Two Genotypes of Streptococcus iniae Are the Causative Agents of Diseased Ornamental Fish, Green Terror Cichlid (Aequidens rivulatus)
Abstract
1. Introduction
2. Materials and Methods
2.1. Fish
2.2. Bacterial Isolation and Parasite Check
2.3. Pathological Section
2.4. Specific Pathogen Detection for Disease Fish
2.5. Physiological and Biochemical Features
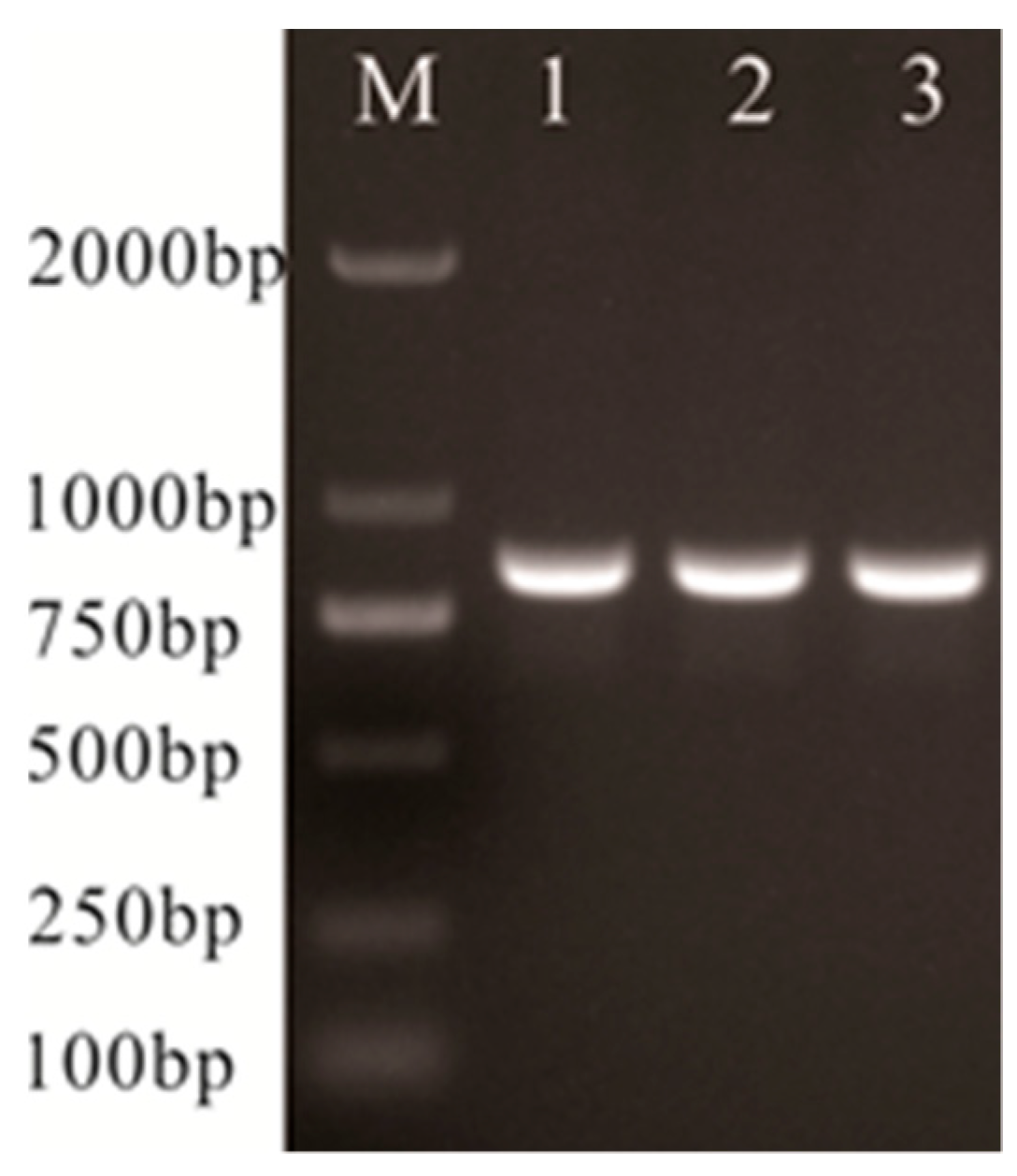
2.6. Lactate Oxidase-Encoding (lctO) and 16S rRNA Gene Sequence Analysis
2.7. Multilocus Sequence Typing (MLST)
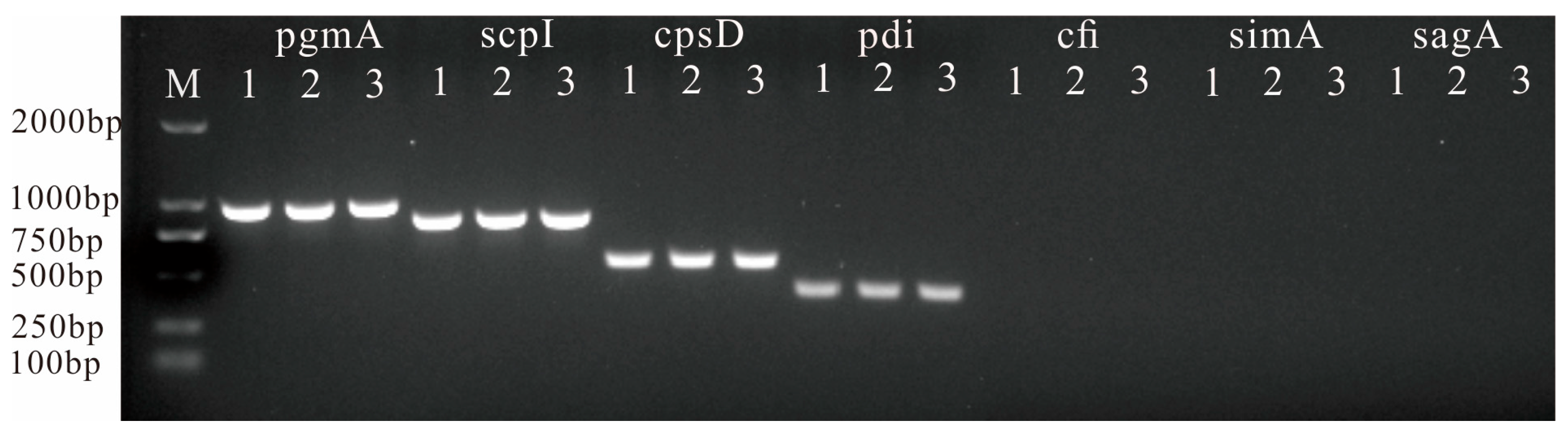
2.8. PCR Tests of Virulence Genes
2.9. Artificial Infection
2.10. Antimicrobial Susceptibility Testing
3. Results
3.1. Clinical Signs and Pathological Features
3.2. Specific Pathogen Detection in Diseased Fish and Isolation of Pathogenic Bacteria
3.3. Identification of Isolated Strains
3.4. MLST
3.5. Detection of Virulence-Related Genes
3.6. Artificial Infection
3.7. Antibiotic Susceptibility Test
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Yu, X.J.; Hao, X.J.; Yang, L.K.; Dang, Z.Q. Development monitoring report of Chinese recreational fishery. China Fish. 2022, 12, 35–40. [Google Scholar]
- Wu, J.; Gao, Q.; Yang, H.; Yang, G.L. Artificial propagation and seed cultivation technology of green terror cichlid Aequidens rivulatus. Fish. Sci. Technol. Info. 2012, 39, 171–173. [Google Scholar]
- Yao, X.L.; Xu, X.L.; Li, H.M.; Zhong, W.H.; Zang, L.; Li, H.; Yang, C.J. Biological characteristics of pathogenic Streptococcus agalactiae isolated from Aequidens rivulatus. Prog. Fish. Sci. 2015, 36, 106–112. [Google Scholar]
- Hao, X.B.; Huang, Y.P.; Hu, Q.; Peng, Y.O.; Sun, L.M.; Gao, J.W.; Song, R. Pathogen isolation, identification and drug susceptibility analysis of Aequidens rivulatus hole-in-the-head disease. Curr. Fish. 2021, 6, 60–63. [Google Scholar]
- Talaat, A.M.; Reimschuessel, R.; Trucksis, M. Identification of mycobacteria infecting fish to the species level using polymerase chain reaction and restriction enzyme analysis. Vet. Microbiol. 1997, 58, 229–237. [Google Scholar] [CrossRef]
- Chase, D.M.; Pascho, R.J. Development of a nested polymerase chain reaction for amplification of a sequence of the p57 gene of Renibacterium salmoninarum that provides a highly sensitive method for detection of the bacterium in salmonid kidney. Dis. Aquat. Org. 1998, 3, 223–229. [Google Scholar] [CrossRef]
