Enhancing Length at First Maturity Estimation Using Machine Learning for Fisheries Resource Management: A Case Study on Small Yellow Croaker (Larimichthys polyactis) in South Korea
Abstract
1. Introduction
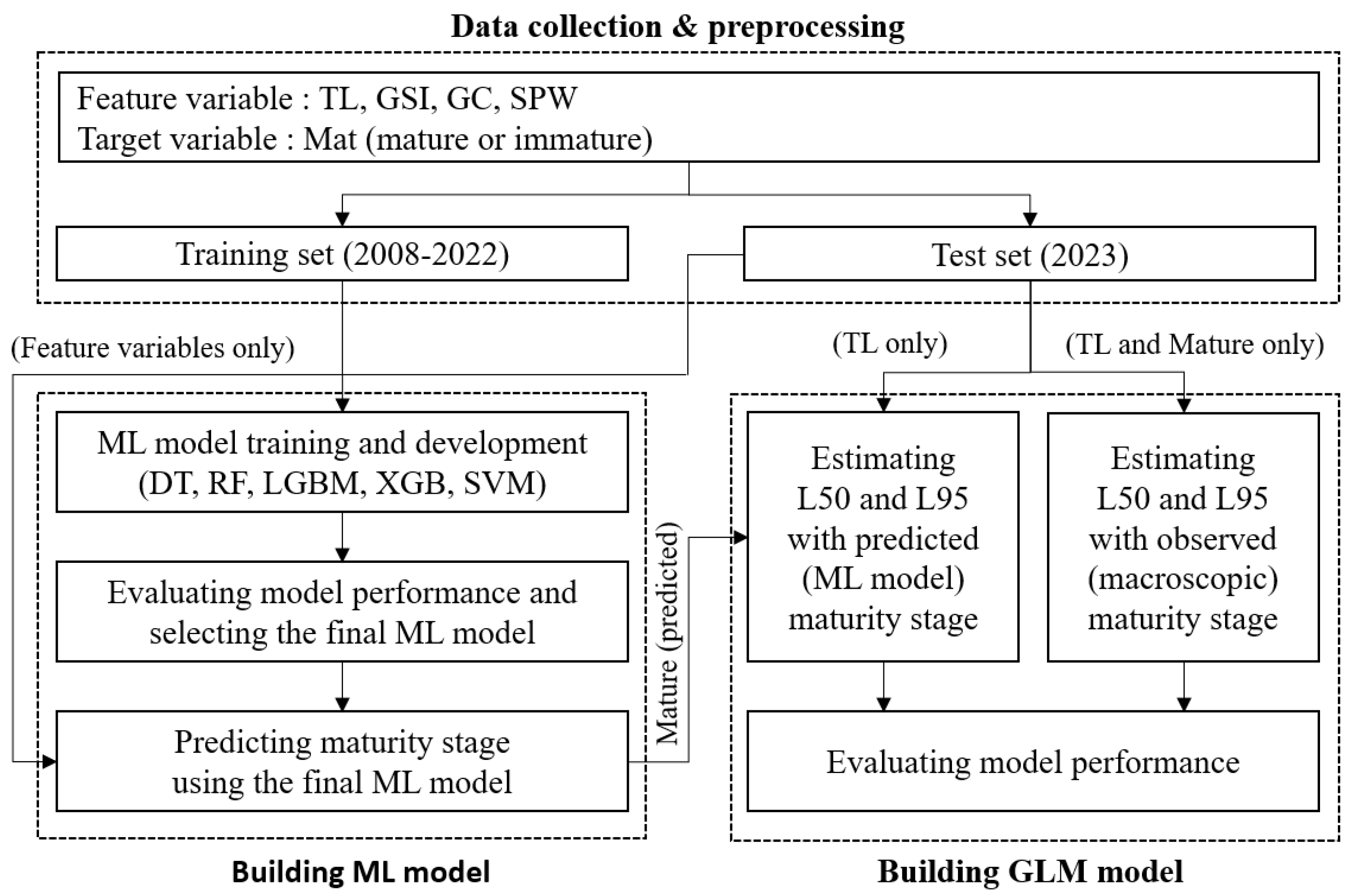
2. Materials and Methods
2.1. Predicting Maturity Stage Using Machine Learning
2.1.1. Data Collection and Preprocessing
- Data Transformation: Categorical variables, such as Month and GC (Gonad Color), were transformed to be suitable for machine learning algorithms. Specifically, The Month variable was converted into an ordered feature called SPW using OrdinalEncoder. It was categorized into three periods: high spawning activity (January to May), post–spawning/recovery (June to September), and off–spawning period (October to December), reflecting the key stages of small yellow croaker’s reproductive cycle. The other categorical feature of GC was transformed using one–hot encoding. Continuous variables, such as TL and GSI were standardized using a StandardScaler to ensure that all input features were on a comparable scale, enhancing the stability and performance of the models [42].
