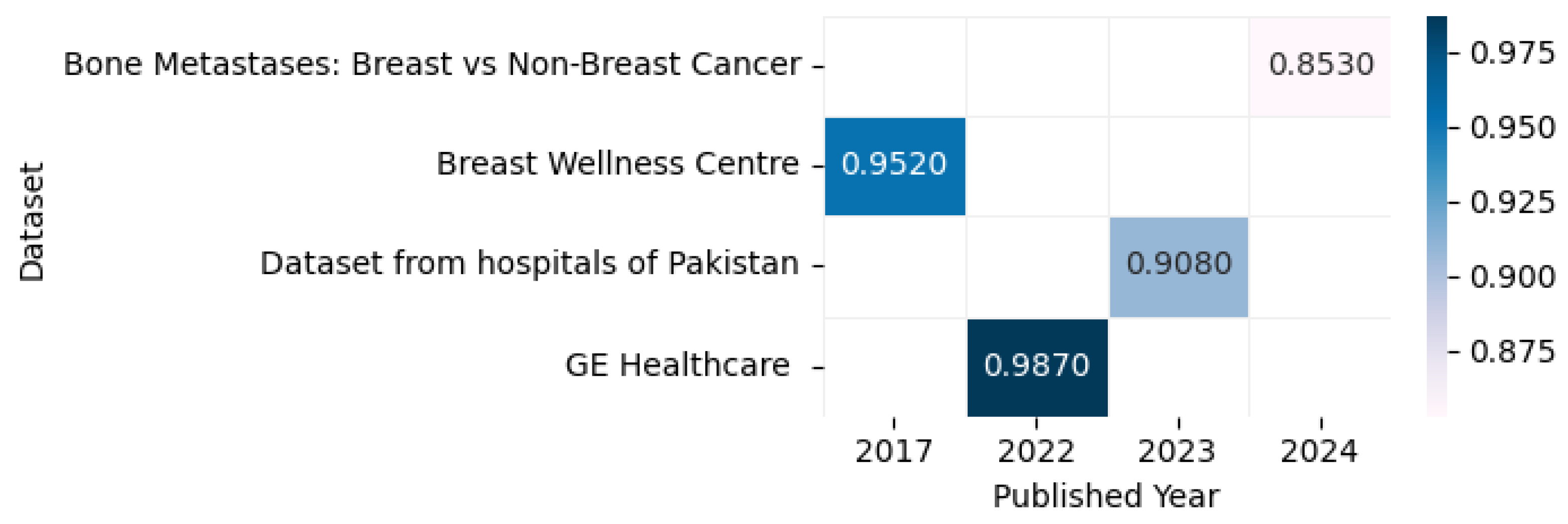Abstract
Breast cancer remains a critical global health challenge, with over 2.1 million new cases annually. This review systematically evaluates recent advancements (2022–2024) in machine and deep learning approaches for breast cancer detection and risk management. Our analysis demonstrates that deep learning models achieve 90–99% accuracy across imaging modalities, with convolutional neural networks showing particular promise in mammography (99.96% accuracy) and ultrasound (100% accuracy) applications. Tabular data models using XGBoost achieve comparable performance (99.12% accuracy) for risk prediction. The study confirms that lifestyle modifications (dietary changes, BMI management, and alcohol reduction) significantly mitigate breast cancer risk. Key findings include the following: (1) hybrid models combining imaging and clinical data enhance early detection, (2) thermal imaging achieves high diagnostic accuracy (97–100% in optimized models) while offering a cost-effective, less hazardous screening option, (3) challenges persist in data variability and model interpretability. These results highlight the need for integrated diagnostic systems combining technological innovations with preventive strategies. The review underscores AI’s transformative potential in breast cancer diagnosis while emphasizing the continued importance of risk factor management. Future research should prioritize multi-modal data integration and clinically interpretable models.
1. Introduction
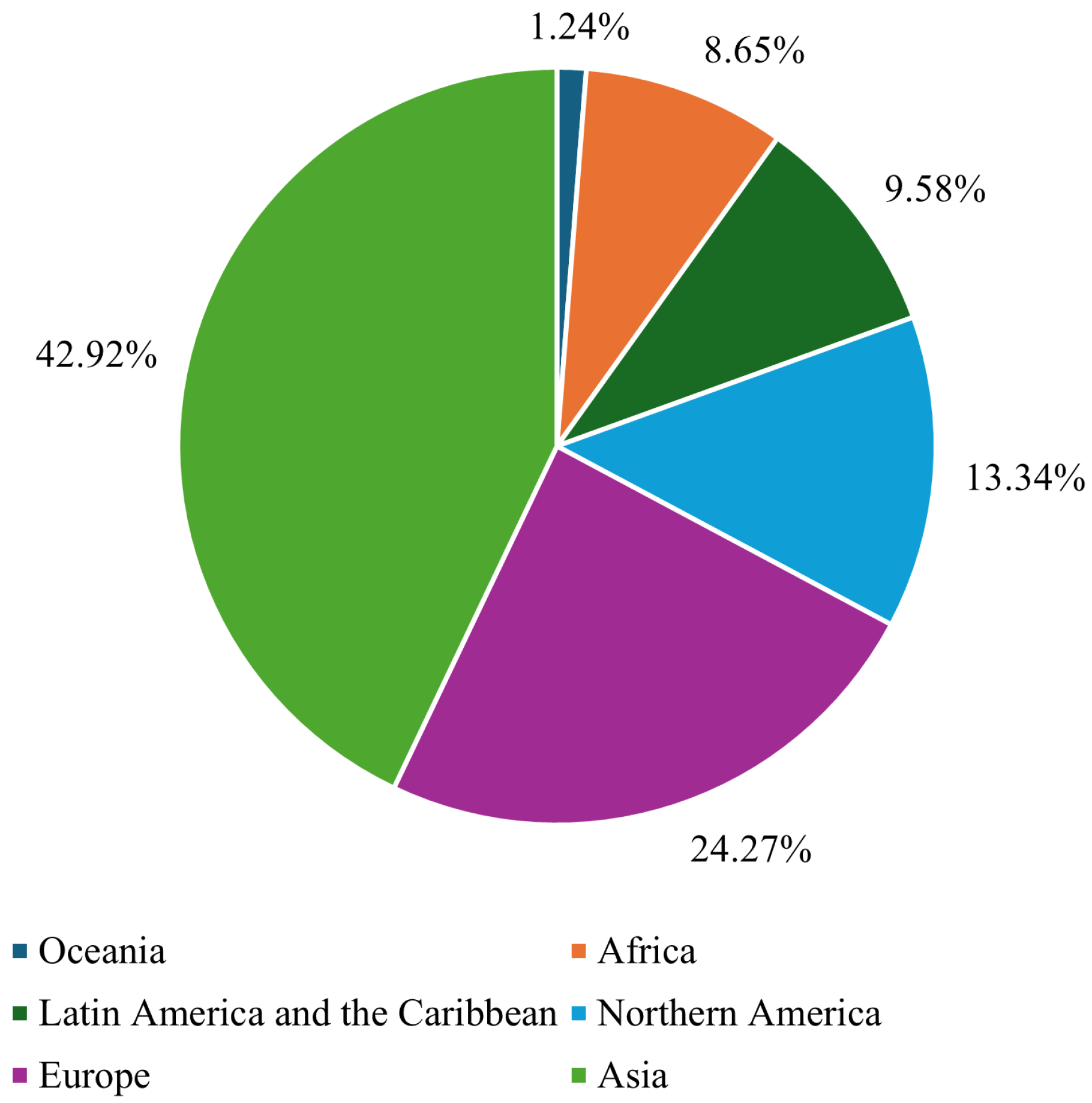
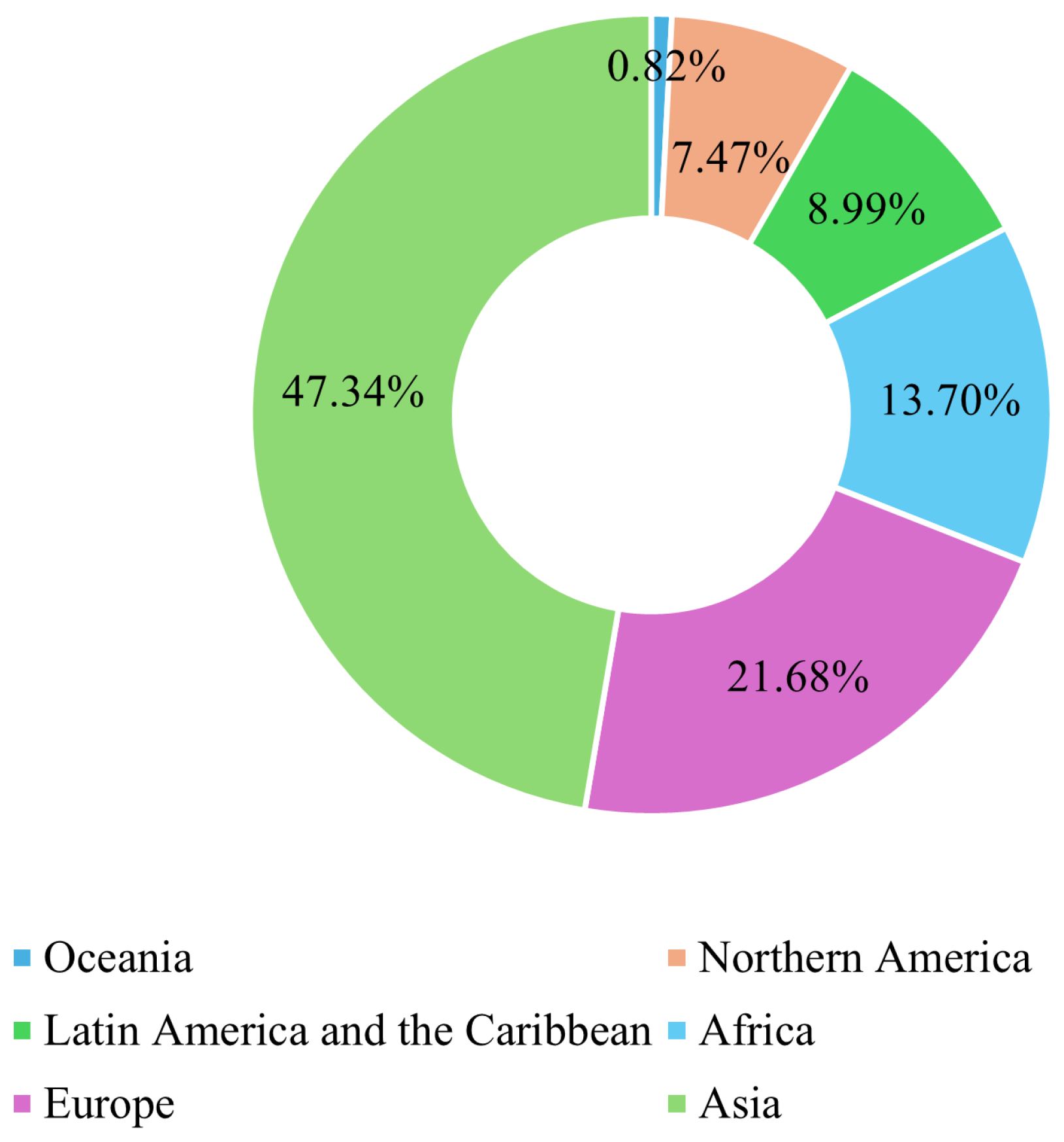
Breast cancer (BC) is the leading form of cancer among women, comprising 25% of global female cancer cases. In 2020, there were roughly 2.3 million new cases and 685,000 deaths worldwide. Early and accurate detection remains critical, as malignant tumors can spread aggressively, significantly reducing the probability of survival if left untreated [1]. Advances in artificial intelligence (AI) and machine learning (ML) have transformed medical image analysis, offering innovative tools for BC diagnosis. Techniques such as convolutional neural networks (CNNs) have shown promise in classifying and diagnosing BC using diverse imaging modalities, including mammography, CT, and ultrasound. By incorporating CAD systems and utilizing deep learning (DL), the fatality rate of breast carcinoma decreased by 40% from the 1980s to 2020, with projections suggesting that up to 2.5 million lives could be saved by 2040 if global mortality rates decline by 2.5% annually [2]. Figure 1 and Figure 2 illustrate the global incidence and mortality rates of BC among females in 2022. Figure 1 shows that Asia has the highest incidence of BC, accounting for 42.92% of cases, followed by Europe at 24.27%. Oceania reports the lowest incidence rate, at just 1.24%. Regarding mortality, 47.34% of female deaths occurred in Asia, while Europe accounted for 21.68%. These statistics demonstrate that over 65% of BC cases and deaths are concentrated in Asia and Europe.