- Lei, Y.; Zhang, H.J.; Wang, J.; Zhang, W.E.; Tang, S.L.; Xiao, Y.; Wang, X.P. Development and Primary Application of a PCR Assay for Detection of Piscirickettsia-like Organisms. J. Guangdong Ocean Univ. 2015, 35, 30–34. [Google Scholar]
- Jiang, Y.Y.; Li, A.X. Establishment of a specific PCR assay to detect Nocardia seriolae. S. China Fish. Sci. 2011, 7, 47–51. [Google Scholar]
- Forsman, M.; Sandstrom, G.; Sjostedt, A. Analysis of 16s ribosomal DNA sequences of Francisella strains and utilization for determination of the phylogeny of the genus and for identification of strains by PCR. Int. J. Syst. Bacteriol. 1994, 44, 38–46. [Google Scholar] [CrossRef]
- Mata, A.I.; Blanco, M.M.; Dominguez, L.; Fernandez-Garayzabal, J.F.; Gibello, A. Development of a PCR assay for Streptococcus iniae based on the lactate oxidase (lctO) gene with potential diagnostic value. Vet. Microbial. 2004, 101, 109–116. [Google Scholar] [CrossRef]
- Lane, D.J. 16S/23S rRNA sequencing. In Nucleic Acid Techniques in Bacterial Systematics; Stackebrandt, E., Goodfellow, M., Eds.; Wiley: New York, NY, USA, 1991. [Google Scholar]
- Irion, S.; Rudenko, O.; Sweet, M.; Chabanet, P.; Barnes, A.C.; Tortosa, P.; Séré, M.G. Molecular investigation of recurrent Streptococcus iniae epizootics affecting coral reef fish on an oceanic island suggests at least two distinct emergence events. Front. Microbial. 2021, 12, 749734. [Google Scholar] [CrossRef]
- Xiong, X.Y.; Huang, G.Q.; Wang, Z.C.; Wen, X. Molecular typing, antibiogram type and detection of virulence genes of Stereptococcus iniae strains isolated from golden pompano (Tranchinotus ovatus) in Guangxi province. J. Fish. China 2018, 42, 586–595. [Google Scholar]
- Baums, C.G.; Hermeyer, K.; Leimbach, S.; Adamekk, M.; Czerny, C.P.; Horstgen-Schwark, G.; Valentin-Weigand, P.; Baumgartner, W.; Steinhagen, D. Establishment of a Model of Streptococcus iniae Meningoencephalitis in Nile Tilapia (Oreochromis niloticus). J. Comp. Path. 2013, 149, 94–102. [Google Scholar] [CrossRef] [PubMed]
- Dong, X.Z.; Cai, M.Y. Manual of Familiar Bacterium Identification; Science Press: Beijing, China, 2001. [Google Scholar]
- Saitou, N.; Nei, M. The neighbor-joining method: A new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 1987, 4, 406–425. [Google Scholar] [PubMed]
- Tamura, K.; Battistuzzi, F.U.; Billing-Ross, P.; Murillo, O.; Filipski, A.; Kumar, S. Estimating divergence times in large molecular phylogenies. Proc. Natl. Acad. Sci. USA 2012, 109, 19333–19338. [Google Scholar] [CrossRef] [PubMed]