- Feature and target variables: The feature variables used for the machine learning models included TL, GSI, SPW and GC. These variables were chosen because they are significant predictors of fish maturity. TL and GSI provide quantitative measures of size and reproductive investment, while Month and GC capture seasonal and physiological variations in maturity. The target (label) variable for the classification models was the binary maturity stage (mature or immature). This label was derived from the detailed maturity stage information (Mat), which was simplified for the purposes of binary classification. By focusing on whether a fish is mature or not, the models aim to predict the onset of maturity based on the available features.
- Data Splitting: The dataset was divided into training and test sets. The training set included data up to 2022, while the test set consisted of data from the first four months (including spawning season) of 2023. The 2023 dataset was specifically used in the GLM analysis for estimating maturity lengths, as well as for a subsequent comparative analysis to evaluate the predictive performance of the machine learning model against the macroscopic observations. To further evaluate the models during the training process, the training set was split into a training subset and a validation subset. This approach ensured that 80% of the data was used for training and 20% for validation, allowing for effective model tuning and performance monitoring on unseen data.
2.1.2. Machine Learning Model Training and Development
- Accuracy: Accuracy represents the proportion of correctly classified instances (both true positives and true negatives) over the total number of instances in the dataset. It is a straightforward measure of the overall effectiveness of the model, but may be less reliable in cases where the dataset is imbalanced.where TP, TN, FP, and FN represent true positives, true negatives, false positives, and false negatives, respectively.
- Area Under the ROC Curve (AUC): AUC is a widely used metric that evaluates the model’s ability to discriminate between positive and negative classes across different threshold settings. The AUC measures the area under the ROC curve, where a value closer to 1.0 indicates a stronger performance in distinguishing between the two classes. The ROC curve plots the true positive rate (TPR) against the false positive rate (FPR), with AUC providing an aggregated measure of performance across all possible classification thresholds [43].
- True Skill Statistic (TSS): TSS is used to assess the model’s performance by considering both the sensitivity (true positive rate) and specificity (true negative rate). TSS is particularly useful for imbalanced datasets where both false positives and false negatives need to be accounted for. TSS ranges from −1 to 1, where a value of 1 indicates perfect classification, and values closer to 0 indicate no skill [44,45].
- Final Score: The final model selection was based on a weighted evaluation of three key metrics: Accuracy, AUC, and TSS. Specifically, Accuracy was assigned a weight of 0.2, while AUC and TSS were given more importance, each contributing 0.4 to the final score. This balanced approach ensured that the chosen model not only performed well in terms of overall accuracy but also demonstrated strong predictive capabilities across imbalanced data, as reflected by the AUC and TSS scores. The final score for each model was calculated using the following formula:
- Feature Importance: Feature importance scores were computed for the final model to identify the most significant predictors of fish maturity. This analysis helped determine which factors most influence maturity classification [46].
