from the corresponding author upon request.
Conflicts of Interest
The authors declare no conflicts of interest.
References
- Hutkins, R.W. Fermented vegetables. Microbiol. Technol. Fermented Foods 2006, 5, 223–259. [Google Scholar]
- Qian, Y.; Tao, Y.; Li, Y.; Guo, S.; Xiang, W.; Liu, G.; Rao, Y. Microbiota succession and chemical composition involved in the radish fermentation process in different containers. Front. Microbiol. 2020, 11, 445. [Google Scholar]
- Hanelt, P.; Buttner, R. Mansfeld’s Encyclopedia of Agricultural and Horticultural Crops: Except Ornamentals; Springer: Berlin/Heidelberg, Germany, 2001; Volume 1. [Google Scholar]
- Tian, Y.; Deng, F. Phytochemistry and biological activity of mustard (Brassica juncea): A review. CyTA-J. Food 2020, 18, 704–718. [Google Scholar] [CrossRef]
- Fan, Y.; Shen, J.; Dong, D. Development status and research prospect of mustard vegetables industry. Agric. Sci. Technol. 2017, 18, 556–564. [Google Scholar]
- Zhang, C.; Zhang, J.; Liu, D. Biochemical changes and microbial community dynamics during spontaneous fermentation of Zhacai, a traditional pickled mustard tuber from China. Int. J. Food Microbiol. 2021, 347, 109199. [Google Scholar] [CrossRef]
- Lingjuan, J.; Yu, C.; Zeyuan, D.; Bing, Z.; Hongyan, L. Evaluation and comparison of physicochemical properties, volatile substances, and microbial communities of leaf mustard (Brassica juncea var. multiceps) under natural and inoculated fermentation. J. Food Sci. 2023, 88, 3255–3273. [Google Scholar]
- Liu, D.; Zhang, C.; Zhang, J.; Xin, X.; Liao, X. Metagenomics reveals the formation mechanism of flavor metabolites during the spontaneous fermentation of potherb mustard (Brassica juncea var. multiceps). Food Res. Int. 2021, 148, 110622. [Google Scholar] [CrossRef]
- Zhang, C.; Chen, J.; Li, X.; Liu, D. Bacterial community and quality characteristics of the fermented potherb mustard (Brassica juncea var. multiceps) under modified atmospheres. Food Res. Int. 2019, 116, 266–275. [Google Scholar] [PubMed]
- Zhao, D.; Ding, X. Studies on the low-salt Chinese potherb mustard (Brassica juncea, Coss.) pickle. I—The effect of a homofermentative l (+)-lactic acid producer Bacillus coagulans on starter culture in the low-salt Chinese potherb mustard pickle fermentation. LWT-Food Sci. Technol. 2008, 41, 474–482. [Google Scholar] [CrossRef]
- Zhao, D.; Tang, J.; Ding, X. Analysis of volatile components during potherb mustard (Brassica juncea, Coss.) pickle fermentation using SPME–GC-MS. LWT-Food Sci. Technol. 2007, 40, 439–447. [Google Scholar] [CrossRef]
- Xiao, M.; Huang, T.; Huang, C.; Hardie, J.; Peng, Z.; Xie, M.; Xiong, T. The microbial communities and flavour compounds of Jiangxi yancai, Sichuan paocai and Dongbei suancai: Three major types of traditional Chinese fermented vegetables. LWT-Food Sci. Technol. 2020, 121, 108865. [Google Scholar] [CrossRef]
- Kim, S.; Park, H.; Kim, J.; Moon, B. Effect of main vegetable ingredient on the glucosinolate, carotenoids, capsaicinoids, chlorophylls, and ascorbic acid content of kimchis. J. Food Compos. Anal. 2022, 110, 104523. [Google Scholar] [CrossRef]
- Kim, Y.T.; Kim, B.K.; Park, K.Y. Antimutagenic and anticancer effects of leaf mustard and leaf mustard kimchi. J. Food Sci. Nutr. 2007, 12, 84–88. [Google Scholar] [CrossRef]
- Lee, M.A.; Choi, J.H.; Choi, Y.S.; Han, D.J.; Kim, H.Y.; Shim, S.Y.; Chung, H.-K.; Kim, C.-J. The antioxidative properties of mustard leaf (Brassica juncea) kimchi extracts on refrigerated raw ground pork meat against lipid oxidation. Meat Sci. 2010, 84, 498–504. [Google Scholar] [CrossRef] [PubMed]