- Tamura, K.; Nei, M.; Kumar, S. Prospects for inferring very large phylogenies by using the neighbor-joining method. Proc. Natl. Acad. Sci. USA 2004, 101, 11030–11035. [Google Scholar] [CrossRef] [PubMed]
- Jolley, K.A.; James, E.B.; Maiden, M.C.J. Open-access bacterial population genomics: BIGSdb software, the PubMLST.org website and their applications [version 1; peer review: 2 approved]. Wellcome. Open. Res. 2018, 3, 124. [Google Scholar] [CrossRef]
- Bauer, A.W.; Kirby, W.M.; Sherris, J.C.; Turck, M. Antibiotic susceptibility testing by a standardized single disk method. Am. J. Clin. Pathol. 1966, 45, 493–496. [Google Scholar] [CrossRef] [PubMed]
- Luo, Z.; Zhang, Z.G.; Hao, S.; Bai, X.H.; Gao, Q.; Feng, S.M. Isolation and Identification of the Pathogen Causing granulomas in internal organs of turbot Scophthalmus maximus. J. Fish. Sci. China 2019, 26, 559–568. [Google Scholar] [CrossRef]
- Luo, Z.; Li, J.; Zhang, Z.G.; Hao, S.; Bai, X.H.; You, H.Z.; Mo, Z.L.; Feng, S.M. Mycobacterium marinum is the causative agent of splenic and renal granulomasin half-smooth tongue sole (Cynoglossus semilaevis Günther) in China. Aquaculture 2018, 490, 203–207. [Google Scholar] [CrossRef]
- Luo, Z.; Hao, S.; Bai, X.H.; Zhang, Z.G.; Ma, Y.F.; Feng, S.M.; Zhang, D.F. Isolation, identification, and genomic analysis of Mycobacterium ulcerans ecovar Liflandii from the farmed Chinese tongue sole, Cynoglossus semilaevis Günther. Aquaculture 2022, 548, 737614. [Google Scholar] [CrossRef]
- Bai, X.H.; Hao, S.; Fu, J.P.; Sun, H.C.; Luo, Z. Identification of Mycobacterium chelonae from Lined Seahorse Hippocampus erectus and Histopathological Analysis. Fishes 2023, 8, 225. [Google Scholar] [CrossRef]
- He, S.Y.; Wei, W.Y.; Liu, T.; Yang, Q.; Xie, H.; He, Q.Y.; Wang, K.Y. Isolation, identification and histopathological study on lethal sarcoidosis of Micropterus salmoides. J. Fish. China 2020, 44, 253–265. [Google Scholar]
- Zhen, C.; Pan, L.D. research progress on pathogen and histopathology of visceral sarcoidosis in fish. Sci. Fish. Farms. 2016, 7, 54–55. [Google Scholar]
- Xiao, P.; Jiang, M.; Liu, Y.; Sun, M.; Zhang, L.; Jie, L.; Li, G.; Mo, Z. Splenic necrosis signs and pathogen detection in cultured half-smooth tongue sole, Cynoglossus semilaevis Günther. J. Fish Dis. 2015, 38, 103–106. [Google Scholar] [CrossRef] [PubMed]
- Ge, M.X.; Fang, H. Renibacterium Salmoninarum and bacterial kidney disease of salmon (summary). J. Hebei. Vocat. Technical. Teach. Coll. 2003, 17, 51–55. [Google Scholar]