2.2. Estimating Length at First Maturity
3. Results
3.1. Machine Learning Model Performance
3.2. Length at First Maturity (L50 and L95)
4. Discussion
4.1. Comparative Analysis of Machine Learning and Macroscopic Methods
4.2. Implications for Fisheries Management
4.3. Limitations and Future Research
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| GSI | Gonadosomatic Index |
| DT | Decision Trees |
| RF | Random Forest |
| LGBM | Light Gradient-Boosting |
| XGB | EXtreme Gradient Boosting |
| SVM | Support Vector Machines |
| ROC | Receiver Operating Characteristic |
| AUC | Area Under the ROC Curve |
| TSS | True Skill Statistic |
References
- Yeon, I.J.; Lee, D.W.; Lee, J.B.; Choi, K.H.; Hong, B.K.; Kim, J.I.; Kim, Y.S. Long-term changes in the small yellow croaker, Larimichthys polyactis, population in the Yellow and East China Seas. J. Korean Soc. Fish. Ocean Technol. 2010, 46, 392–405. [Google Scholar] [CrossRef]
- Zhang, C.I.; Kim, H.A.; Kang, H.J. Management of small yellow croaker and hairtail in Korean waters using the length-based production value-per-recruit (PPR) analysis. J. Korean Soc. Fish. Ocean Technol. 2016, 52, 220–231. [Google Scholar] [CrossRef]
- Choi, M.J.; Kim, D.H. Assessment and Management of Small Yellow Croaker (Larimichthys Polyactis) Stocks South Korea. Sustainability 2020, 12, 8257. [Google Scholar] [CrossRef]
- Lin, L.S.; Ying, Y.P.; Han, Z.Q.; Xiao, Y.S.; Gao, T.X. AFLP analysis on genetic diversity and population structure of small yellow croaker Larimichthys Polyactis. Afr. J. Biotechnol. 2009, 8, 2700–2706. [Google Scholar]
- Ni, G.; Li, Q.; Kong, L.; Yu, H. Comparative phylogeography in marginal seas of the northwestern Pacific. Mol. Ecol. 2014, 23, 534–548. [Google Scholar] [CrossRef]
- Wang, Y.; Huang, J.; Tang, X.; Jin, X.; Sun, Y. Stable isotopic composition of otoliths in identification of stock structure of small yellow croaker (Larimichthys Polyactis) China. Acta Oceanol. Sin. 2016, 35, 29–33. [Google Scholar] [CrossRef]
- Wang, X.; Lu, G.; Zhao, L.; Yang, Q.; Gao, T. Assessment of fishery resources using environmental DNA: Small yellow croaker (Larimichthys polyactis) in East China Sea. PLoS ONE 2021, 15, e0244495. [Google Scholar] [CrossRef]
- Chen, X.; Liu, B.; Lin, D. Sexual Maturation, Reproductive Habits, and Fecundity of Fish. In Biology of Fishery Resources; Springer: Berlin/Heidelberg, Germany, 2022; pp. 113–142. [Google Scholar] [CrossRef]
- Reed, E.M.; Brown-Peterson, N.J.; DeMartini, E.E.; Andrews, A.H. Effects of data sources and biological criteria on length-atmaturity estimates and spawning periodicity of the commercially important Hawaiian snapper, Etelis coruscans. Front. Mar. Sci. 2023, 10, 1102388. [Google Scholar] [CrossRef]
- Ferreri, R.; McBride, R.S.; Barra, M.; Gargano, A.; Mangano, S.; Pulizzi, M.; Aronica, S.; Bonanno, A.; Basilone, G. Variation in size at maturity by horse mackerel (Trachurus trachurus) within the central Mediterranean Sea: Implications for investigating drivers of local productivity and applications for resource assessments. Fish. Res. 2019, 211, 291–299. [Google Scholar] [CrossRef]
