Identification and Expression Analysis of the BTB/POZ Gene Family in Solanum tuberosum
Abstract
:1. Introduction
2. Materials and Methods
2.1. Test Materials
2.2. Search and Identification of Potato BTB Family Member Genes
2.3. Analysis of Basic Physical and Chemical Properties of Potato BTB Family Proteins
2.4. Homology Analysis of BTB Family Proteins
2.5. Analysis of BTB Motifs, Gene Structures, and Domain in Potato
2.6. Analysis of Chromosomal Location, Gene Duplication
2.7. Collinearity Analysis of the BTB Family
2.8. Gene Expression Analysis of Potato BTB Family Members
2.9. Expression Verification of Screened Genes
3. Results
3.1. Physicochemical Properties and Subcellular Localization of Potato BTB Family
3.2. Phylogenetic Analysis of Potato BTB Protein
3.3. Analysis of the Conserved Motif, Gene Architecture, and Domains in Potato BTB Family Proteins
3.4. Chromosome Localization and Collinearity Analysis of Potato BTB Gene
3.5. Collinearity Analysis of BTB Family Genes between Potato and A. thaliana
3.6. Expression Profile Analysis of Potato BTB Family Genes
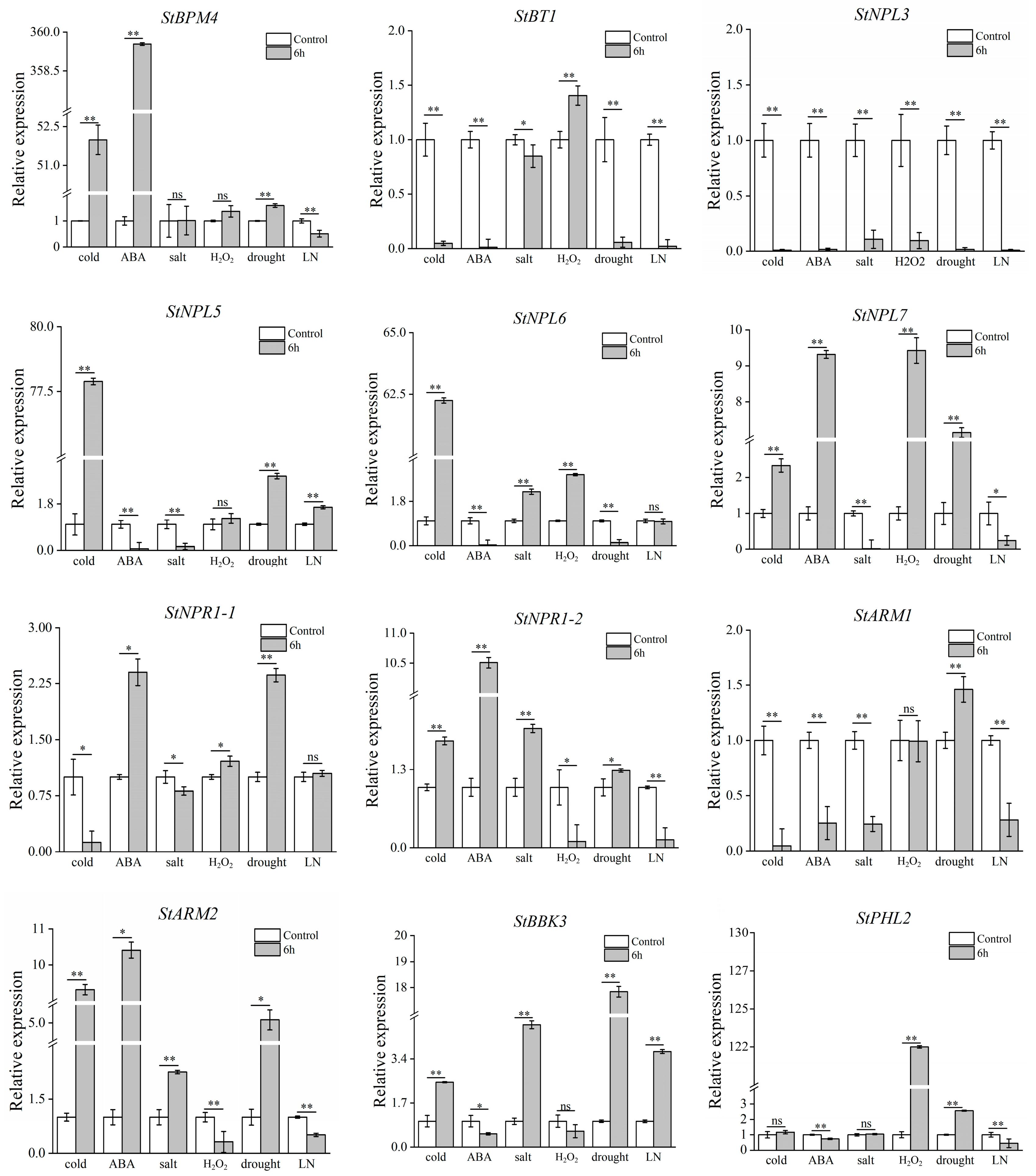
3.7. Validation of Potato BTB Family Gene Expression under Abiotic Stress
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Bardwell, V.J.; Treisman, R. The POZ domain: A conserved protein-protein interaction motif. Genes Dev. 1994, 8, 1664–1677. [Google Scholar] [CrossRef] [PubMed]
- Xu, L.; Wei, Y.; Reboul, J.; Vaglio, P.; Shin, T.-H.; Vidal, M.; Elledge, S.J.; Harper, J.W. BTB proteins are substrate-specific adaptors in an SCF-like modular ubiquitin ligase containing CUL-3. Nature 2003, 425, 316–321. [Google Scholar] [CrossRef] [PubMed]
- Chaharbakhshi, E.; Jemc, J.C. Broad-complex, tramtrack, and bric-à-brac (BTB) proteins: Critical regulators of development. Genesis 2016, 54, 505–518. [Google Scholar] [CrossRef] [PubMed]
- Perez-Torrado, R.; Yamada, D.; Defossez, P.A. Born to bind: The BTB protein-protein interaction domain. BioEssays 2006, 28, 1194–1202. [Google Scholar] [CrossRef] [PubMed]