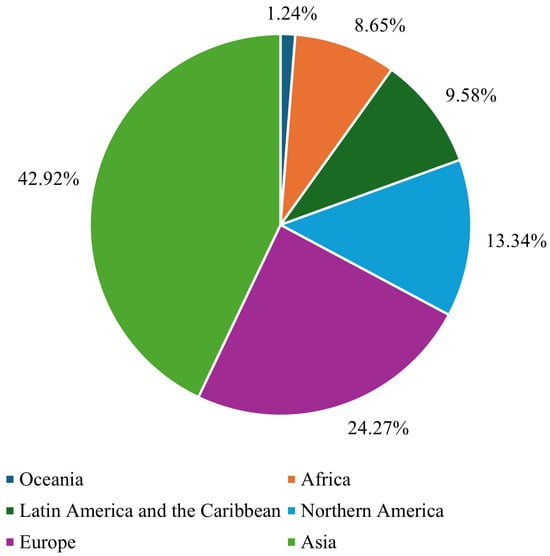
Figure 1.
Global breast cancer incidence rate in females in 2022 [3].
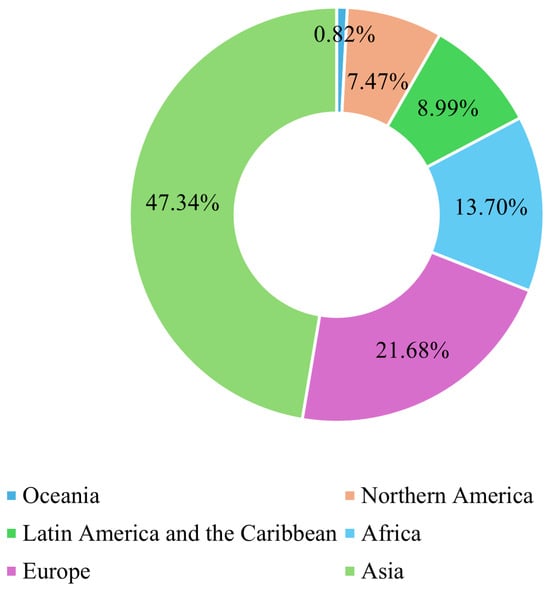
Figure 2.
Global breast cancer mortality rate in females in 2022 [3].
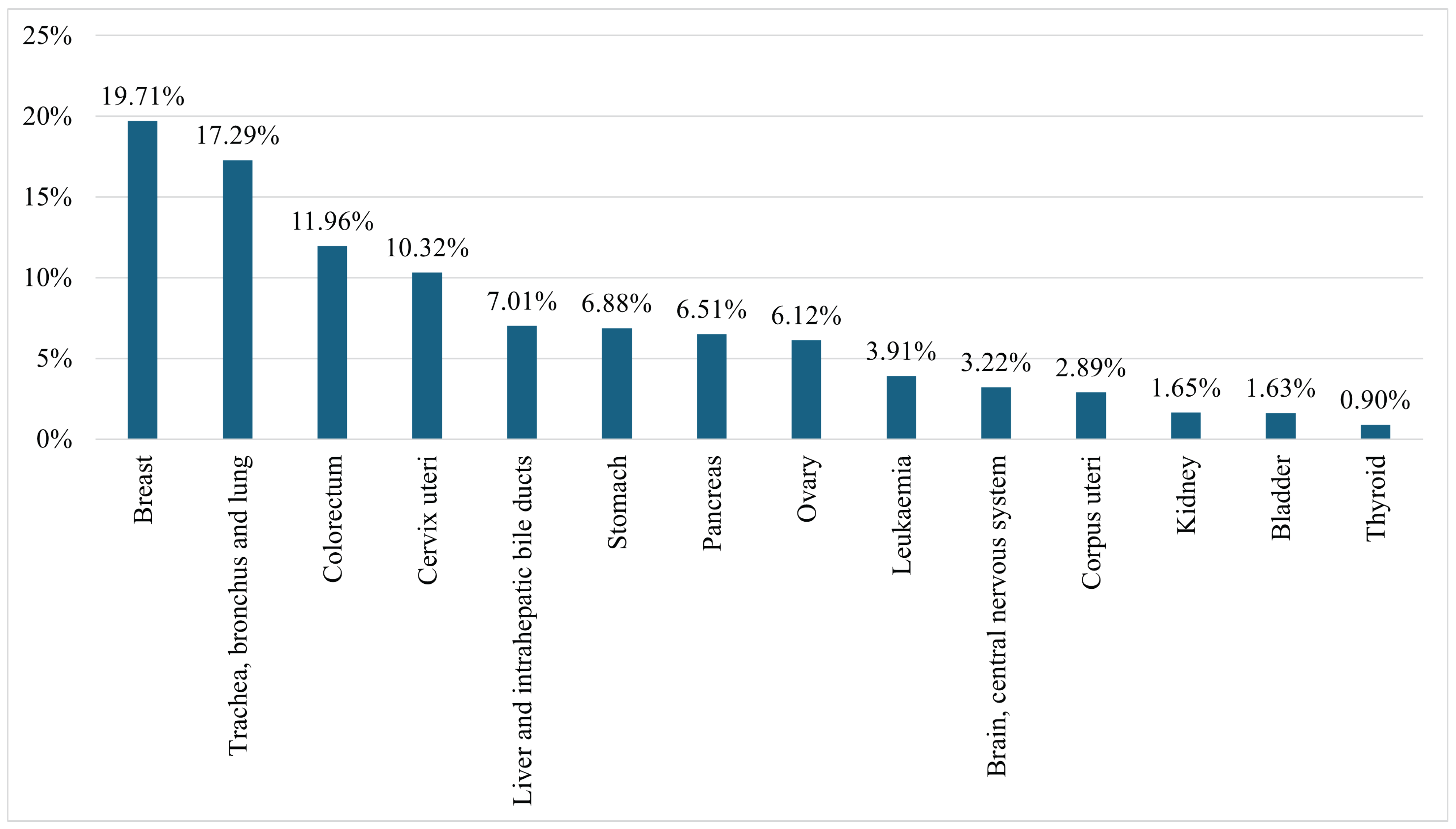
Figure 3 presents the mortality rates of females from the fourteen leading cancers worldwide in 2022, including ‘Breast’; ‘Trachea, bronchus, and lung’; ‘Colorectum’; ‘Cervix uteri’; ‘Liver and intrahepatic bile ducts’; ‘Stomach’; ‘Pancreas’; ‘Ovary’; ‘Leukaemia’; ‘Brain, central nervous system’; ‘Corpus uteri’; ‘Kidney’; ‘Bladder’; and ‘Thyroid’. The figure highlights that 19.71% of female cancer-related deaths are due to BC, making it the leading cause of cancer-related mortality among women [3].
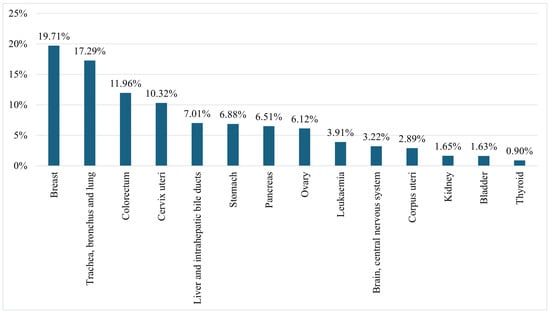
Figure 3.
Female mortality rates from all cancers worldwide in 2022 [3].
1.1. Motivation
Breast cancer remains a significant global health concern, particularly among women, where it stands as one of the most prevalent and life-threatening forms of cancer. Despite advances in medical imaging and therapeutic strategies, there are still persistent challenges to early and accurate diagnosis, representing a major hurdle to effective treatment.
While DL models achieve high accuracy, their ‘black-box’ nature remains a barrier to clinical trust. Recent advancements in explainable AI (XAI) techniques, such as Grad-CAM and SHAP, are critical for bridging this gap. This review evaluates not only performance metrics but also the interpretability of state-of-the-art models, highlighting their clinical applicability.
Below are the key motivations for this study.
- A1.
- Breast cancer affects millions annually, with increasing prevalence across diverse age groups. Traditional diagnostic techniques such as mammography are often inadequate, particularly in dense breast tissues, leading to missed or delayed diagnoses.
- A2.
- DL techniques have revolutionized medical imaging by enabling precise analysis of complex, high-dimensional datasets. These methods reduce human error, increase diagnostic consistency, and improve efficiency.
- A3.
- Existing screening tools have inherent strengths and weaknesses. Investigating these methods to identify the most effective approaches for breast cancer detection remains critical for advancing diagnostic quality.
- A4.
- Innovations such as transfer learning, hybrid DL models, and explainable AI have enhanced diagnostic accuracy while addressing trust and interpretability concerns, essential factors for clinical integration.
- A5.
- By integrating advanced technologies into clinical practice, we can increase early detection rates, potentially saving countless lives and transforming breast cancer treatment outcomes.
1.2. Contributions
This paper offers a comprehensive review of advancements in BC detection, with a strong focus on the transformative impact of machine learning and DL. The key contributions are as follows:
- A1.
- The global BC trends and risk factors are examined to inform preventive strategies.
- A2.
- We present a visualization of current trends in BC research, review the impacts of early detection and technology on mortality rates, and highlight the potential of lifestyle changes to reduce the incidence of breast cancer in high-risk groups.