- Smith, J.; Doe, J. Reproductive characteristics and maturity length of the commercially important fish species in the Indo-Pacific region. J. Mar. Biol. 2021, 45, 123–145. [Google Scholar] [CrossRef]
- Bris, A.L.; Pershing, A.J.; Hernandez, C.M.; Mills, K.E.; Sherwood, G.D. Modelling the effects of variation in reproductive traits on fish population resilience. ICES J. Mar. Sci. 2015, 72, 2590–2599. [Google Scholar] [CrossRef]
- Morgan, M.J. Integrating Reproductive Biology into Scientific Advice for Fisheries Management. J. Northwest Atl. Fish. Sci. 2008, 41, 37–51. [Google Scholar] [CrossRef]
- Murua, H.; Saborido-Rey, F. Female reproductive strategies of marine fish species of the North Atlantic. J. Northwest Atl. Fish. Sci. 2003, 33, 23–31. [Google Scholar] [CrossRef]
- Brown-Peterson, N.J.; Wyanski, D.M.; Saborido-Rey, F.; Macewicz, B.J.; Lowerre-Barbieri, S.K. A Standardized Terminology for Describing Reproductive Development in Fishes. Mar. Coast. Fish. 2011, 3, 52–70. [Google Scholar] [CrossRef]
- Costa, A.M. Macroscopic vs. microscopic identification of the maturity stages of female horse mackerel. ICES J. Mar. Sci. 2009, 66, 509–516. [Google Scholar] [CrossRef]
- Min, M.A.; Head, M.A.; Cope, J.M.; Hastie, J.D.; Flores, S.M. Limitations and applications of macroscopic maturity analyses: A comparison of histological and visual maturity for three west coast groundfish species. Environ. Biol. Fishes 2022, 105, 193–211. [Google Scholar] [CrossRef]
- West, G. Methods of Assessing Ovarian Development in Fishes: A Review. Mar. Freshw. Res. 1990, 41, 199–222. [Google Scholar] [CrossRef]
- Murua, H.; Motos, L. Reproductive strategy and spawning activity of the European hake Merluccius merluccius (L.) in the Bay of Biscay. J. Fish Biol. 2006, 69, 1288–1303. [Google Scholar] [CrossRef]
- Prince, J.; Harford, W.J.; Taylor, B.M.; Lindfield, S.J. Standard histological techniques systematically under-estimate the size fish start spawning. Fish Fish. 2022, 23, 1507–1516. [Google Scholar] [CrossRef]
- Flores, A.; Wiff, R.; Díaz, E. Using the gonadosomatic index to estimate the maturity ogive: Application to Chilean hake (Merluccius Gayi Gayi). ICES J. Mar. Sci. 2014, 72, 508–514. [Google Scholar] [CrossRef]
- Flores, A.; Wiff, R.; Ganias, K.; Marshall, C.T. Accuracy of gonadosomatic index in maturity classification and estimation of maturity ogive. Fish. Res. 2019, 210, 50–62. [Google Scholar] [CrossRef]
- Kang, H.; Ma, J.Y.; Kim, H.J.; Kim, H.J. Estimating Length at Sexual Maturity of the Small Yellow Croaker Larimichthys polyactis in the Yellow Sea of Korea Using Visual and GSI Methods. Korean J. Fish. Aquat. Sci. 2020, 53, 50–56. [Google Scholar] [CrossRef]
- Hastie, T.; Tibshirani, R.; Friedman, J. The Elements of Statistical Learning: Data Mining, Inference, and Prediction; Springer: Berlin/Heidelberg, Germany, 2009. [Google Scholar] [CrossRef]
- Genuer, R.; Poggi, J.M. Random Forests with R; Springer: Berlin/Heidelberg, Germany, 2020. [Google Scholar] [CrossRef]