- Morimoto, K.; Ohama, N.; Kidokoro, S.; Mizoi, J.; Takahashi, F.; Todaka, D.; Mogami, J.; Sato, H.; Qin, F.; Kim, J.S.; et al. BPM-CUL3 E3 ligase modulates thermotolerance by facilitating negative regulatory domain-mediated degradation of DREB2A in Arabidopsis. Proc. Natl. Acad. Sci. USA 2017, 114, E8528–E8536. [Google Scholar] [CrossRef] [PubMed]
- Bonchuk, A.; Balagurov, K.; Georgiev, P. BTB domains: A structural view of evolution, multimerization, and protein-protein interactions. Bioessays 2023, 45, e2200179. [Google Scholar] [CrossRef] [PubMed]
- Collins, T.; Stone, J.R.; Williams, A.J. All in the Family: The BTB/POZ, KRAB, and SCAN Domains. Mol. Cel. Biol. 2023, 21, 3609–3615. [Google Scholar] [CrossRef] [PubMed]
- Huang, C.-J.; Chen, C.-Y.; Chen, H.-H.; Tsai, S.-F.; Choo, K.-B. TDPOZ, a family of bipartite animal and plant proteins that contain the TRAF (TD) and POZ/BTB domains. Gene 2004, 324, 117–127. [Google Scholar] [CrossRef] [PubMed]
- Ellmeier, W.; Taniuchi, I. The Role of BTB-Zinc Finger Transcription Factors During T Cell Development and in the Regulation of T Cell-mediated Immunity. Curr. Top. Microbiol. 2014, 381, 21–49. [Google Scholar]
- Boyle, P.; Le Su, E.; Rochon, A.; Shearer, H.L.; Murmu, J.; Chu, J.Y.; Fobert, P.R.; Després, C. The BTB/POZ Domain of the Disease Resistance Protein NPR1 Interacts with the Repression Domain of TGA2 to Negate Its Function. Plant Cell 2009, 21, 3700–3713. [Google Scholar] [CrossRef]
- Mandadi, K.K.; Misra, A.; Ren, S.X.; McKnight, T.D. BT2, a BTB Protein, Mediates Multiple Responses to Nutrients, Stresses, and Hormones in Arabidopsis. Plant Physiol. 2009, 150, 1930–1939. [Google Scholar] [CrossRef]
- Weber, H.; Bernhardt, A.; Dieterle, M.; Hano, P.; Mutlu, A.l.; Estelle, M.; Genschik, P.; Hellmann, H. Arabidopsis AtCUL3a and AtCUL3b Form Complexes with Members of the BTB/POZ-MATH Protein Family. Plant Physiol. 2005, 137, 83–93. [Google Scholar] [CrossRef] [PubMed]
- Araus, V.; Vidal, E.A.; Puelma, T.; Alamos, S.; Mieulet, D.; Guiderdoni, E.; Gutiérrez, R.A. Members of BTB gene family regulate negatively nitrate uptake and nitrogen use efficiency in Arabidopsis thaliana and Oryza sativa. Plant Physiol. 2016, 171, 1523–1532. [Google Scholar] [CrossRef]