- Liu, C.; Chang, O.Q.; Zhang, D.F.; Li, K.B.; Wang, F.; Lin, M.H.; Shi, C.B.; Jiang, L.; Wang, Q.; Bergmann, S.M. Aeromonas schubertii as a cause of multi-organ necrosis in internal organs of Nile tilapia, Oreochromis niloticus. J. Fish Dis. 2012, 35, 335–342. [Google Scholar] [CrossRef]
- Qiu, Y.Y.; Zheng, L.; Mao, Z.J.; Chen, X.X. Isolation and identification of the causative agent and histopathology observation of white-spots disease in internal organs of Larimichthys crocea. Microbiol. China 2012, 39, 361–370. [Google Scholar]
- He, X.X.; Liu, K.M.; Wang, X.Y.; Hao, S.; Jia, L.; Shao, P.; Luo, Z. Isolation, identification and histopathology of pathogen of intussusception disease in half-smooth tongue sole Cynoglossus semilaevis. J. Dalian Ocean Univ. 2022, 37, 19–25. [Google Scholar]
- Kim, M.S.; Jin, J.W.; Han, H.J.; Choi, H.S.; Hong, S.; Cho, J.Y. Genotype and virulence of Streptococcus iniae isolated from diseased olive flounder Paralichthys olivaceus in Korea. Fish Sci. 2014, 80, 1277–1284. [Google Scholar] [CrossRef]
- Pierezan, F.; Shahin, K.; Hechman, T.l.; Ang, J.; Byrne, B.A.; Soto, E. Outbreaks of severe myositis in cultured white sturgeon (Acipenser transmontanus L.) associated with Streptococcus iniae. J. Fish Dis. 2020, 43, 485–490. [Google Scholar] [CrossRef] [PubMed]
- Chen, C.Y.; Chao, C.B.; Bowser, P.R. Comparative histopathology of Streptococcus iniae and Streptococcus agalactiae-infected tilapia. Bull. Eur. Ass. Fish Pathol. 2007, 27, 2–9. [Google Scholar]
- Yu, X.L.; Chen, M.; Li, C.; Li, P.P.; Lei, A.Y.; Zhong, M.N.; Liang, W.W. Channel catfish Ictalurus punctatus outbreak infected by bacterium Streptococcus iniae. J. Dalian Fish. Univ. 2008, 23, 185–191. [Google Scholar]
- Elaamri, F. First report of Streptococcus iniae in red porgy (Pagrus pagrus L.). J. Fish Dis. 2010, 33, 901–905. [Google Scholar] [CrossRef]
- Xu, W.; Shi, H.; Wang, W.; Zhang, D.Y.; Xu, W.J.; Chai, X.J. Isolation and identification of Streptococcus iniae in Larimichthys polyactis. Chin. J. Prev. Vet. Med. 2022, 44, 725–730. [Google Scholar]
- Pretto-Giordano, L.G.; Scarpassa, J.A.; Barbosa, A.R.; Altrão, C.S.; Ribeiro, C.G.G.; Vilas-Boas, L.A. Streptococcus iniae: An unusual important pathogen fish in Brazil. J. Aquac. Res. Dev. 2015, 6, 9. [Google Scholar] [CrossRef]
- Luo, Z.; Xu, J.; Han, J.G.; Xu, Y.X.; Ma, W.T.; Feng, S.M. Isolation and identification of Pathogenetic Streptococcus iniae from Selenotoca multifasciata. J. Huazhong Agric. Uni. 2012, 31, 95–99. [Google Scholar]