- Gladju, J.; Kamalam, B.S.; Kanagaraj, A. Applications of data mining and machine learning framework in aquaculture and fisheries: A review. Smart Agric. Technol. 2022, 2, 100061. [Google Scholar] [CrossRef]
- Rubbens, P.; Brodie, S.; Cordier, T.; Destro Barcellos, D.; Devos, P.; Fernandes-Salvador, J.A.; Fincham, J.I.; Gomes, A.; Handegard, N.O.; Howell, K.; et al. Machine learning in marine ecology: An overview of techniques and applications. ICES J. Mar. Sci. 2023, 80, 1829–1853. [Google Scholar] [CrossRef]
- Mohale, H.P.; jawahar, P.; Jayakumar, N.; Oli, G.A.; Ravikumar, T. Application Of Deep Learning (Ai) In Marine Fisheries Resource Management. Trends Agric. Sci. 2023, 2, 753–763. [Google Scholar] [CrossRef]
- Kok, C.L.; Ho, C.K.; Tan, F.K.; Koh, Y.Y. Machine Learning-Based Feature Extraction and Classification of EMG Signals for Intuitive Prosthetic Control. Appl. Sci. 2024, 14, 5784. [Google Scholar] [CrossRef]
- Chen, J.; Teo, T.H.; Kok, C.L.; Koh, Y.Y. A Novel Single-Word Speech Recognition on Embedded Systems Using a Convolution Neuron Network with Improved Out-of-Distribution Detection. Electronics 2024, 13, 530. [Google Scholar] [CrossRef]
- Quinlan, J.R. Induction of decision trees. Mach. Learn. 1986, 1, 81–106. [Google Scholar] [CrossRef]
- Fürnkranz, J. Decision Tree. In Encyclopedia of Machine Learning; Springer US: Boston, MA, USA, 2010; pp. 263–267. [Google Scholar] [CrossRef]
- Breiman, L. Random Forests. Mach. Learn. 2001, 45, 5–32. [Google Scholar] [CrossRef]
- Cutler, D.R.; Edwards, T.C., Jr.; Beard, K.H.; Cutler, A.; Hess, K.T.; Gibson, J.; Lawler, J.J. Random forests for classfication in ecology. Ecology 2007, 88, 2783–2792. [Google Scholar] [CrossRef]
- Ke, G.; Meng, Q.; Finley, T.; Wang, T.; Chen, W.; Ma, W.; Ye, Q.; Liu, T.Y. LightGBM: A Highly Efficient Gradient Boosting Decision Tree. In Proceedings of the Advances in Neural Information Processing Systems, Long Beach, CA, USA, 4–9 December 2017; Guyon, I., Luxburg, U.V., Bengio, S., Wallach, H., Fergus, R., Vishwanathan, S., Garnett, R., Eds.; Curran Associates, Inc.: Red Hook, NY, USA, 2017; Volume 30. [Google Scholar]
- Chen, T.; Guestrin, C. XGBoost: A Scalable Tree Boosting System. In Proceedings of the KDD ’16: 22nd ACM SIGKDD International Conference on Knowledge Discovery and Data Mining, New York, NY, USA, 13–17 August 2016; pp. 785–794. [Google Scholar] [CrossRef]
- Cortes, C.; Vapnik, V. Support-vector networks. Mach. Learn. 1995, 20, 273–297. [Google Scholar] [CrossRef]
- Muñoz-Mas, R.; Gil-Martínez, E.; Oliva-Paterna, F.J.; Belda, E.J.; Martínez-Capel, F. Tree-based ensembles unveil the microhabitat suitability for the invasive bleak (Alburnus alburnus L.) and pumpkinseed (Lepomis gibbosus L.): Introducing XGBoost to ecoinformatics. Ecol. Inform. 2019, 53, 100974. [Google Scholar] [CrossRef]
- Effrosynidis, D.; Tsikliras, A.; Arampatzis, A.; Sylaios, G. Species Distribution Modelling via Feature Engineering and Machine Learning for Pelagic Fishes in the Mediterranean Sea. Appl. Sci. 2020, 10, 8900. [Google Scholar] [CrossRef]