- Oh, S.; Tsukamoto, C.; Kim, K.; Choi, M. Investigation of glucosinolates, and the antioxidant activity of D olsan leaf mustard kimchi extract using HPLC and LC-PDA-MS/MS. J. Food Biochem. 2017, 41, e12366. [Google Scholar] [CrossRef]
- Lim, H.S.; Yoo, E.J.; Choi, M.R. Changes of physiological activity of mustard leaf during its fermentation period. J. Microbiol. Biotechnol. 2000, 10, 43–47. [Google Scholar]
- Wang, Z.; Shao, Y. Effects of microbial diversity on nitrite concentration in pao cai, a naturally fermented cabbage product from China. Food Microbiol. 2018, 72, 185–192. [Google Scholar] [CrossRef] [PubMed]
- Yang, X.; Hu, W.; Xiu, Z.; Jiang, A.; Yang, X.; Saren, G.; Ji, Y.; Guan, Y.; Feng, K. Microbial community dynamics and metabolome changes during spontaneous fermentation of northeast sauerkraut from different households. Front. Microbiol. 2020, 11, 1878. [Google Scholar] [CrossRef] [PubMed]
- Peng, Q.; Jiang, S.; Chen, J.; Ma, C.; Huo, D.; Shao, Y.; Zhang, J. Unique microbial diversity and metabolic pathway features of fermented vegetables from Hainan, China. Front. Microbiol. 2018, 9, 399. [Google Scholar] [CrossRef]
- Hernández-Carrión, M.; Sanz, T.; Hernando, I.; Llorca, E.; Fiszman, S.M.; Quiles, A. New formulations of functional white sauces enriched with red sweet pepper: A rheological, microstructural and sensory study. Eur. Food Res. Technol. 2015, 240, 1187–1202. [Google Scholar] [CrossRef]
- Yang, X.; Hu, W.; Xiu, Z.; Jiang, A.; Yang, X.; Saren, G.; Ji, Y.; Feng, K. Microbial dynamics and volatilome profiles during the fermentation of Chinese northeast sauerkraut by Leuconostoc mesenteroides ORC 2 and Lactobacillus plantarum HBUAS 51041 under different salt concentrations. Food Res. Int. 2020, 130, 108926. [Google Scholar] [CrossRef] [PubMed]
- Gaowa, S.; Feng, K.; Li, Y.; Long, Y.; Hu, W. Effect of alginate-based edible coating containing thyme essential oil on quality and microbial safety of fresh-cut potatoes. Horticulturae 2023, 9, 543. [Google Scholar] [CrossRef]
- Sarengaowa; Wang, L.; Liu, Y.; Yang, C.; Feng, K.; Hu, W. Screening of Essential Oils and Effect of a chitosan-based edible coating containing cinnamon oil on the quality and microbial safety of fresh-cut potatoes. Coatings 2022, 12, 1492. [Google Scholar] [CrossRef]
- Walters, W.; Hyde, E.R.; Berglyons, D.; Ackermann, G.; Humphrey, G.; Parada, A.; Gilbert, J.A.; Jansson, J.K.; Caporaso, J.G.; Fuhrman, J.A.; et al. Improved bacterial 16S rRNA gene (V4 and V4-5) and fungal internal transcribed spacer marker gene primers for microbial community surveys. Msystems 2016, 1, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Liou, J.S.; Huang, C.H.; Wang, C.L.; Lee, A.Y.; Mori, K.; Tamura, T.; Watanabe, M.; Blom, J.; Huang, L.; Watanabe, K. Lactobacillus suantsaii sp. nov., isolated from suan-tsai, a traditional Taiwanese fermented mustard green. Int. J. Syst. Evol. Microbiol. 2019, 69, 1484–1489. [Google Scholar] [CrossRef] [PubMed]
- Cao, J.; Yang, J.; Hou, Q.; Xu, H.; Zheng, Y.; Zhang, H.; Zhang, L. Assessment of bacterial profiles in aged, home-made Sichuan paocai brine with varying titratable acidity by PacBio SMRT sequencing technology. Food Control 2017, 78, 14–23. [Google Scholar] [CrossRef]
- Liu, A.; Liu, G.; Huang, C.; Shen, L.; Li, C.; Liu, Y.; Liu, S.; Hu, B.; Chen, H. The bacterial diversity of ripened Guang’yuan Suancai and in vitro evaluation of potential probiotic lactic acid bacteria isolated from Suancai. LWT-Food Sci. Technol. 2017, 85, 175–180. [Google Scholar] [CrossRef]
- Liu, X.; Kuda, T.; Takahashi, H.; Kimura, B. Bacterial and fungal microbiota of spontaneously fermented Chinese products, Rubing milk cake and Yan-cai vegetable pickles. Food Microbiol. 2018, 72, 106. [Google Scholar] [CrossRef] [PubMed]