- A3.
- This review examines various BC imaging techniques, including mammograms, ultrasound, MRI, CT scans, histopathology, and thermal imaging. We discuss their advantages and disadvantages and present examples of specific cases.
- A4.
- We explore the use of structured data (CSV) for BC detection, focusing on feature engineering, preprocessing, and algorithmic approaches.
- A5.
- The study presents a summary of hybrid frameworks combining deep feature extraction and machine learning classifiers to enhance diagnostic accuracy.
- A6.
- We report the level of accuracy achieved to date by various models using different datasets.
2. Background Study
Breast cancer is a major global health issue, with the World Health Organization (WHO) reporting 7.8 million women being diagnosed in the previous three years, making it the most prevalent cancer worldwide. Breast cancer accounts for 4–5.5% of all new cancer cases annually, significantly contributing to morbidity rates [4,5]. In the United States alone, over 250,000 individuals are diagnosed with BC each year; the disease accounts for 41,690 deaths, making it the most frequent and fatal cancer type among women. BC represents approximately 25% of all cancer diagnoses in women globally [6]. Early diagnosis is critical, as patients diagnosed in the early stages have a five-year survival rate of about 99% [7,8].
Breast cancer typically begins in the ducts or lobules and manifests through symptoms such as lumps, changes in breast size or shape, and abnormal discharge. Traditional diagnostic methods, including mammography, histopathological analysis, and other imaging techniques, are widely employed but are time-consuming, requiring skilled professionals, and are prone to human error. For example, mammograms rely on radiologists for interpretation, while histopathological analysis demands experienced pathologists to accurately classify tissues [9,10,11,12]. Breast cancer was first identified in Egypt around 1600 BC and has been responsible for approximately 15% of female deaths [13,14]. While BC affects men, women, and transgender individuals, its prevalence is significantly higher among females. The risk for transgender individuals, particularly the impact of gender-affirming hormonal therapy (GAHT) on BC, remains unclear, and guidelines for screening in this group are still undefined [15,16].
The WHO projects that BC cases will increase to 2.7 million by 2040. The National Breast Cancer Coalition (NBCC) has reported a rise in the lifetime risk of developing invasive breast cancer among women in the US since 1975, with 287,850 new cases identified in women and 2710 in men. Additionally, the American Chemical Society (ACS) notes that a woman’s likelihood of being diagnosed with invasive breast cancer increased from 9.09% to 12.9% during the same period [17,18,19,20]. Early and accurate BC detection is essential for improving survival rates, but early-stage diagnosis remains challenging. Diagnostic tools such as mammography, magnetic resonance imaging (MRI), computed tomography (CT), and ultrasonography are critical, with mammography being the most widely used method. Mammography has a sensitivity range of 77–95% and a specificity range of 92–95%, yet its accuracy can be affected by dense breast tissue. To address these limitations, advancements in technology, particularly machine learning (ML), are being utilized to enhance diagnostic precision, offering significant promise for early detection and personalized treatment [15].
In India, breast cancer ranks among the top five most common cancers, with new cases rising from 159,417 in 2017 to 178,361 in 2020, comprising nearly 26% of all cancer cases in women [14].
The remainder of the paper is organized as follows: Section 3 presents the research questions and includes a comparison table of our review with existing review papers on breast cancer detection. Section 4 discusses the most popular datasets used in breast cancer detection research, while Section 5 details the machine learning techniques applied for breast cancer detection. Section 6 provides statistics on various cancers, focusing on mortality and incidence rates for breast cancer across different categories. Section 7 outlines current diagnostic techniques for breast cancer and highlights key disease-associated factors. Section 8 analyzes different breast cancer detection approaches, followed by Section 9, which examines their applications, strengths, and weaknesses. Section 10 presents an overview of publication trends across journals. Finally, Section 11 concludes the paper, and Section 12 proposes future research directions to advance breast cancer detection and management.
3. Research Methodology
This section outlines the research methodology used to conduct a comprehensive review of breast cancer detection techniques using machine learning and deep learning approaches.
3.1. Study Selection Strategy
To ensure a rigorous and unbiased review, we implemented a structured selection process for identifying relevant studies:
3.1.1. Search Strategy
- SS1.
- Conducted searches across major academic databases including IEEE Xplore, SpringerLink, ScienceDirect, Nature, Wiley Online Library, and MDPI.
- SS2.
- Used keywords: (“breast cancer detection” OR “breast cancer diagnosis”) AND (“machine learning” OR “deep learning” OR “artificial intelligence”) combined with modality-specific terms (“mammography”, “ultrasound”, “MRI”, “CT”, “Histopathological”, “Thermal”, etc.).
3.1.2. Inclusion Criteria
- IC1.
- Primary focus on studies published between 2023 and 2024.
- IC2.
- Included a few foundational papers from 2019 to 2022 that introduced significant methodological advances.
- IC3.
- Included studies were required to report quantitative performance metrics such as accuracy, AUC, sensitivity, and specificity.
3.1.3. Exclusion Criteria
- EC1.
- Non-English publications, conference abstracts, and editorials.
- EC2.
- Duplicate or overlapping studies.
3.2. Research Questions
The article addresses ten key research questions, as outlined below. Additionally, Table 1 provides a comparison of existing review papers on breast cancer detection with our own, highlighting the main differences and contributions. The key research questions explored in this review include the following:

Table 1.
Comparative analysis of recently published review papers and our review of literature on breast cancer detection.
- Q1.
- What are the incidence and death rates for breast cancer across various categories?
- Q2.
- What diagnostic methods are currently used for breast cancer detection?
- Q3.
- What are the key risk factors for breast cancer?
- Q4.
- How can lifestyle changes help reduce the risk of breast cancer?
- Q5.
- Which breast cancer datasets have been used in the development of machine and deep learning models?
- Q6.
- What are the reasons for adopting machine and deep learning techniques in breast cancer detection?
- Q7.
- What are the different medical imaging modalities used for breast cancer classification, and what is the highest accuracy achieved by various models on these modalities to date?
- Q8.
- What are the various CSV datasets employed for breast cancer classification, and what is the best accuracy attained by different models on these datasets so far?
- Q9.
- What are the strengths, weaknesses, and applications of imaging modalities in breast cancer detection techniques?
- Q10.
- What are the benefits, drawbacks, and use cases of breast cancer detection techniques based on CSV datasets?
4. Overview of Breast Cancer Datasets
Researchers utilize diverse breast cancer datasets across multiple modalities—including X-ray, ultrasound, MRI, histopathology, thermography, and structured clinical records—for developing and validating detection algorithms. These datasets vary significantly in scale (from hundreds to thousands of samples), content complexity (from basic images to comprehensive multimodal clinical data), and accessibility (ranging from open public repositories to restricted institutional databases). For systematic evaluation, Table 2 provides a short description for the modalities, detailing key characteristics such as sample size, benign/malignant class distribution, annotation quality, and access protocols, enabling researchers to identify optimal resources for their specific computational analysis needs.

Table 2.
Comprehensive overview of multimodal breast cancer datasets.
5. Machine Learning Techniques
The three main categories of machine learning are supervised learning, unsupervised learning, and reinforcement learning. Supervised learning trains models using labeled data to map inputs to known outputs, making it ideal for predictive tasks like classification and regression. The process involves analyzing training data, computing statistical features, and establishing input–output relationships. While effective for accurate predictions, it requires large labeled datasets and significant training resources. Unsupervised learning discovers hidden patterns in unlabeled data through techniques like clustering and association rule mining. Valuable for exploratory analysis, it requires less preprocessing than supervised methods but can yield more challenging-to-interpret results. Reinforcement learning trains autonomous agents through environmental interaction and reward feedback, excelling in dynamic domains like robotics and game AI. This powerful approach demands substantial computation and careful reward mechanism design. Table 3 summarizes the key characteristics of machine learning algorithms, including their types, categorizations, approaches, advantages, and limitations.

Table 3.
Machine learning algorithm taxonomy (type, categorization, approach, advantages, limitations).
6. Cancer Statistics and Incidence and MortalityRates
This section presents a statistical analysis of data for cancer rankings from 1990 to 2021. We highlight the mortality rates for both males and females, along with the global death rate per 100,000 individuals. Additionally, the section includes the incidence and death rates for both sexes categorized by age groups (20+ years and under 20 years) specifically for Bangladesh. Table 4 presents the year-wise rankings of six major cancers—Breast, Lung, Stomach, Cervical, Colorectal, and Brain—from 1990 to 2021, obtained from the Institute for Health Metrics and Evaluation (IHME) [62]. In the table, lower values indicate a higher impact and higher values indicate a lower impact. The rankings highlight significant changes in the incidence of these cancers over three decades. Breast Cancer ranked third in 1990 and 2000, worsened to become the most impactful cancer by 2011, and it had retained its top position by 2021, underscoring its increasing global prevalence and burden. Lung Cancer consistently ranked among the top two, taking the first position in 2000 and 2010 before being overtaken by Breast Cancer. Stomach Cancer, initially the most impactful cancer (ranked first) in 1990, had declined to fifth by 2021, reflecting advancements in prevention and treatment. Cervical Cancer remained stable at fourth until 2011 before worsening to third in 2021, while Brain Cancer consistently held the sixth position, indicating a lower comparative global impact. Meanwhile, Colorectal Cancer worsened from fifth in 1990 to fourth by 2021, reflecting its global increase. These patterns illustrate the evolving global cancer landscape, with BC emerging as the most impactful over time, particularly during e period 2011 to 2021. The worsening of BC highlights its growing prevalence and significance, underscoring the urgent need for enhanced research, early detection, and effective treatment strategies.

Table 4.
Year-wise ranking of the six top cancers in the world [62].