- Luo, Y.; Deng, Y.T.; Zhao, F.; Tan, A.P.; Zhang, M.C.; Jiang, L. Comparative on characteristics and pathogenicity of Nocardia seriolae isoalted from 9 fishes. Microbiol. China 2021, 48, 2733–2749. [Google Scholar]

 ), necrosis (
), necrosis ( ), and accumulation of red blood cells (★). (D) Spleen, showing oval nodules interconnected by fibroblasts (
), and accumulation of red blood cells (★). (D) Spleen, showing oval nodules interconnected by fibroblasts ( ). (E) Spleen, showing granulomas characterized by necrotic centers surrounded by a thin fibrous capsule (
). (E) Spleen, showing granulomas characterized by necrotic centers surrounded by a thin fibrous capsule ( ), a decreased number of red blood cells, and extensive cell necrosis (★). (G) Kidney, showing renal tubular epithelial cell granular degeneration (
), a decreased number of red blood cells, and extensive cell necrosis (★). (G) Kidney, showing renal tubular epithelial cell granular degeneration ( ) and necrosis (★). (H) Anterior kidney, showing extensive cell necrosis (★), and many macrophages in the outer layer of the granuloma. (I) Spleen from the fish artificially infected with strain HG-2021-1, showing small circular nodules (★).
) and necrosis (★). (H) Anterior kidney, showing extensive cell necrosis (★), and many macrophages in the outer layer of the granuloma. (I) Spleen from the fish artificially infected with strain HG-2021-1, showing small circular nodules (★).
 ), necrosis (
), necrosis ( ), and accumulation of red blood cells (★). (D) Spleen, showing oval nodules interconnected by fibroblasts (
), and accumulation of red blood cells (★). (D) Spleen, showing oval nodules interconnected by fibroblasts ( ). (E) Spleen, showing granulomas characterized by necrotic centers surrounded by a thin fibrous capsule (
). (E) Spleen, showing granulomas characterized by necrotic centers surrounded by a thin fibrous capsule ( ), a decreased number of red blood cells, and extensive cell necrosis (★). (G) Kidney, showing renal tubular epithelial cell granular degeneration (
), a decreased number of red blood cells, and extensive cell necrosis (★). (G) Kidney, showing renal tubular epithelial cell granular degeneration ( ) and necrosis (★). (H) Anterior kidney, showing extensive cell necrosis (★), and many macrophages in the outer layer of the granuloma. (I) Spleen from the fish artificially infected with strain HG-2021-1, showing small circular nodules (★).