- Guan, Q.; Zheng, W.; Huang, T.; Xiao, Y.; Xiong, T. Comparison of microbial communities and physiochemical characteristics of two traditionally fermented vegetables. Food Res. Int. 2020, 128, 108755. [Google Scholar] [CrossRef]
- Zhu, Y.; Zhang, F.; Zhang, C.; Yang, L.; Fan, G.; Xu, Y.; Sun, B.; Li, X. Dynamic microbial succession of Shanxi aged vinegar and its correlation with flavor metabolites during different stages of acetic acid fermentation. Sci. Rep. 2018, 8, 8612. [Google Scholar] [CrossRef]
- Liang, T.; Xie, X.; Wu, L.; Li, L.; Li, H.; Xi, Y.; Feng, Y.; Xue, L.; Chen, M.; Chen, X.; et al. Microbial communities and physiochemical properties of four distinctive traditionally fermented vegetables from North China and their influence on quality and safety. Foods 2022, 11, 21. [Google Scholar] [CrossRef] [PubMed]
- He, Z.; Chen, H.; Wang, X.; Lin, X.; Ji, C.; Li, S.; Liang, H. Effects of different temperatures on bacterial diversity and volatile flavor compounds during the fermentation of suancai, a traditional fermented vegetable food from northeastern China. LWT-Food Sci. Tech. 2020, 118, 108773. [Google Scholar] [CrossRef]
- Zhang, J.; Song, H.S.; Zhang, C.; Kim, Y.; Roh, S.W.; Liu, D. Culture-independent analysis of the bacterial community in Chinese fermented vegetables and genomic analysis of lactic acid bacteria. Arch. Microbiol. 2021, 203, 4693–4703. [Google Scholar] [CrossRef]
- Tsuji, A.; Takei, Y.; Nishimura, T.; Azuma, Y. Identification of new Halomonas strains from food-related environments. Microbes Environ. 2022, 37, ME21052. [Google Scholar] [CrossRef] [PubMed]
- Korena, K.; Krzyzankova, M.; Florianova, M.; Karasova, D.; Babak, V.; Strakova, N.; Juricova, H. Microbial succession in the cheese ripening process—Competition of the starter cultures and the microbiota of the cheese plant environment. Microorganisms 2023, 11, 1735. [Google Scholar] [CrossRef] [PubMed]
- Xu, X.X.; Wu, B.B.; Zhao, W.T.; Lao, F.; Chen, F.; Liao, X.; Wu, I.H. Shifts in autochthonous microbial diversity and volatile metabolites during the fermentation of chili pepper (Capsicum frutescens L.). Food Chem. 2020, 335, 127512. [Google Scholar] [CrossRef] [PubMed]
- Liu, C.; Xue, W.J.; Ding, H.; An, C.; Ma, S.J.; Liu, Y. Probiotic potential of lactobacillus strains isolated from fermented vegetables in Shaanxi, China. Front. Microbiol. 2022, 12, 774903. [Google Scholar] [CrossRef] [PubMed]
- Larissa, B.; Anna, P.; Lubov, S.; Marina, E.; Yulia, N.; Olga, N.; Oksana, S.; Liudmila, T.; Valery, R. Nucleolytic enzymes from the marine bacterium Cobetia amphilecti KMM 296 with antibiofilm activity and biopreservative effect on meat products. Food Control 2017, 78, 270–278. [Google Scholar]
- Carolina, N.; Nestor, G.; Karen, A.; Ana, P.; Dely, R.C.; Martha, Y.L. Texture properties of miniature chihuahua-type cheese manufactured with different strains of Lactococcus lactis isolated from plants and raw milk cheese. J. Texture Stud. 2014, 45, 487–494. [Google Scholar]
- Skonberg, D.I.; Fader, S.; Perkins, L.B.; Perry, J.J. Lactic acid fermentation in the development of a seaweed sauerkraut-style product: Microbiological, physicochemical, and sensory evaluation. J. Food Sci. 2021, 86, 334–342. [Google Scholar] [CrossRef]
- Al-Sahlany, S.T.G.; Niamah, A.K. Bacterial viability, antioxidant stability, antimutagenicity and sensory properties of onion types fermentation by using probiotic starter during storage. Nutr. Food Sci. 2022, 52, 901–916. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).