- He, F.; Kong, D.; Feng, Z.; Xu, Y.; Yuan, Q.; Liu, D.; Wang, X.; Feng, X.; Li, F. Genome-Wide Identification of the NPR1-like Gene Family in Solanum tuberosum and Functional Characterization of StNPR1 in Resistance to Ralstonia solanacearum. Genes 2023, 14, 1170. [Google Scholar] [CrossRef]
- Woodger, F.J.; Jacobsen, J.V.; Gubler, F. GMPOZ, a BTB/POZ Domain Nuclear Protein, is a Regulator of Hormone Responsive Gene Expression in Barley Aleurone. Plant Cell Physiol. 2004, 45, 945–950. [Google Scholar] [CrossRef]
- Spooner, D.M. The potato: Evolution, biodiversity and genetic resources. J.G. Hawkes. Am. Potato J. 1990, 67, 733–735. [Google Scholar] [CrossRef]
- Zierer, W.; Rüscher, D.; Sonnewald, U.; Sonnewald, S. Tuber and Tuberous Root Development. Annu. Rev. Plant Biol. 2021, 72, 551–580. [Google Scholar] [CrossRef] [PubMed]
- Du, L.; Poovaiah, B.W. A Novel Family of Ca2+/Calmodulin-Binding Proteins Involved in Transcriptional Regulation: Interaction with fsh/Ring3 Class Transcription Activators. Plant Mol. Biol. 2004, 54, 549–569. [Google Scholar] [CrossRef] [PubMed]
- Shalmani, A.; Huang, Y.B.; Chen, Y.B.; Muhammad, I.; Li, B.B.; Ullah, U.; Jing, X.Q.; Bhanbhro, N.; Liu, W.T.; Li, W.Q.; et al. The highly interactive BTB domain targeting other functional domains to diversify the function of BTB proteins in rice growth and development. Int. J. Biol. Macromol. 2021, 192, 1311–1324. [Google Scholar] [CrossRef]
- Li, J.H.; Su, X.X.; Wang, Y.L.; Yang, W.; Pan, Y.; Su, C.G.; Zhang, X.G. Genome-wide identification and expression analysis of the BTB domain-containing protein gene family in tomato. Genes Genom. 2018, 40, 1–15. [Google Scholar] [CrossRef]
- Kumar, S.; Stecher, G.; Tamura, K. MEGA7: Molecular Evolutionary Genetics Analysis Version 7.0 for Bigger Datasets. Mol. Biol. Evol. 2016, 33, 1870–1874. [Google Scholar] [CrossRef] [PubMed]
- Xie, J.M.; Chen, Y.R.; Cai, G.J.; Cai, R.L.; Hu, Z.; Wang, H. Tree Visualization by One Table (tvBOT): A web application for visualizing, modifying and annotating phylogenetic trees. Nucleic Acids Res. 2023, 51, W587–W592. [Google Scholar] [CrossRef] [PubMed]