) and necrosis (★). (H) Anterior kidney, showing extensive cell necrosis (★), and many macrophages in the outer layer of the granuloma. (I) Spleen from the fish artificially infected with strain HG-2021-1, showing small circular nodules (★).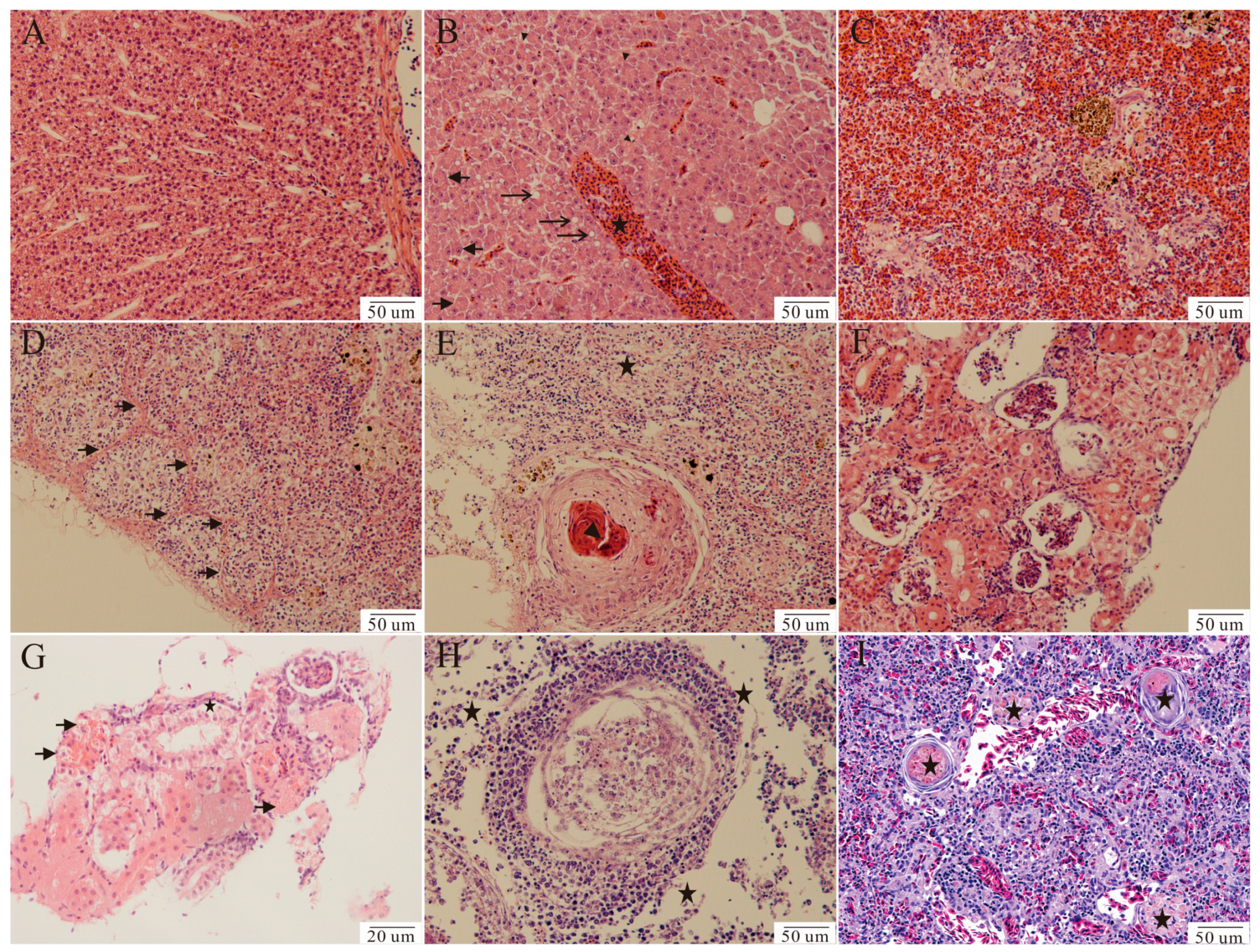


| Primer | Sequence (5′–3′) | Length (bp) | Application |
|---|---|---|---|
| T39 | GCGAACGGGTGAGTAACACG | 936 | Test Mycobacteria spp. by nested PCR [5] |
| T13 | TGCACACAGGCCACAAGGGA | ||
| preT43 | AATGGGCGCAAGCCTGATG | 300–312 | |
| T531 | ACCGCTACACCAGGAAT | ||
| P3 | AGCTTCGCAAGGTGAAGGG | 383 | Test R. salmoninarum by nested PCR [6] |
| M21 | GCAACAGGTTTATTTGCCGGG | ||
| P4 | ATTCTTCCACTTCAACAGTACAAGG | 320 | |
| M38 | CATTATCGTTACACCCGAAACC | ||
| PS-F | CTAGGAGATGAGCCCGCGTTG | 390 | Test Piscirickettsia by PCR [7] |
| PS-R | ATTTCA CATCCAACTTAATCT | ||
| Noc-f | CACCTACGAAAATCCCATTTGGT | 156 | Test Nocardia seriolae by PCR [8] |
| Noc-r | CATCGGATTGGATTCAAGGACCTTGA | ||
| F5 | CCTTTTTGAGTTTCGCTCC | 1133 | Test Francisella spp. by PCR [9] (FORSMAN) |
| F11 | TACCAGTTGGAAACGACTGT | ||
| lctO-F | AAGGGGAAATCGCAAGTGCC | 870 | Clone lctO gene [10] |
| lctO-R | ATATCTGATTGGGCCGTCTAA | ||
| 27F | AGAGTTTGATCCTGGCTCAG | 1480–1494 | Clone 16S rRNA gene [11] |
| 1492R | TACGGCTACCTTGTTACGCTT | ||
| dnaN-F | GCACATGTTAATTCGCCAGAGG | 404 | Multilocus sequence typing [12] |
| dnaN-R | CAGCACCAACTCTGATAATTTTCCA | ||
| mutL-F | CCAACCAAGCAGGAAGTTCG | 545 | |
| mutL-R | CGTTCTTGAGCTGCGTGTTG | ||
| mutM-F | CAGAGTAGATGGTTTGACCC | 410 | |
| mutM-R | TGCCCTGTATGATGCCTATC | ||
| mutS-F | TTTAACTGGCGCATCCCCAT | 448 | |
| mutS-R | TGGATCTTGCAACAGGTGAGT | ||
| mutX-F | TGGCCATTGGTTTCATCAAGG | 547 | |
| mutX-R | CGTAATCCCCTTCCCACGTT | ||
| recD2-F | AGGGCTTCCTAGTGCTACCA | 563 | |
| recD2-R | ACTCGCTTTGCCCATCAAGA | ||
| rnhC-F | GGAATCGCTGTTGTTGCAAGT | 582 | |
| rnhC-R | TTGAGTGTTTGCGAAGTGGC | ||
| yfhQ-F | AGGCCAGGTGATTTCAACCA | 511 | |
| yfhQ-R | CAGGAGAAACCCAGGCCATT | ||