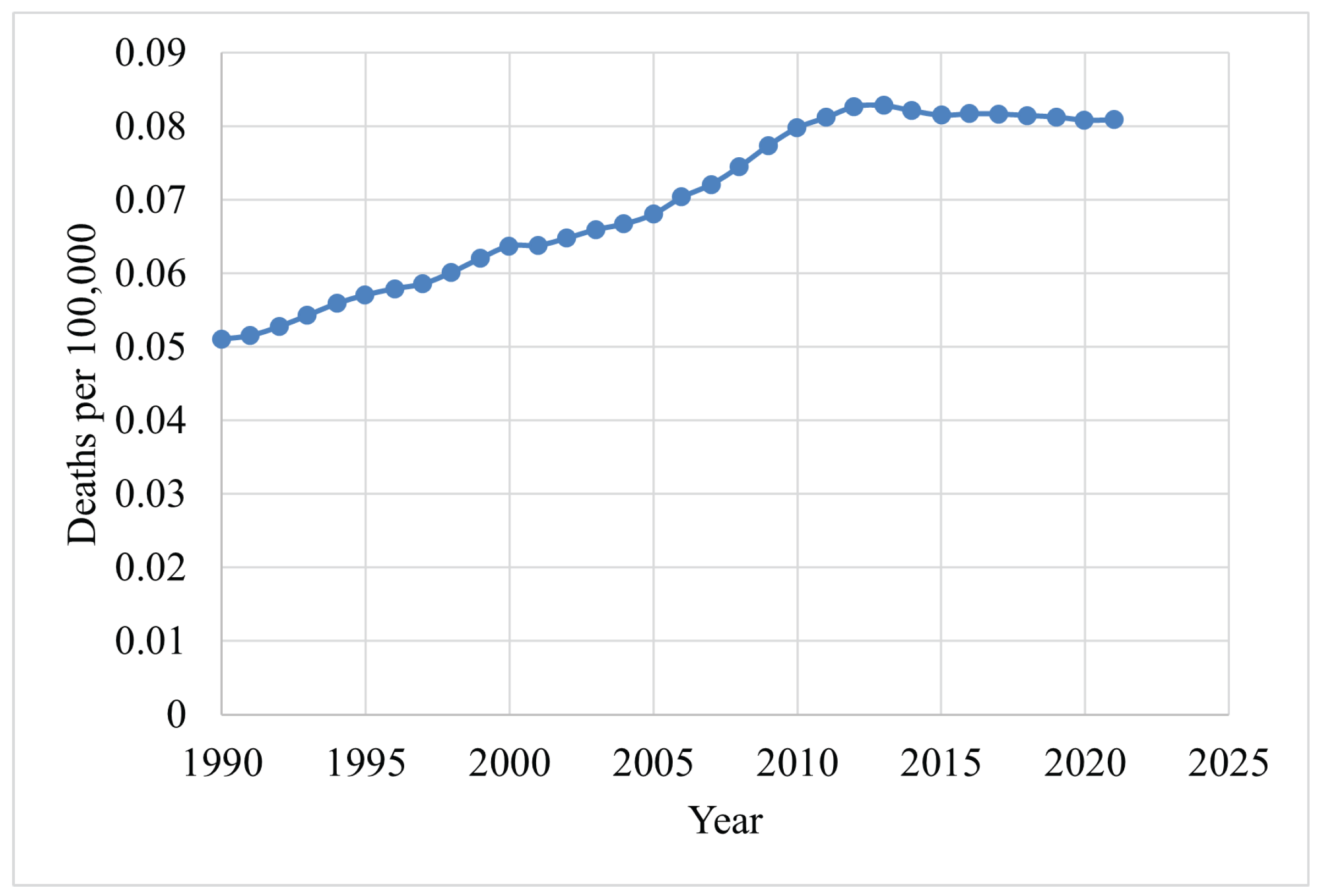
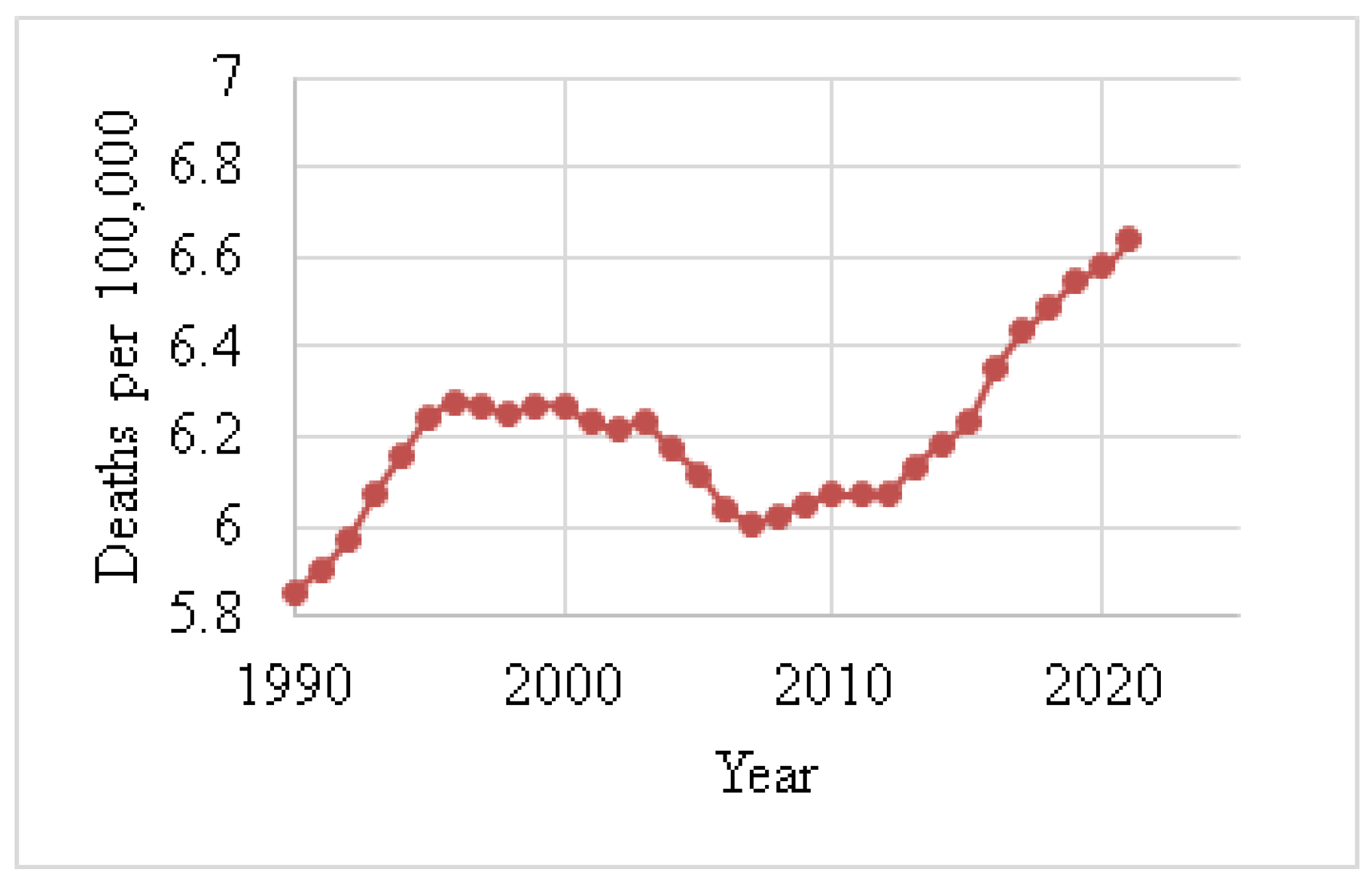
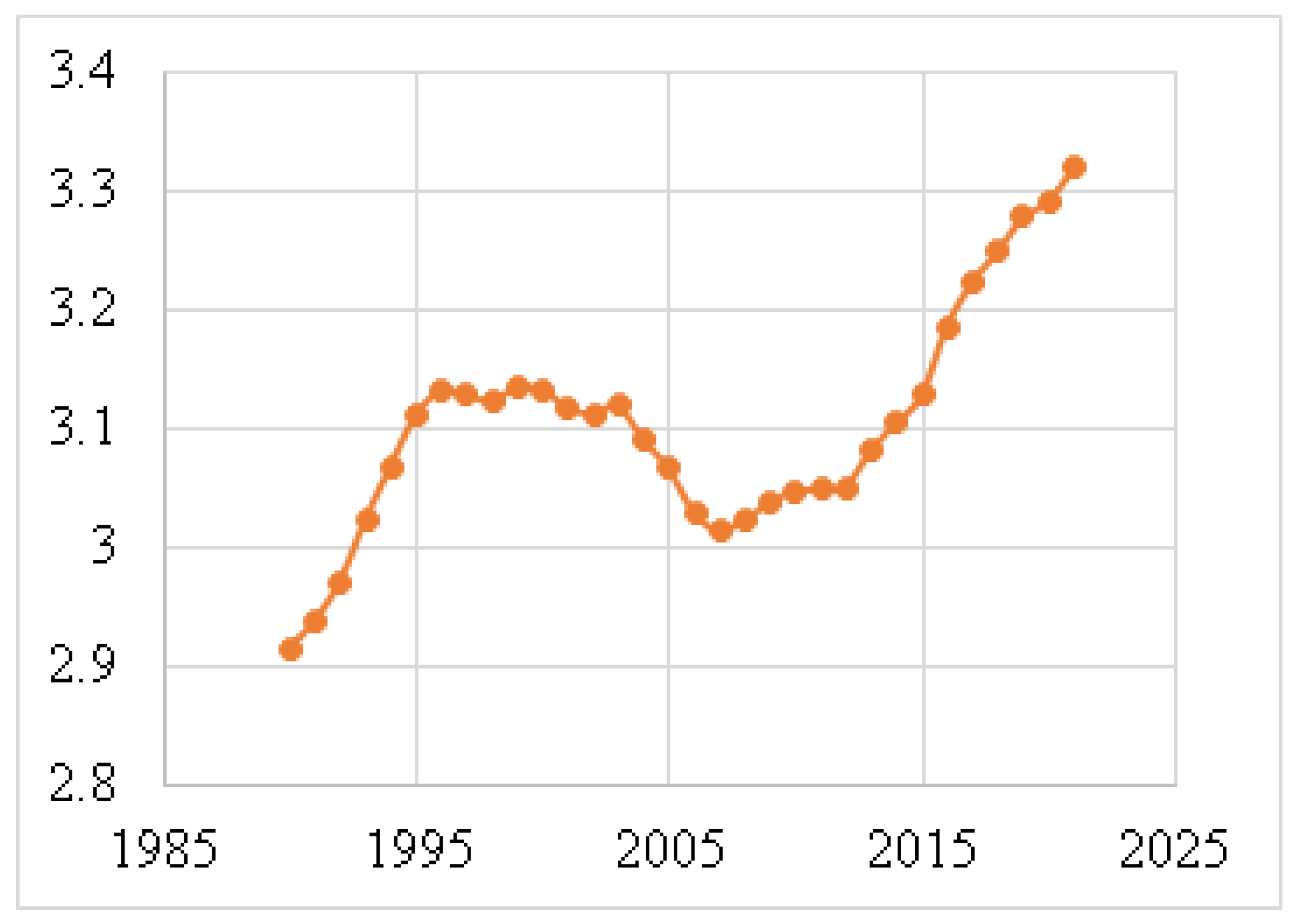
Figure 4 shows the death rates for males, and Figure 5 depicts the death rates for females per 100,000 individuals aged 15–49 between 1990 and 2021, using data obtained from the Institute for IHME [62]. The data reveal that the average death rate for females was 6.6401, while for males this was 0.0829. This indicates that the female death rate for BC is 98.75% higher than that of males during this period. Figure 6 illustrates the global death rates per 100,000 individuals. The data demonstrate an increase from 2.9141 in 1990 to 3.3183 in 2021, marking a 12.181% increase over this period using data provided by the same institute.
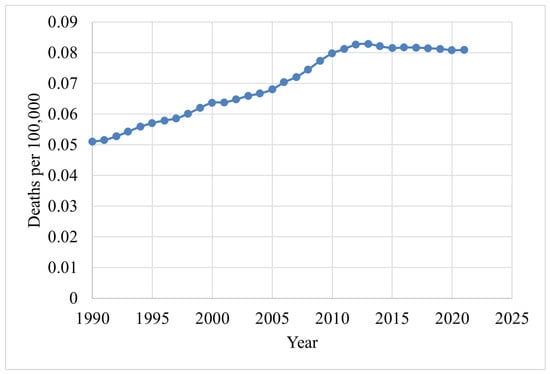
Figure 4.