- Chen, C.; Chen, H.; Zhang, Y.; Thomas, H.R.; Frank, M.H.; He, Y.; Xia, R. TBtools: An Integrative Toolkit Developed for Interactive Analyses of Big Biological Data. Mol. Plant 2020, 13, 1194–1202. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Coleman-Derr, D.; Chen, G.; Gu, Y.Q. OrthoVenn: A web server for genome wide comparison and annotation of orthologous clusters across multiple species. Nucleic Acids Res. 2015, 43, W78–W84. [Google Scholar] [CrossRef] [PubMed]
- The Potato Genome Sequencing Consortium. Genome sequence and analysis of the tuber crop potato. Nature 2011, 475, 189–195. [Google Scholar] [CrossRef] [PubMed]
- Livak, K.J.; Schmittgen, T.D. Analysis of Relative Gene Expression Data Using Real-Time Quantitative PCR and the 2−ΔΔCT Method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef] [PubMed]
- Leprince, O.A.P.D.C.D.G.L.D. The BTB/POZ domain: A new protein-protein interaction motif common to DNA- and actin-binding proteins. Cell Growth Differ. 1995, 6, 1193–1198. [Google Scholar]
- Ahmad, K.F.; Engel, C.K.; Privé, G.G. Crystal structure of the BTB domain from PLZF. Proc. Natl. Acad. Sci. USA 1998, 95, 12123–12128. [Google Scholar] [CrossRef] [PubMed]
- Zollman, S.; Godt, D.; Privé, G.G.; Couderc, J.L.; Laski, F.A. The BTB domain, found primarily in zinc finger proteins, defines an evolutionarily conserved family that includes several developmentally regulated genes in Drosophila. Proc. Natl. Acad. Sci. USA 1994, 91, 10717–10721. [Google Scholar] [CrossRef]
- Lin, L.Y.; Evans, S.E.; Fairall, L.; Schwabe, J.W.R.; Wagner, S.D.; Muskett, F.W. Backbone resonance assignment of the BCL6-BTB/POZ domain. Biomol. NMR Assign. 2018, 12, 47–50. [Google Scholar] [CrossRef]
- Robert, H.S.; Quint, A.; Brand, D.; Vivian-Smith, A.; Offringa, R. BTB and TAZ domain scaffold proteins perform a crucial function in Arabidopsis development. Plant J. 2009, 58, 109–121. [Google Scholar] [CrossRef] [PubMed]
- Stogios, P.J.; Downs, G.S.; Jauhal, J.J.S.; Nandra, S.K.; Privé, G.G. Sequence and structural analysis of BTB domain proteins. Genome Biol. 2005, 6, R82. [Google Scholar] [CrossRef] [PubMed]