| pgmA-F | AGACGGGGTCACAGACTACAT | 949 | Test virulence genes by PCR [13,14] |
| pgmA-R | AGGAGCACTTTGACGGAATT | ||
| cfi-F | GTGCCTCAACATCAAACA | 328 | |
| cfi-R | TAGCAAATCCCATATCAA | ||
| scpI-F | GCAACGGGTTGTCAAAAATC | 822 | |
| scpI-R | GAGCAAAAGGAGTTGCTTGG | ||
| simA-F | AATTCGCTCAGCAGGTCTTG | 994 | |
| simA-R | AACCATAACCGCGATAGCAC | ||
| pdi-F | TTTCGACGACAGCATGATTG | 381 | |
| pdi-R | GCTAGCAAGGCCTTCATTTG | ||
| sagA-F | AGGAGGTAAGCGTTATGTTAC | 190 | |
| sagA-R | AAGAAGTGAATTACTTTGG | ||
| cpsD-F | TGGTGAAGGAAAGTCAACCAC | 534 | |
| cpsD-R | TCTCCGTAGGAACCGTAAGC |
| Items | HG-2021-1 | HG-2021-3 | Items | HG-2021-1 | HG-2021-3 |
|---|---|---|---|---|---|
| Gram stain | + | + | Vogus–Proskauer | − | − |
| 10 °C growth | − | − | Catalase | − | − |
| 45 °C growth | − | − | Methyl red reaction | + | + |
| Motility | − | − | Esculin hydrolysis | + | + |
| Mannitol | − | − | Sucrose | + | + |
| Glucose | + | + | 6.5% NaCl growth | − | − |
| Sorbitol | − | − | Glucose gas production | − | − |
| Oxidase | − | − | Hemolysis | α | α |
| Urease | − | − | Arginine dihydrolase | + | + |
| D-trehalose | + | + | D-maltose | + | + |
| a-galactosidase | − | − | β-glucuronidase | + | + |
| Phosphatase | + | − | D-raffinose | − | + |
| D-amygdalin | − | + | D-xylose | − | + |
| Isolates | Bacterial Concentration (CFU/mL) | Amount of Death Post-Infection in Different Days | Cumulative Mortality (%) | LC50 (CFU/mL) | |||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 10 | 11 | 12 | 13 | 14 | ||||
| HG-2021-1 | 3.3 × 108 | 0 | 0 | 5 | 8 | 12 | 18 | 26 | 30 | 30 | 30 | 30 | 30 | 30 | 30 | 100 | 1.8 × 106 |
| 3.3 × 107 | 0 | 0 | 3 | 6 | 11 | 18 | 24 | 27 | 28 | 28 | 28 | 28 | 28 | 28 | 93.3 | ||
| 3.3 × 106 | 0 | 0 | 1 | 6 | 8 | 12 | 15 | 16 | 17 | 18 | 18 | 18 | 18 | 18 | 60.0 | ||
| 3.3 × 105 | 0 | 0 | 0 | 0 | 1 | 2 | 4 | 6 | 6 | 7 | 7 | 7 | 7 | 7 | 23.3 | ||
| HG-2021-3 | 3.1 × 108 | 0 | 0 | 4 | 5 | 8 | 14 | 23 | 27 | 27 | 27 | 27 | 27 | 27 | 27 | 90.0 | 6.6 × 106 |