Male death rates (1990–2021) due to BC [62].
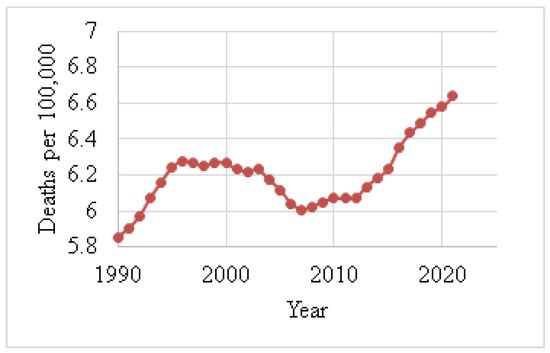
Figure 5.
Female death rates (1990–2021) due to BC [62].
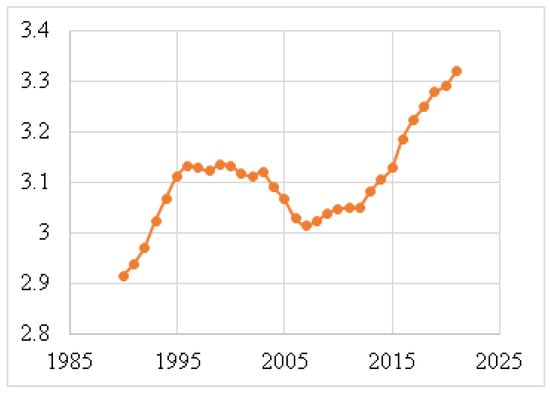
Figure 6.
Global death rates per 100,000 individuals (1990–2021) [62].
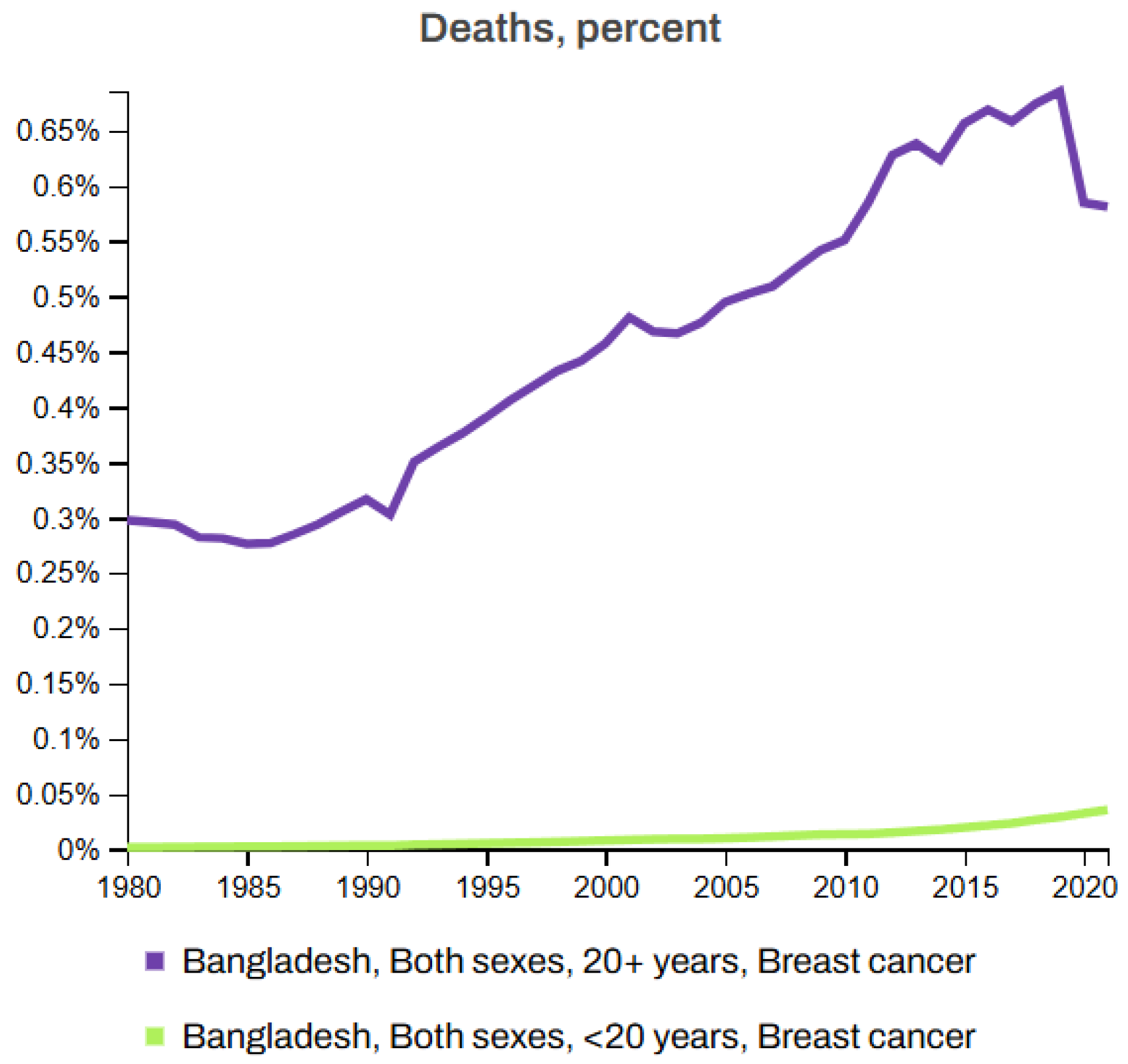
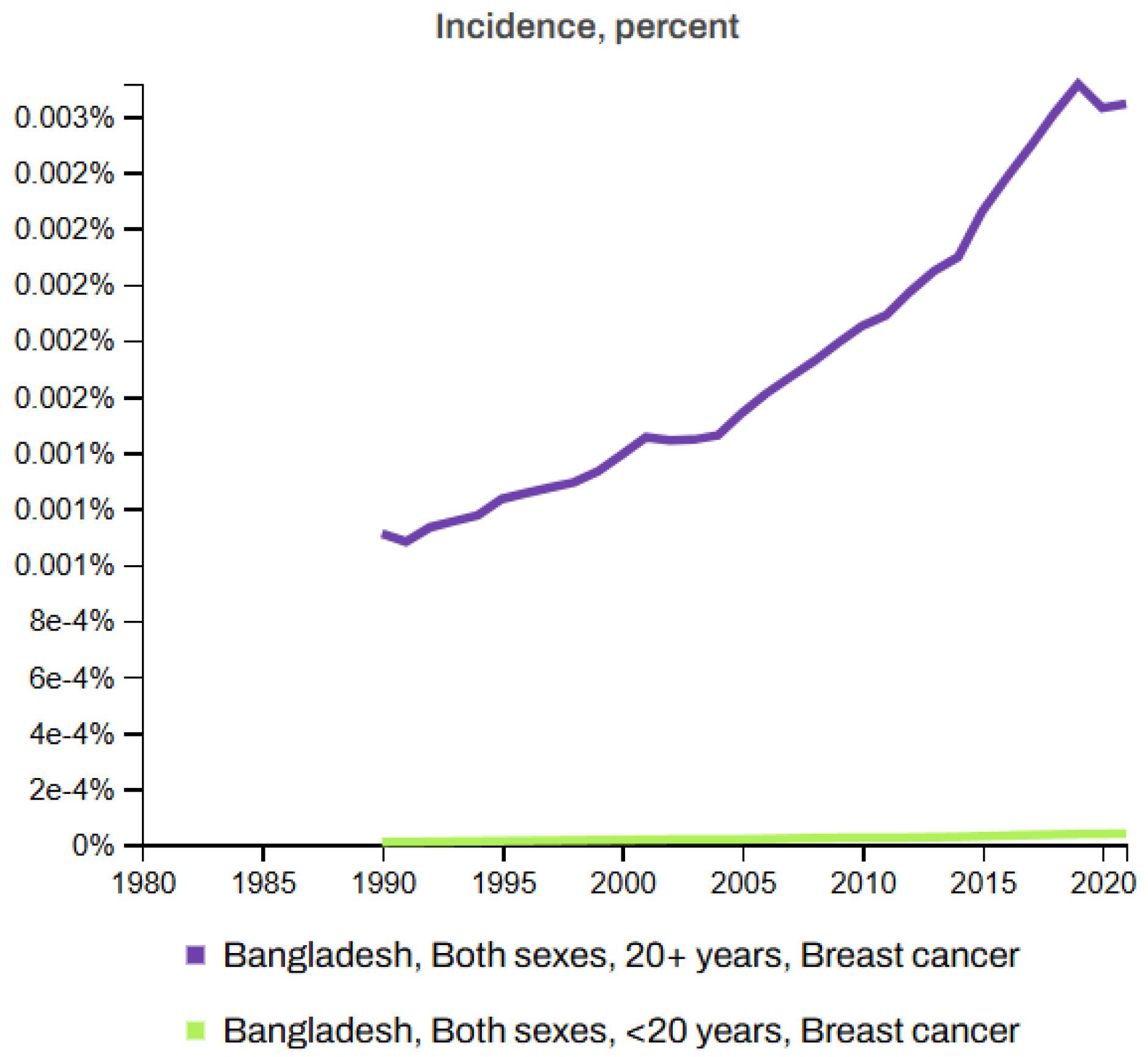
Figure 7 and Figure 8 depict the death rates and incidence rates of BC in Bangladesh from 1980 to 2021 categorized by age groups (20+ years and under 20 years) using data provided by the Institute for IHME, Seattle, United States [63]. Both figures clearly show a consistent increase in the mortality and incidence rates over time.
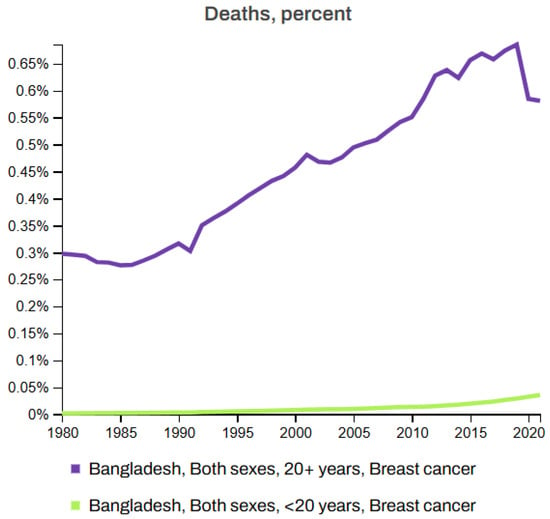
Figure 7.
Breast cancer death rate in Bangladesh (1980–2021) [63].