- Gingerich, D.J.; Hanada, K.; Shiu, S.H.; Vierstra, R.D. Large-scale, lineage-specific expansion of a bric-a-brac/tramtrack/broad complex ubiquitin-ligase gene family in rice. Plant Cell 2007, 19, 2329–2348. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Li, Y.; Jia, H.X.; Li, J.B.; Huang, J.; Lu, M.Z.; Hu, J.J. The heat shock factor gene family in: A genome-wide survey and expression profiling during development and abiotic stresses. Front. Plant Sci. 2015, 6, 748. [Google Scholar] [CrossRef]
- Jiang, L.; Yang, S.-L.; Xie, L.-F.; Puah, C.S.; Zhang, X.-Q.; Yang, W.-C.; Sundaresan, V.; Ye, D. VANGUARD1 Encodes a Pectin Methylesterase That Enhances Pollen Tube Growth in the Arabidopsis Style and Transmitting Tract. Plant Cell 2005, 17, 584–596. [Google Scholar] [CrossRef]
- Sasca, D.; Yun, H.Y.; Giotopoulos, G.; Szybinski, J.; Evan, T.; Wilson, N.K.; Gerstung, M.; Gallipoli, P.; Green, A.R.; Hills, R.; et al. Cohesin-dependent regulation of gene expression during differentiation is lost in cohesin-mutated myeloid malignancies. Blood 2019, 134, 2195–2208. [Google Scholar] [CrossRef] [PubMed]
- Sankoff, D. Gene and genome duplication. Curr. Opin. Genet. Dev. 2001, 11, 681–684. [Google Scholar] [CrossRef] [PubMed]
- Bowers, J.E.; Chapman, B.A.; Rong, J.; Paterson, A.H. Unravelling angiosperm genome evolution by phylogenetic analysis of chromosomal duplication events. Nature 2003, 422, 433–438. [Google Scholar] [CrossRef]
- Lagarde, D.; Basset, M.; Lepetit, M.; Conejero, G.; Gaymard, F.; Astruc, S.; Grignon, C. Tissue-specific expression of Arabidopsis AKT1 gene is consistent with a role in K+ nutrition. Plant J. 2002, 9, 195–203. [Google Scholar] [CrossRef]
- Govers, F.; Harmsen, H.; Heidstra, R.; Michielsen, P.; Prins, M.; van Kammen, A.; Bisseling, T. Characterization of the pea ENOD12B gene and expression analyses of the two ENOD12 genes in nodule, stem and flower tissue. Mol. Gener. Genet. MGG 1991, 228, 160–166. [Google Scholar] [CrossRef]
- Hasanuzzaman, M.; Nahar, K.; Alam, M.M.; Roychowdhury, R.; Fujita, M. Physiological, Biochemical, and Molecular Mechanisms of Heat Stress Tolerance in Plants. Int. J. Mol. Sci. 2013, 14, 9643–9684. [Google Scholar] [CrossRef]