| 3.1 × 107 | 0 | 0 | 3 | 3 | 7 | 7 | 11 | 12 | 14 | 21 | 21 | 21 | 21 | 21 | 70.0 | ||
| 3.1 × 106 | 0 | 0 | 1 | 2 | 4 | 7 | 8 | 8 | 10 | 11 | 13 | 13 | 13 | 13 | 43.3 | ||
| 3.1 × 105 | 0 | 0 | 0 | 0 | 1 | 2 | 2 | 3 | 3 | 3 | 4 | 4 | 4 | 4 | 13.3 | ||
| PBS | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | |
| Antibiotics | Sensitivity | Antibiotics | Sensitivity | ||
|---|---|---|---|---|---|
| HG-2021-1 | HG-2021-3 | HG-2021-1 | HG-2021-3 | ||
| Tetracycline | I | I | Ampicillin | S | S |
| Neomycin | R | R | Norfloxacin | R | R |
| Streptomycin | S | S | Ciprofloxacin | R | R |
| Polymyxin B | R | R | Florfenicol | S | S |
| Doxycycline | R | R | Vancomycin | R | R |
| Kanamycin | I | I | Roxithromycin | S | S |
| Tobramycin | R | R | Gentamycin | R | R |
| Levofloxacin | R | R | Erythromycin | S | S |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Luo, Z.; Bai, X.; Hao, S.; Wang, M.; Wu, Y.; Sun, H. Two Genotypes of Streptococcus iniae Are the Causative Agents of Diseased Ornamental Fish, Green Terror Cichlid (Aequidens rivulatus). Fishes 2024, 9, 140. https://doi.org/10.3390/fishes9040140
Luo Z, Bai X, Hao S, Wang M, Wu Y, Sun H. Two Genotypes of Streptococcus iniae Are the Causative Agents of Diseased Ornamental Fish, Green Terror Cichlid (Aequidens rivulatus). Fishes. 2024; 9(4):140. https://doi.org/10.3390/fishes9040140
Chicago/Turabian StyleLuo, Zhang, Xiaohui Bai, Shuang Hao, Mengyu Wang, Yongjiang Wu, and Hanchang Sun. 2024. "Two Genotypes of Streptococcus iniae Are the Causative Agents of Diseased Ornamental Fish, Green Terror Cichlid (Aequidens rivulatus)" Fishes 9, no. 4: 140. https://doi.org/10.3390/fishes9040140
APA StyleLuo, Z., Bai, X., Hao, S., Wang, M., Wu, Y., & Sun, H. (2024). Two Genotypes of Streptococcus iniae Are the Causative Agents of Diseased Ornamental Fish, Green Terror Cichlid (Aequidens rivulatus). Fishes, 9(4), 140. https://doi.org/10.3390/fishes9040140