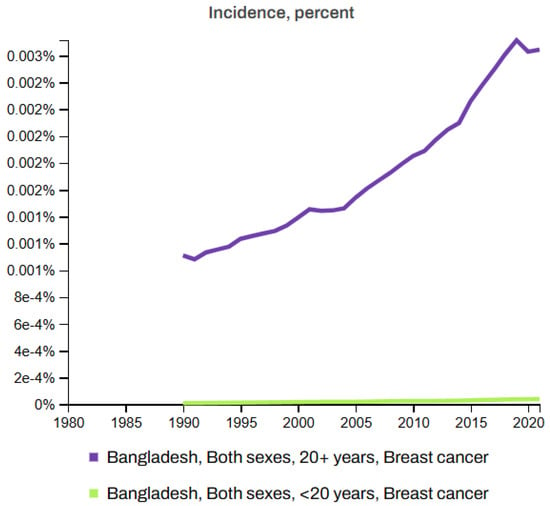
Figure 8.
Incidence rates of breast cancer in Bangladesh (1980–2021) [63].
The integration of ML and DL technologies has emerged as a transformative approach, enabling rapid and more accurate diagnosis. Research has shown that ML techniques such as CNNs can achieve up to 90% accuracy in classifying mammographic images as benign, malignant, or normal. Similarly, artificial neural networks have demonstrated high efficiency in analyzing ultrasound images to distinguish between benign and malignant cases. Some studies have reported that ML techniques can improve the prediction accuracy of BC recurrence by up to 25% compared to traditional methods [24,64,65]. These advancements in ML and DL offer the potential to address the limitations of traditional diagnostics by automating image interpretation, enhancing predictive analytics, and tailoring individual treatment plans. This review summarizes commonly used ML and DL algorithms and methodologies used in BC research, providing insights into their applications, strengths, and prospects.
7. Breast Cancer Diagnosis and Classification
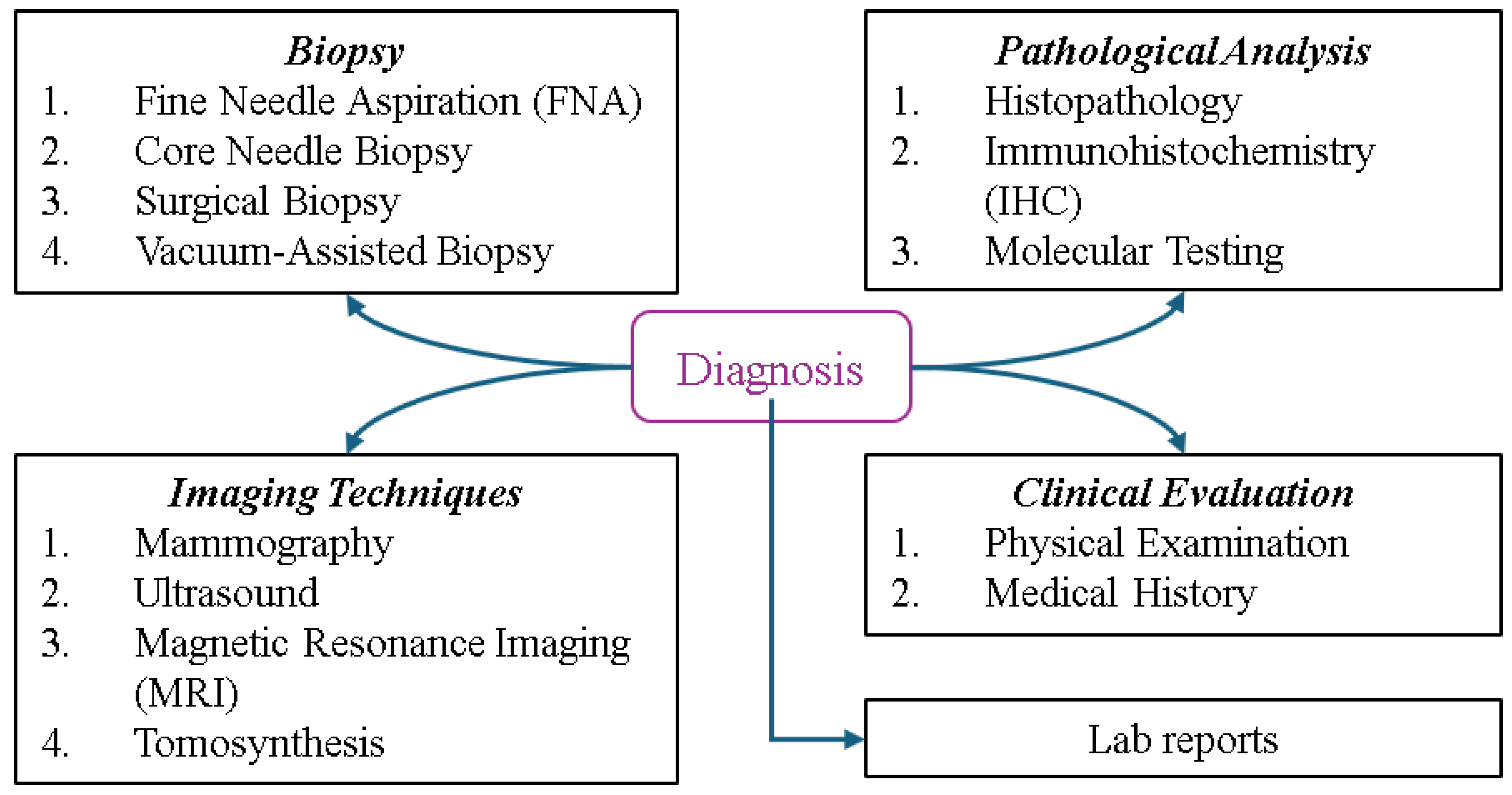
Determining the type and stage of breast cancer is crucial for patients, as this directly impacts treatment planning and prognosis, helping physicians develop optimal strategies and predict the chance of survival. Figure 9 illustrates the types of BC diagnosis, spanning multiple categories. Biopsy methods such as fine needle aspiration (FNA) for extracting fluid or cells, core needle biopsy for removing small tissue samples, Surgical Biopsy for excising part or all of a suspicious mass, and vacuum-assisted biopsy for obtaining larger samples play key roles in diagnosis. Imaging techniques include mammography, an X-ray of the breast; ultrasound, which uses sound waves to differentiate masses; MRI, a technology that provides detailed images of soft tissues, and tomosynthesis, a 3D mammography employed for enhanced clarity. Pathological analysis involves histopathology to examine tissue, immunohistochemistry (IHC) to identify protein markers such as HER2 or hormone receptors, and molecular testing to detect genetic mutations or biomarkers. Clinical evaluation includes a physical examination to check for abnormalities and a review of the patient’s medical history, while lab reports involve tests such as tumor markers (e.g., CA 15-3, CEA) for cancer indicators and a complete blood count (CBC) to assess general health. These methods provide a comprehensive framework for accurate identification and evaluation [2,66].
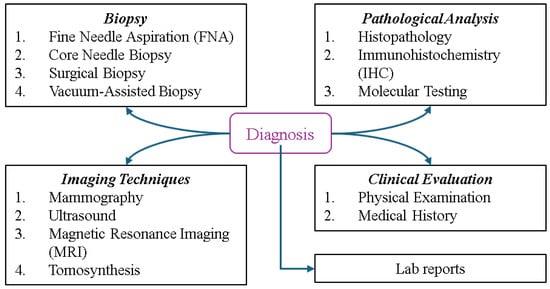
Figure 9.
Overview of breast cancer diagnostic methods.
7.1. Breast Cancer Variants
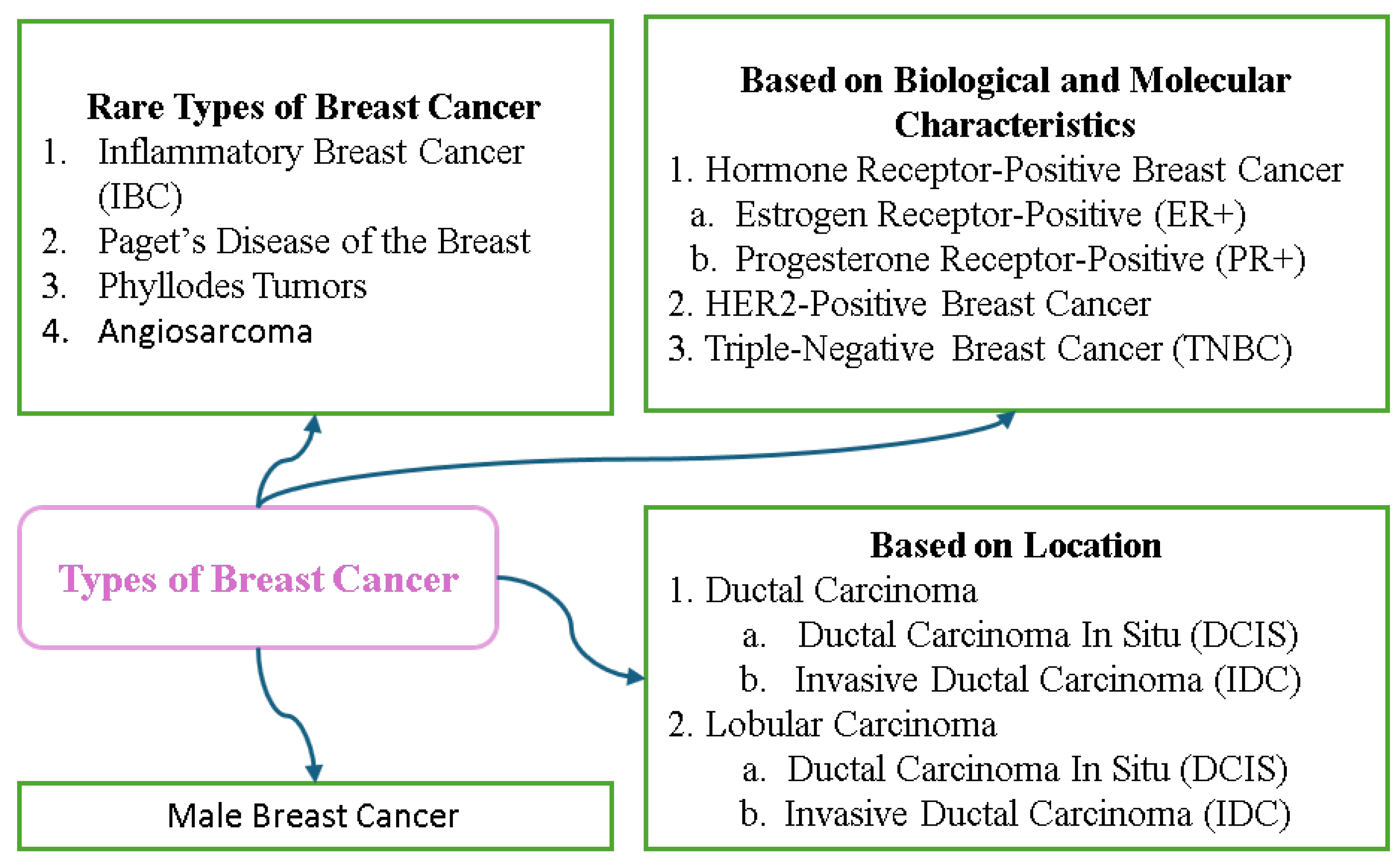
Breast cancer can be categorized according to the location, biological characteristics, and molecular subtype. Non-invasive types such as Ductal Carcinoma In Situ (DCIS) and Lobular Carcinoma In Situ (LCIS) are confined to their point of origin and serve as warning signs for potential invasive cancers. Invasive types include Invasive Ductal Carcinoma (IDC), the most common form, and Invasive Lobular Carcinoma (ILC) that often responds to hormone therapy. Rare types include Inflammatory Breast Cancer, a highly aggressive form that causes redness and swelling, and Paget’s Disease that affects the nipple and areola. Breast cancer is also classified into molecular subtypes based on the hormone receptor (ER, PR) and HER2 status; these include Luminal A, Luminal B, HER2-enriched, and Triple-Negative, and these subtypes can be used to guide treatment strategies. Assignment of stages from 0 to 4 and grading further assess the progression and behavior of the cancer, with stages indicating the degree of spread and grades reflecting growth patterns. Understanding the meanings of the classifications is crucial for diagnosis and treatment planning. Figure 10 displays the different types of breast cancer.