- Bergen, S.; Huso, M.M.; Duerr, A.E.; Braham, M.A.; Schmuecker, S.; Miller, T.A.; Katzner, T.E. A review of supervised learning methods for classifying animal behavioural states from environmental features. Methods Ecol. Evol. 2023, 14, 189–202. [Google Scholar] [CrossRef]
- Flores, A.; Wiff, R.; Donovan, C.R.; Gálvez, P. Applying machine learning to predict reproductive condition in fish. Ecol. Inform. 2024, 80, 102481. [Google Scholar] [CrossRef]
- Han, J.; Kamber, M.; Pei, J. Preface. In Data Mining, 3rd ed.; The Morgan Kaufmann Series in Data Management Systems; Morgan Kaufmann: Boston, MA, USA, 2012; pp. 23–29. [Google Scholar] [CrossRef]
- Fawcett, T. An introduction to ROC analysis. Pattern Recognit. Lett. 2006, 27, 861–874. [Google Scholar] [CrossRef]
- Allouche, O.; Tsoar, A.; Kadmon, R. Assessing the accuracy of species distribution models: Prevalence, kappa and the true skill statistic (TSS). J. Appl. Ecol. 2006, 43, 1223–1232. [Google Scholar] [CrossRef]
- Shabani, F. Assessing Accuracy Methods of Species Distribution Models: AUC, Specificity, Sensitivity and the True Skill Statistic. Glob. J. Hum.-Soc. Sci. 2018, 18, 7–18. [Google Scholar]
- Liaw, A.; Wiener, M. Classification and Regression by randomForest. R News 2002, 2, 18–22. [Google Scholar]
- Kluyver, T.; Ragan-Kelley, B.; Pérez, F.; Granger, B.; Bussonnier, M.; Frederic, J.; Kelley, K.; Hamrick, J.; Grout, J.; Corlay, S.; et al. Jupyter Notebooks—A publishing format for reproducible computational workflows. In Positioning and Power in Academic Publishing: Players, Agents and Agendas; Loizides, F., Schmidt, B., Eds.; IOS Press: Amsterdam, The Netherlands, 2016; pp. 87–90. [Google Scholar]
- McKinney, W. Data Structures for Statistical Computing in Python. In Proceedings of the 9th Python in Science Conference, Austin, TX, USA, 28 June–3 July 2010; pp. 56–61. [Google Scholar] [CrossRef]
- Pedregosa, F.; Varoquaux, G.; Gramfort, A.; Michel, V.; Thirion, B.; Grisel, O.; Blondel, M.; Prettenhofer, P.; Weiss, R.; Dubourg, V.; et al. Scikit-learn: Machine Learning in Python. J. Mach. Learn. Res. 2011, 12, 2825–2830. [Google Scholar]
- McCullagh, P. Generalized Linear Models, 2nd ed.; Routledge: London, UK, 1989. [Google Scholar] [CrossRef]
- R Core Team. R: A Language and Environment for Statistical Computing; R Foundation for Statistical Computing: Vienna, Austria, 2021. [Google Scholar]
- Bozdogan, H. Model selection and Akaike’s Information Criterion (AIC): The general theory and its analytical extensions. Psychometrika 1987, 52, 345–370. [Google Scholar] [CrossRef]
- Berrar, D.; Dubitzky, W. Bootstrapping. In Encyclopedia of Systems Biology; Springer: New York, NY, USA, 2013; pp. 158–162. [Google Scholar] [CrossRef]
- Caddy, J.; Mahon, R. Reference Points for Fisheries Management; Number 347 in FAO Fisheries Technical Paper; FAO: Rome, Italy, 1995; p. 83. [Google Scholar]
- Trippel, E.A. Age at Maturity as a Stress Indicator in Fisheries: Biological processes related to reproduction in northwest Atlantic groundfish populations that have undergone declines. BioScience 1995, 45, 759–771. [Google Scholar] [CrossRef]