- Juranić, M.; Srilunchang, K.-o.; Krohn, N.G.; Leljak-Levanić, D.; Sprunck, S.; Dresselhaus, T. Germline-Specific MATH-BTB Substrate Adaptor MAB1 Regulates Spindle Length and Nuclei Identity in Maize. Plant Cell 2012, 24, 4974–4991. [Google Scholar] [CrossRef]
- Xu, C.; Park, S.J.; Van Eck, J.; Lippman, Z.B. Control of inflorescence architecture in tomato by BTB/POZ transcriptional regulators. Genes Dev. 2016, 30, 2048–2061. [Google Scholar] [CrossRef]
- Zhao, Q.; Ren, Y.-R.; Wang, Q.-J.; Wang, X.-F.; You, C.-X.; Hao, Y.-J. Ubiquitination-Related MdBT Scaffold Proteins Target a bHLH Transcription Factor for Iron Homeostasis. Plant Physiol. 2016, 172, 1973–1988. [Google Scholar] [CrossRef]
- Wan, X.; Peng, L.; Xiong, J.; Li, X.; Wang, J.; Li, X.; Yang, Y. AtSIBP1, a Novel BTB Domain-Containing Protein, Positively Regulates Salt Signaling in Arabidopsis thaliana. Plants 2019, 8, 573. [Google Scholar] [CrossRef]
- Sreenivasulu, N.; Harshavardhan, V.T.; Govind, G.; Seiler, C.; Kohli, A. Contrapuntal role of ABA: Does it mediate stress tolerance or plant growth retardation under long-term drought stress? Gene 2012, 506, 265–273. [Google Scholar] [CrossRef]
- Hibberd, J.M.; Weber, A.P.M. Plant metabolism and physiology. Curr. Opin. Plant Biol. 2012, 15, 225–227. [Google Scholar] [CrossRef]
- Li, X.; Jiang, H.; Liu, F.; Cai, J.; Dai, T.; Cao, W.; Jiang, D. Induction of chilling tolerance in wheat during germination by pre-soaking seed with nitric oxide and gibberellin. Plant Growth Regul. 2013, 71, 31–40. [Google Scholar] [CrossRef]
- Qiao, S.; Feng, Y.; Yan, J.; Li, K.; Xu, H. Overexpression of tomato SlTpx improves salt stress tolerance in transgenic tobacco plants by scavenging H2O2. Plant Cell Tiss. Org. 2022, 151, 321–333. [Google Scholar] [CrossRef]
- Srikanth, B.; Rao, I.S.; Surekha, K.; Subrahmanyam, D.; Voleti, S.R.; Neeraja, C.N. Enhanced expression of OsSPL14 gene and its association with yield components in rice (Oryza sativa) under low nitrogen conditions. Gene 2016, 576, 441–450. [Google Scholar] [CrossRef]