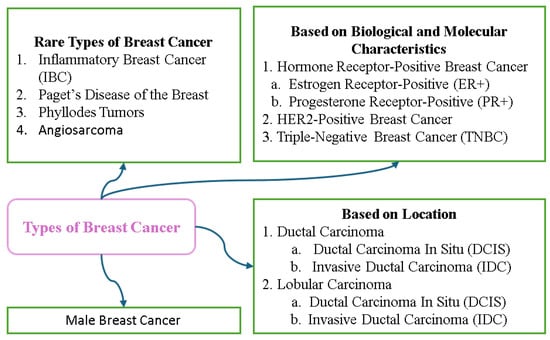
Figure 10.
Types of breast cancer.
7.2. Key Risk Factors Associated with Breast Cancer
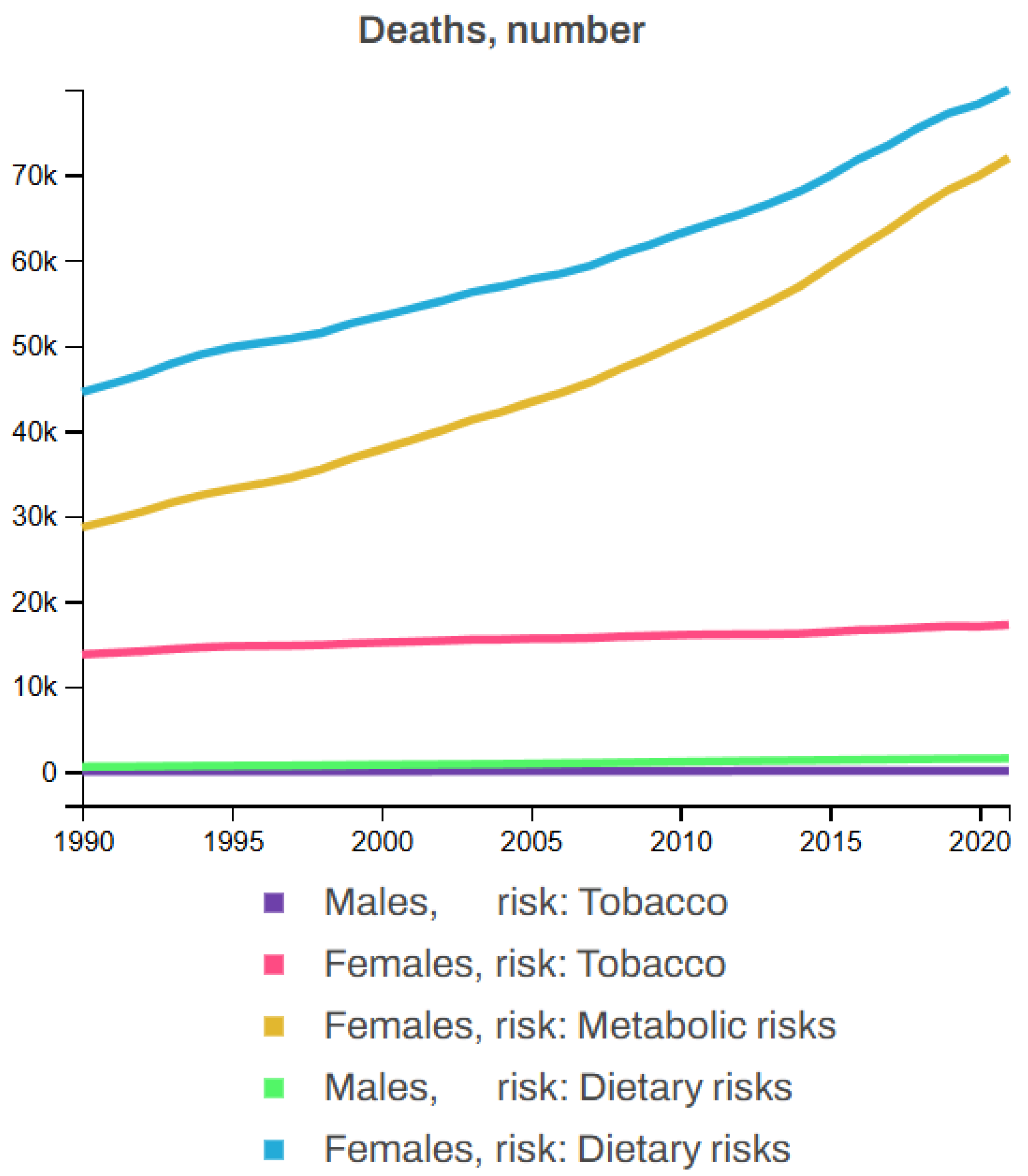
Breast cancer is influenced by several non-modifiable risk factors, including gender, age, family history, genetics, hormonal factors, and personal history, with women being at a significantly higher risk, particularly after the age of 50. A family history of BC, especially in first-degree relatives, along with known genetic mutations such as BRCA1 and BRCA2, further increases the likelihood of developing the disease. Hormonal factors such as early menstruation, late menopause, or hormone replacement therapy also contribute to this risk. Additionally, individuals who have had BC in one breast are at higher risk. Beyond these, other risk factors including tobacco use (smoking, chewing tobacco, and secondhand smoke), poor dietary habits (low intake of fruits, vegetables, whole grains, and seafood omega-3 fatty acids, along with a high intake of red meat, processed meat, sugar-sweetened beverages, and trans fats), and metabolic factors (high levels of fasting plasma glucose and LDL cholesterol, high blood pressure and body mass index, low bone mineral density, and kidney dysfunction) also play significant roles in the development of BC. Figure 11 illustrates the statistics of BC deaths linked to various risk factors, showing that, from 1990 to 2021, tobacco contributed to 3.92% of female and 0.88% of male deaths; dietary factors accounted for 11.87% of male and 12.62% of female deaths, while metabolic risk factors were responsible for 10.48% of female deaths but none for males among all BC patients [62,63].
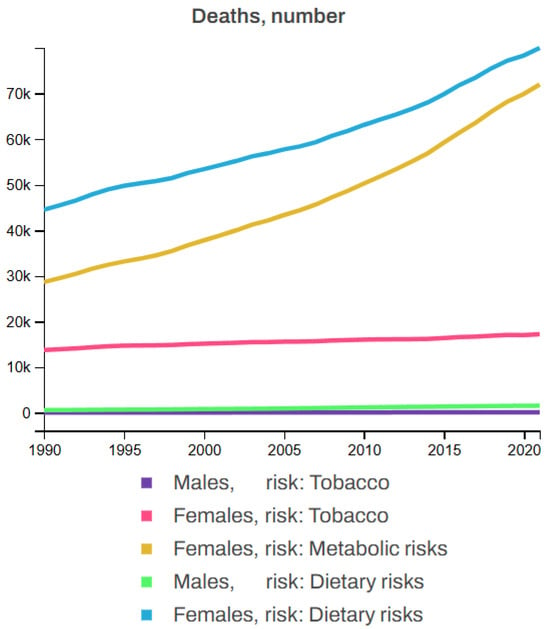
Figure 11.
Global breast cancer mortality by risk factor (1990–2021) [63].
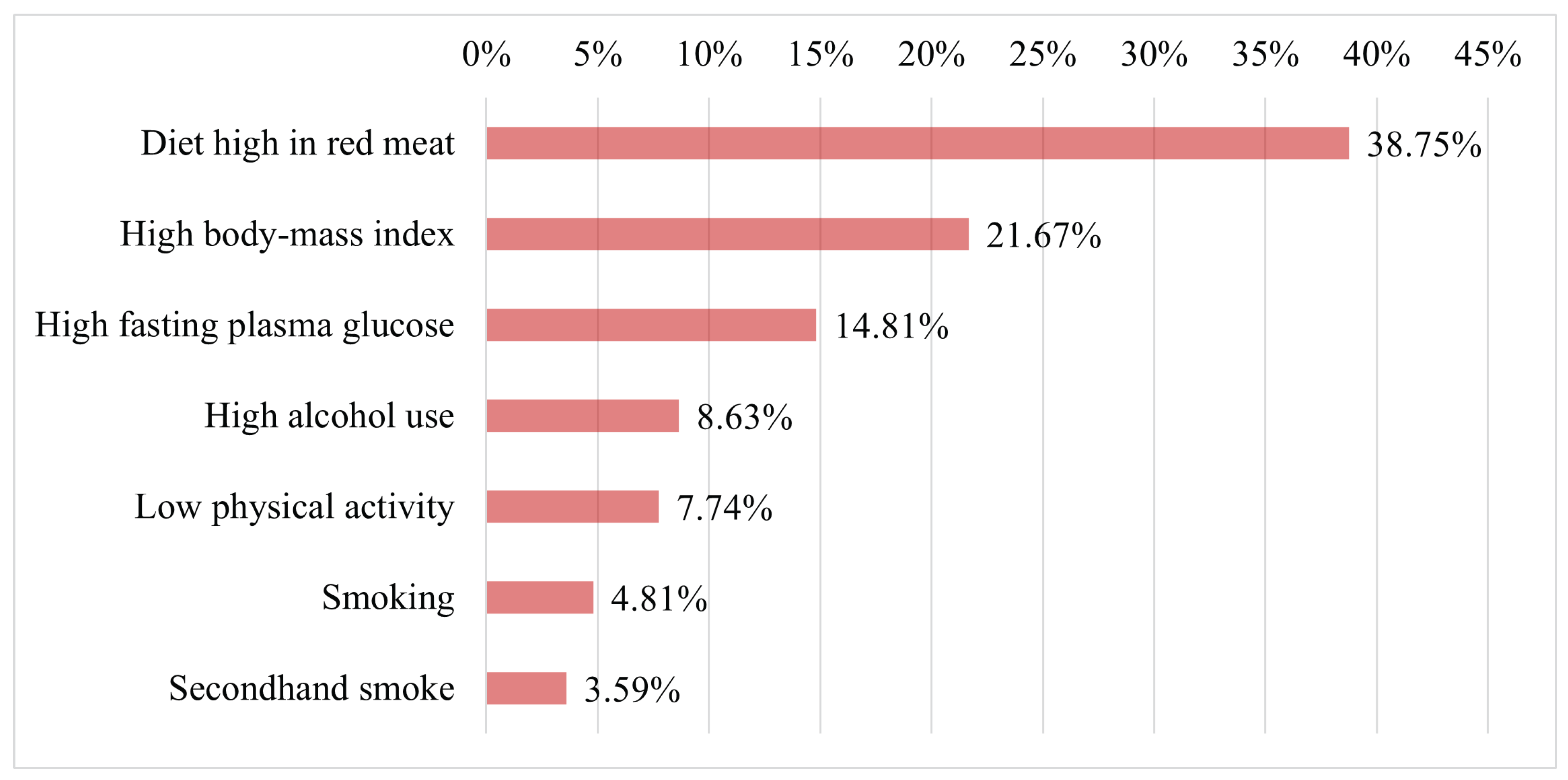
Figure 12 presents the seven key factors associated with the occurrence of breast cancer in females of all ages in 2021. The figure highlights that high consumption of red meat was the leading factor associated with BC, followed by high body mass index and high fasting plasma glucose as the second and third most significant factors, respectively. Furthermore, the figure shows that other critical factors contributing to BC risk include high alcohol use, low physical activity, smoking, and exposure to secondhand smoke. Therefore, avoiding these risk factors could significantly reduce the likelihood of developing BC in females.