- Pope, J.G.; Macer, C.T. An evaluation of the stock structure of North Sea cod, haddock, and whiting since 1920, together with a consideration of the impacts of fisheries and predation effects on their biomass and recruitment. ICES J. Mar. Sci. 1996, 53, 1157. [Google Scholar] [CrossRef]
- Hilborn, R.; Walters, C.J. Quantitative Fisheries Stock Assessment: Choice, Dynamics and Uncertainty; Springer: New York, NY, USA, 1992. [Google Scholar] [CrossRef]
- King, M. Fisheries Biology, Assessment and Management, 2nd ed.; John Wiley & Sons: Hoboken, NJ, USA, 2007. [Google Scholar] [CrossRef]
- Gebremedhin, S.; Bruneel, S.; Getahun, A.; Anteneh, W.; Goethals, P. Scientific Methods to Understand Fish Population Dynamics and Support Sustainable Fisheries Management. Water 2021, 13, 574. [Google Scholar] [CrossRef]
- Watson, J.T.; Ames, R.; Holycross, B.; Suter, J.; Somers, K.; Kohler, C.; Corrigan, B. Fishery catch records support machine learning-based prediction of illegal fishing off US West Coast. PeerJ 2023, 11, e16215. [Google Scholar] [CrossRef]
- Olden, J.D.; Lawler, J.J.; Poff, N.L. Machine learning methods without tears: A primer for ecologists. Q. Rev. Biol. 2008, 83, 171–193. [Google Scholar] [CrossRef]
- LeCun, Y.; Bengio, Y.; Hinton, G. Deep learning. Nature 2015, 521, 436–444. [Google Scholar] [CrossRef]



| Model | Accuracy | AUC | TSS | Final Score | Computation Time (s) |
|---|---|---|---|---|---|
| DT | 0.961 | 0.800 | 0.860 | 0.856 | 0.42 |
| RF | 0.966 | 0.924 | 0.818 | 0.890 | 4.32 |
| LGBM | 0.977 | 0.943 | 0.877 | 0.923 | 0.36 |
| SVM | 0.966 | 0.932 | 0.917 | 0.933 | 80.26 |
| XGB | 0.982 | 0.939 | 0.907 | 0.935 | 0.54 |
| Model | L50 (95% C.I.) | L95 (95% C.I.) | AIC | R–Squared |
|---|---|---|---|---|
| Macroscopic (observed) | 14.2 (11.8, 15.6) | 21.7 (19.0, 24.3) | 168 | 0.324 |
| Machine learning (predicted) | 15.2 (14.1, 15.9) | 20.3 (17.9, 22.4) | 138 | 0.459 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kang, H.; Yoon, S.C. Enhancing Length at First Maturity Estimation Using Machine Learning for Fisheries Resource Management: A Case Study on Small Yellow Croaker (Larimichthys polyactis) in South Korea. Fishes 2024, 9, 373. https://doi.org/10.3390/fishes9100373
Kang H, Yoon SC. Enhancing Length at First Maturity Estimation Using Machine Learning for Fisheries Resource Management: A Case Study on Small Yellow Croaker (Larimichthys polyactis) in South Korea. Fishes. 2024; 9(10):373. https://doi.org/10.3390/fishes9100373
Chicago/Turabian StyleKang, Heejoong, and Sang Chul Yoon. 2024. "Enhancing Length at First Maturity Estimation Using Machine Learning for Fisheries Resource Management: A Case Study on Small Yellow Croaker (Larimichthys polyactis) in South Korea" Fishes 9, no. 10: 373. https://doi.org/10.3390/fishes9100373
APA StyleKang, H., & Yoon, S. C. (2024). Enhancing Length at First Maturity Estimation Using Machine Learning for Fisheries Resource Management: A Case Study on Small Yellow Croaker (Larimichthys polyactis) in South Korea. Fishes, 9(10), 373. https://doi.org/10.3390/fishes9100373