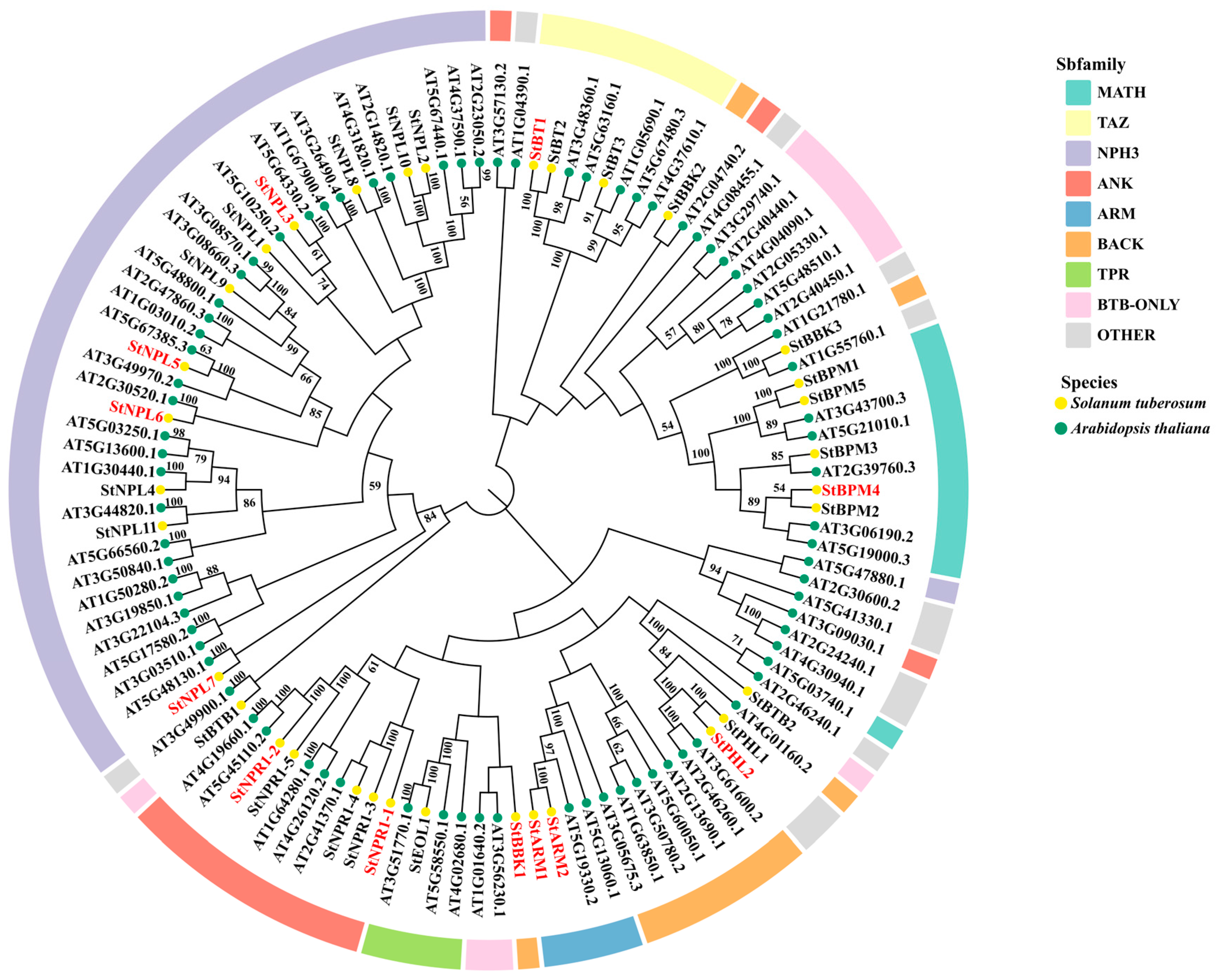
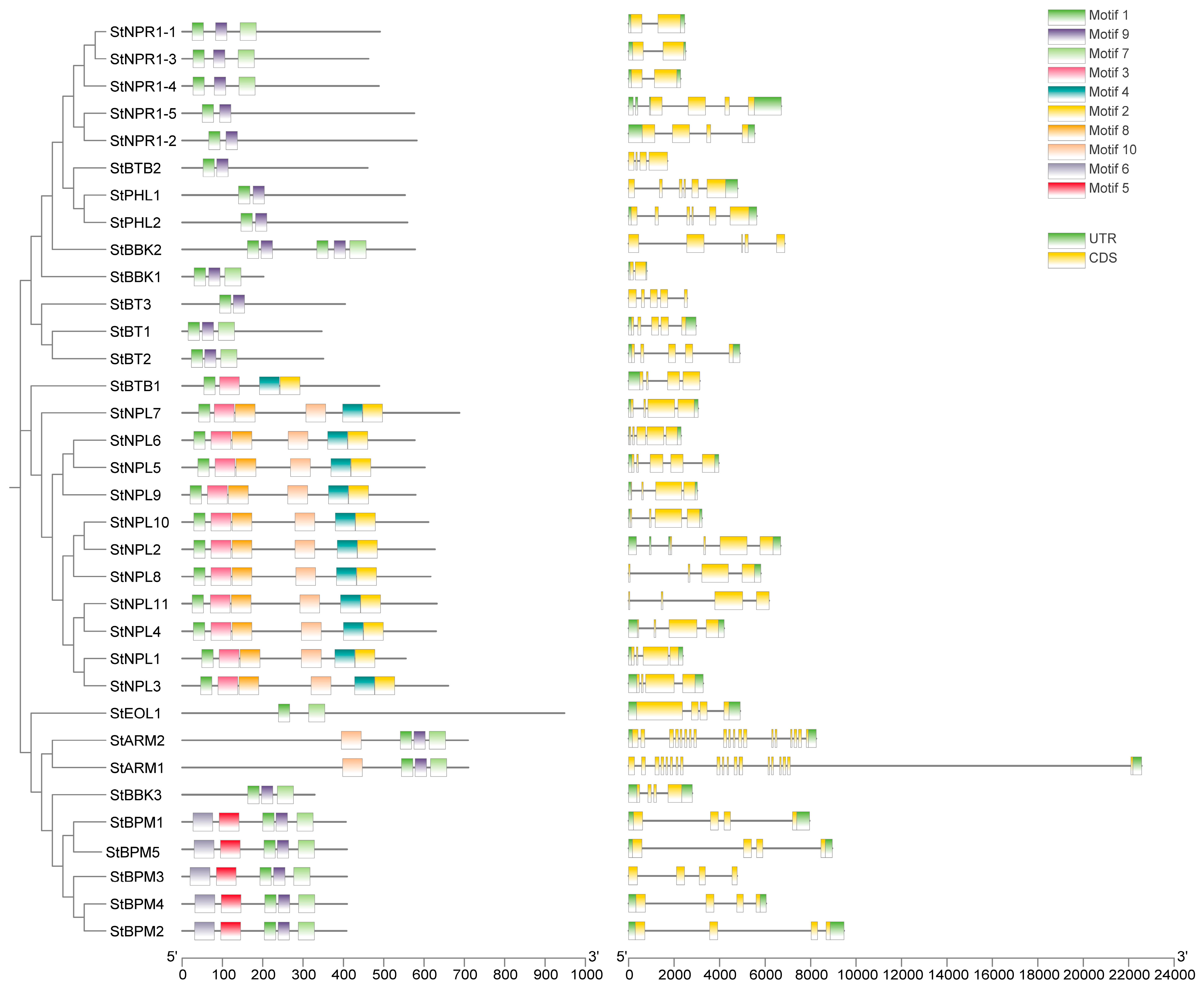
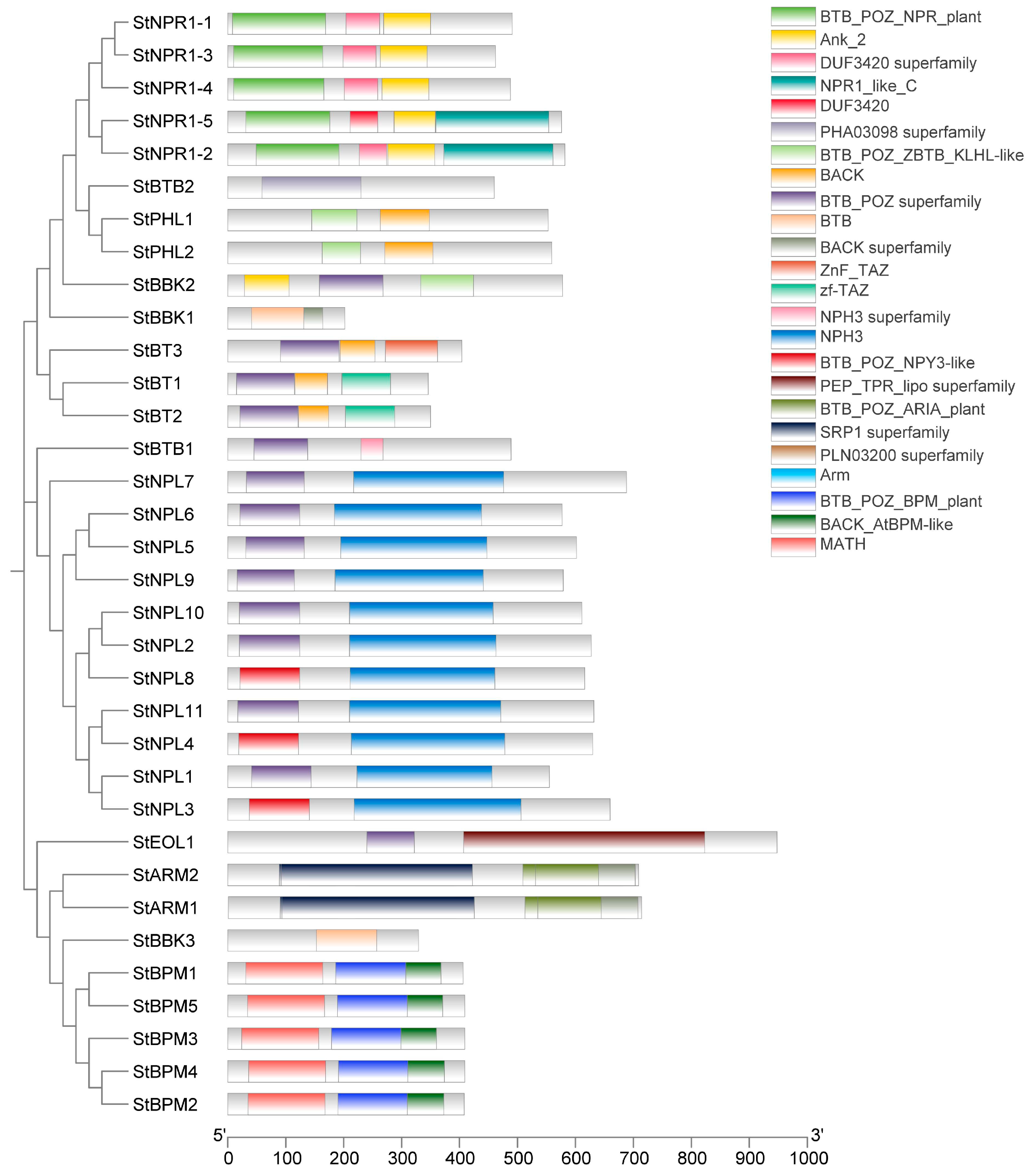

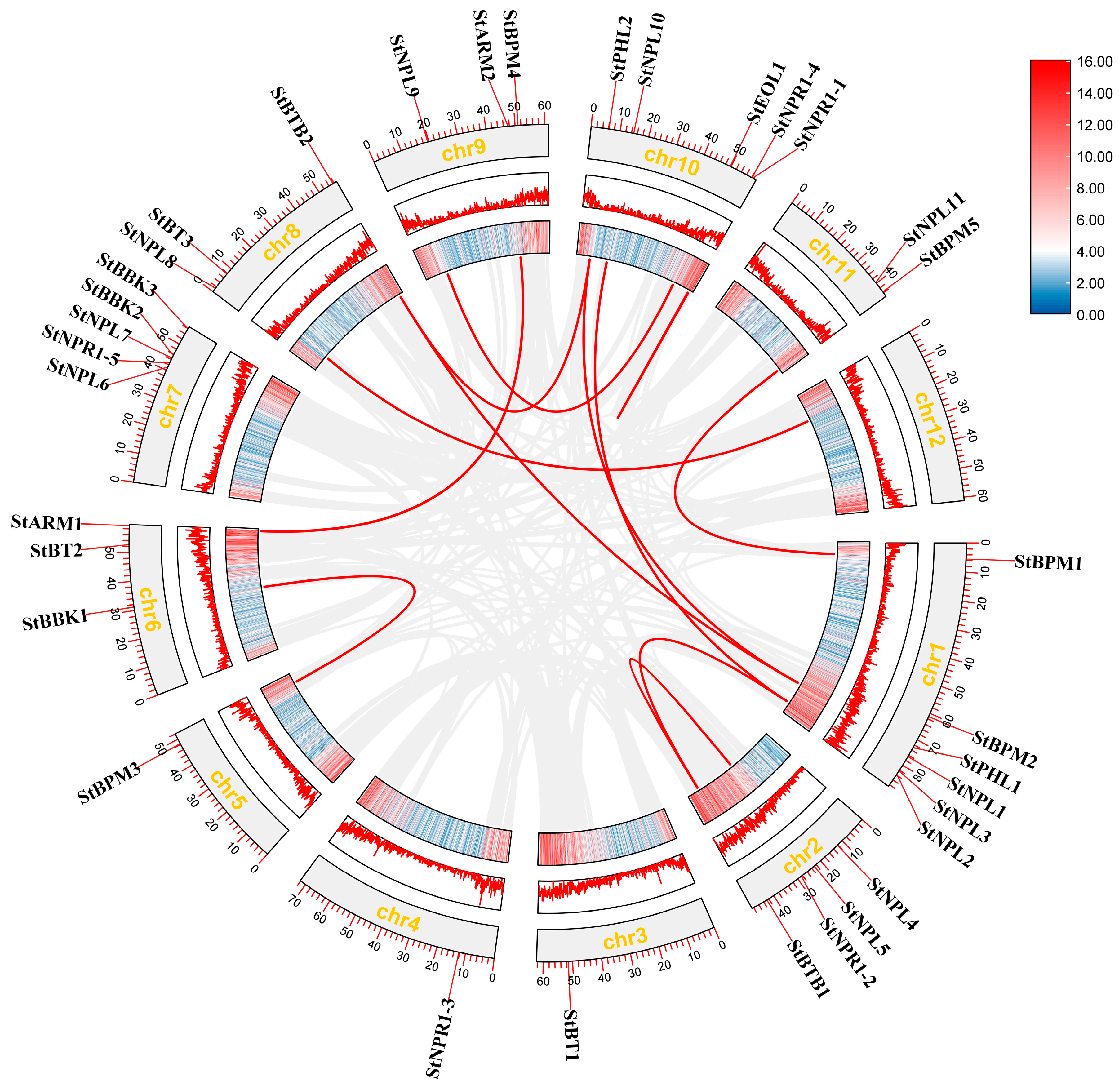


| ID | Sequence ID | Number of Amino Acids | Molecular Weight | pI | Instability Index | Aliphatic Index | Grand Average of Hydropathicity | Localization Prediction |
|---|---|---|---|---|---|---|---|---|
| PGSC0003DMT400058456 | StBPM1 | 405 | 44,974.76 | 6.07 | 40.55 | 81.33 | −0.291 | Nuc |
| PGSC0003DMT400011115 | StBPM2 | 407 | 45,182.59 | 6.02 | 35.53 | 90.79 | −0.111 | Nuc |
| PGSC0003DMT400069942 | StBPM3 | 408 | 45,178.55 | 5.86 | 42.19 | 86.03 | −0.162 | Nuc |
| PGSC0003DMT400009802 | StBPM4 | 408 | 45,235.32 | 6.02 | 32.49 | 83.16 | −0.181 | Nuc |
| PGSC0003DMT400070468 | StBPM5 | 408 | 44,966.77 | 5.66 | 35.64 | 82.48 | −0.218 | Nuc |
| PGSC0003DMT400039384 | StBT1 | 345 | 39,521.12 | 9.18 | 61.43 | 84.75 | −0.43 | Cyt/Nuc |
| PGSC0003DMT400069392 | StBT2 | 349 | 40,831.39 | 8.81 | 61.42 | 84.64 | −0.473 | Cyt/Nuc |
| PGSC0003DMT400064939 | StBT3 | 403 | 45,248.57 | 8.89 | 48.9 | 91.14 | −0.124 | Cyt/Nuc |
| PGSC0003DMT400073584 | StNPL1 | 554 | 63,575.64 | 8.98 | 40.07 | 96.19 | −0.24 | Nuc |
| PGSC0003DMT400066161 | StNPL2 | 626 | 69,014.65 | 6.78 | 34.07 | 85.99 | −0.303 | Cel/Nuc |
| PGSC0003DMT400083737 | StNPL3 | 659 | 74,285.54 | 7.02 | 44.17 | 86.15 | −0.408 | Nuc |
| PGSC0003DMT400008211 | StNPL4 | 629 | 69,867.04 | 5.74 | 45.44 | 87.14 | −0.284 | Cel/Nuc |
| PGSC0003DMT400033866 | StNPL5 | 601 | 67,427.69 | 6.18 | 41.26 | 95.21 | −0.184 | Cel/Nuc |
| PGSC0003DMT400031245 | StNPL6 | 576 | 64,374.17 | 7.17 | 33.72 | 94.69 | −0.137 | Nuc |
| PGSC0003DMT400015699 | StNPL7 | 687 | 78,154.78 | 8.39 | 55.12 | 90.52 | −0.286 | Cel/Cyt/Nuc |
| PGSC0003DMT400022573 | StNPL8 | 615 | 69,021.61 | 8.92 | 42.21 | 92.07 | −0.256 | Cel/Cyt/Nuc |
| PGSC0003DMT400010359 | StNPL9 | 578 | 64,675.52 | 5.86 | 42.78 | 96.31 | −0.108 | Nuc |
| PGSC0003DMT400013928 | StNPL10 | 610 | 67,523.02 | 8.38 | 33.78 | 87.38 | −0.291 | Cel/Nuc |
| PGSC0003DMT400001154 | StNPL11 | 631 | 70,448.94 | 6.79 | 44.55 | 91.17 | −0.128 | Cel/Cyt/Nuc |
| PGSC0003DMT400021076 | StNPR1-1 | 575 | 64,579.71 | 5.66 | 44.6 | 92.42 | −0.345 | Cyt |
| PGSC0003DMT400054660 | StNPR1-2 | 581 | 64,360.48 | 6.03 | 37.36 | 89.12 | −0.213 | Cyt |
| PGSC0003DMT400027591 | StNPR1-3 | 461 | 50,646.89 | 6.03 | 50.55 | 94.32 | −0.116 | Cyt |
| PGSC0003DMT400076710 | StNPR1-4 | 487 | 53,415.76 | 6.14 | 52.27 | 91.48 | −0.199 | Cyt |
| PGSC0003DMT400002412 | StNPR1-5 | 490 | 54,288.74 | 6.28 | 54.16 | 90.35 | −0.234 | Cyt |
| PGSC0003DMT400051860 | StARM1 | 709 | 78,098.49 | 5.79 | 45.24 | 104.01 | −0.11 | Cyt |
| PGSC0003DMT400033530 | StARM2 | 708 | 78,339.6 | 5.86 | 47.84 | 104.31 | −0.131 | Nuc |
| PGSC0003DMT400071906 | StBBK1 | 201 | 22,742.1 | 5.05 | 29.77 | 100 | −0.138 | Nuc |
| PGSC0003DMT400016452 | StBBK2 | 577 | 65,471.59 | 4.84 | 50.44 | 89.64 | −0.268 | Cyt |
| PGSC0003DMT400057328 | StBBK3 | 328 | 37,372.73 | 5.65 | 41.26 | 96.43 | −0.179 | Nuc |
| PGSC0003DMT400041081 | StEOL1 | 947 | 107,692.49 | 5.68 | 44.71 | 80.7 | −0.361 | Nuc |
| PGSC0003DMT400055245 | StBTB1 | 488 | 54,586.29 | 8.34 | 54.18 | 91.91 | −0.3 | Nuc |
| PGSC0003DMT400032077 | StBTB2 | 459 | 52,632.78 | 5.94 | 42.77 | 95.19 | −0.027 | Cyt/Nuc |
| PGSC0003DMT400000261 | StPHL1 | 552 | 62,965.09 | 5.18 | 55.2 | 74.09 | −0.387 | Cyt/Nuc |
| PGSC0003DMT400049926 | StPHL2 | 558 | 63,419.06 | 5.39 | 50.71 | 76.99 | −0.291 | Cyt/Nuc |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Feng, H.; Lu, Y.; Ren, B.; Yang, S.; Liu, Y.; Lu, L.; Li, L. Identification and Expression Analysis of the BTB/POZ Gene Family in Solanum tuberosum. Horticulturae 2024, 10, 543. https://doi.org/10.3390/horticulturae10060543
Feng H, Lu Y, Ren B, Yang S, Liu Y, Lu L, Li L. Identification and Expression Analysis of the BTB/POZ Gene Family in Solanum tuberosum. Horticulturae. 2024; 10(6):543. https://doi.org/10.3390/horticulturae10060543
Chicago/Turabian StyleFeng, Haoyue, Yifei Lu, Bi Ren, Shimin Yang, Yongjian Liu, Liming Lu, and Liqin Li. 2024. "Identification and Expression Analysis of the BTB/POZ Gene Family in Solanum tuberosum" Horticulturae 10, no. 6: 543. https://doi.org/10.3390/horticulturae10060543