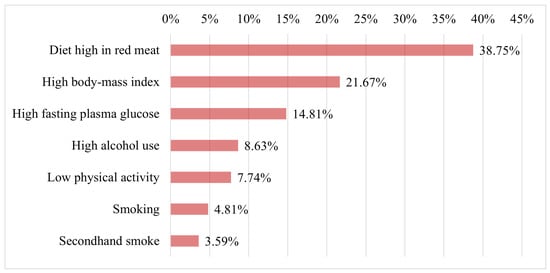
Figure 12.
Key afctors contributing to breast cancer risk in females in 2021 [62].
Beyond hormonal and lifestyle factors, genetic mutations significantly influence breast cancer risk. BRCA1/2 mutations impair DNA repair, conferring up to 65% lifetime risk and association with triple-negative subtypes. TP53 and PTEN mutations dysregulate cell-cycle control and PI3K signaling, respectively, while PROC variants may promote metastatic survival via coagulation pathways [67,68].
8. Breast Cancer Detection Approaches
The diagnosis of breast cancer employs both traditional methods, such as imaging (mammograms, ultrasounds, and MRIs) and biopsies, as well as advanced computational techniques like machine learning and deep learning (DL). Deep learning, particularly methods using convolutional neural networks (CNNs), has shown exceptional performance in analyzing medical images and tissue samples. Emerging approaches, including liquid biopsies, radiomics, and AI-powered multimodal systems, are further transforming the accuracy and efficiency of BC diagnosis.
8.1. Mammogram Image-Based Breast Cancer Detection
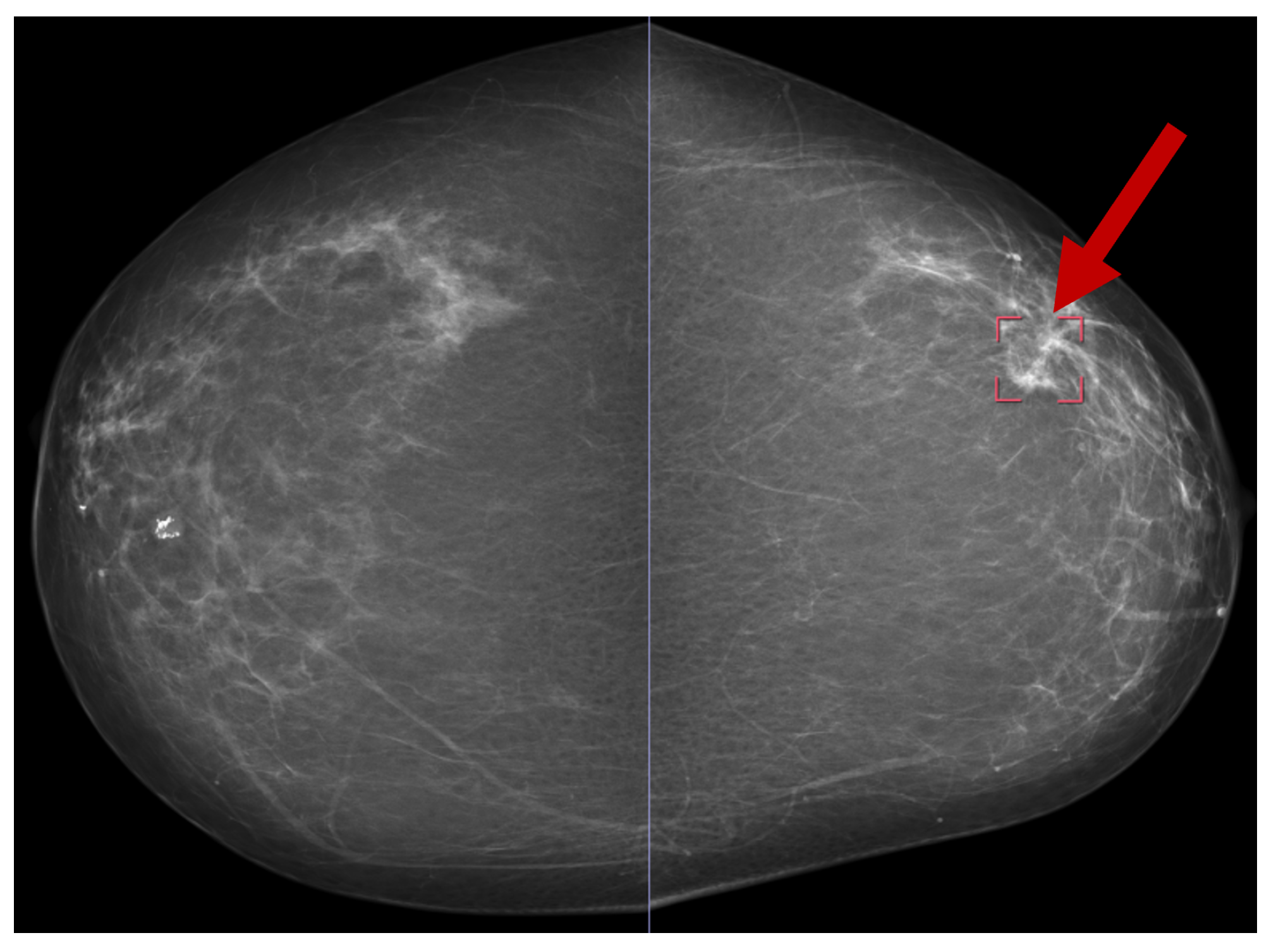
Several publicly available datasets are widely used for mammogram image-based BC detection, including the Curated Breast Imaging Subset of DDSM (CBIS-DDSM) [69], the Digital Database for Screening Mammography (DDSM) [70], the Mammographic Image Analysis Society (MIAS) dataset [71], and the INbreast dataset [30]. Figure 13 shows a sample mammogram image with a marked areaindicated by the red square box on the right side highlighting a potential abnormality for further analysis.
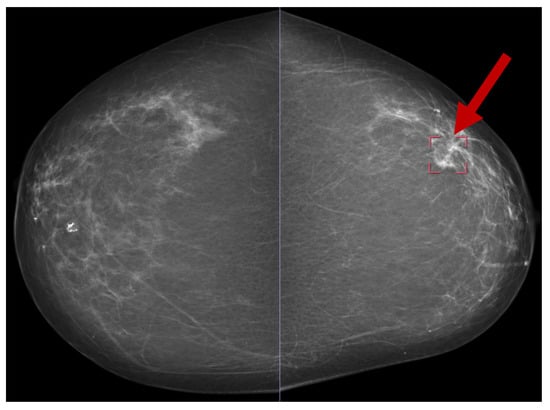
Figure 13.
A sample mammogram image [72].
The approach proposed by Zhao et al. involved identifying eligible patients, preparing image data, extracting labels and BI-RADS scores, and applying AlexNet, VGG, ResNet, and DenseNet models, with DenseNet achieving the best performance. The evaluation was conducted under three settings: General (BI-RADS 1–5), False Positive Reduction (BI-RADS 4–5), and Difficult Cases (BI-RADS 0) [73]. Jamil et al. proposed a BC detection technique using mammography images, achieving an accuracy of 0.9719. Their approach applied three techniques in sequence: Log Ratio, Gabor Filter, and Fuzzy C-Means (FCM) [74]. Nemade et al. proposed an ensemble-based model for BC classification, utilizing VGG16, InceptionV3, and VGG19 as base learners, with an artificial neural network for classification. The model achieved an accuracy of 98.10%, a specificity of 99.12%, and a sensitivity of 97.01% [75].
Another study conducted five experiments using different preprocessing techniques. The researchers applied six classifiers—SVM, Random Forest, ANN, KNN, Naive Bayes, and Decision Tree—and achieved 100% for all metrics (accuracy, sensitivity, specificity, and F1-score) with SVM and ANN. This was achieved when preprocessing was performed using contrast limited adaptive histogram equalization (CLAHE) and unsharp masking (USM) on the mini-MIAS dataset [76].
Yaqub et al. developed a two-stage system for BC diagnosis using mammogram images, where the first stage involves image segmentation through the Atrous Convolution-based Attentive and Adaptive Trans-Res-UNet (ACA-ATRUNet) model, and the second stage utilizes the Atrous Convolution-based Attentive and Adaptive Multi-scale DenseNet (ACA-AMDN) model for cancer detection, with hyperparameter optimization carried out using the Modified Mussel Length-based Eurasian Oystercatcher Optimization (MML-EOO) algorithm. They achieved an accuracy of 89.13%, a specificity of 0.8937, a precision of 0.8226, and an F1-score of 0.8536 for the MIAS mammography dataset, and accuracy, precision, and F1-score values of 89.061%, 80.348%, and 84.424%, respectively, for the CBIS-DDSM BC image dataset [77]. In [78], EfficientNet achieved the highest accuracy of 0.9829 on the MaMaTT2 dataset. For the DDSM dataset, a CNN classifier with different fine-tuning achieved an accuracy of 0.9996, a sensitivity of 1.0, a precision of 0.9992, and an F1-score of 0.9996 [79]. Additionally, the Inception Recurrent Residual Convolutional Neural Network (IRRCNN) achieved an accuracy of 0.9676 on the BreakHis dataset (factor ×40) and 0.9711 on the BC Classification Challenge 2015 dataset [10].
Table 5 summarizes recently published research on mammography image-based studies.

Table 5.
Comparison of recent mammography-based breast cancer detection techniques.
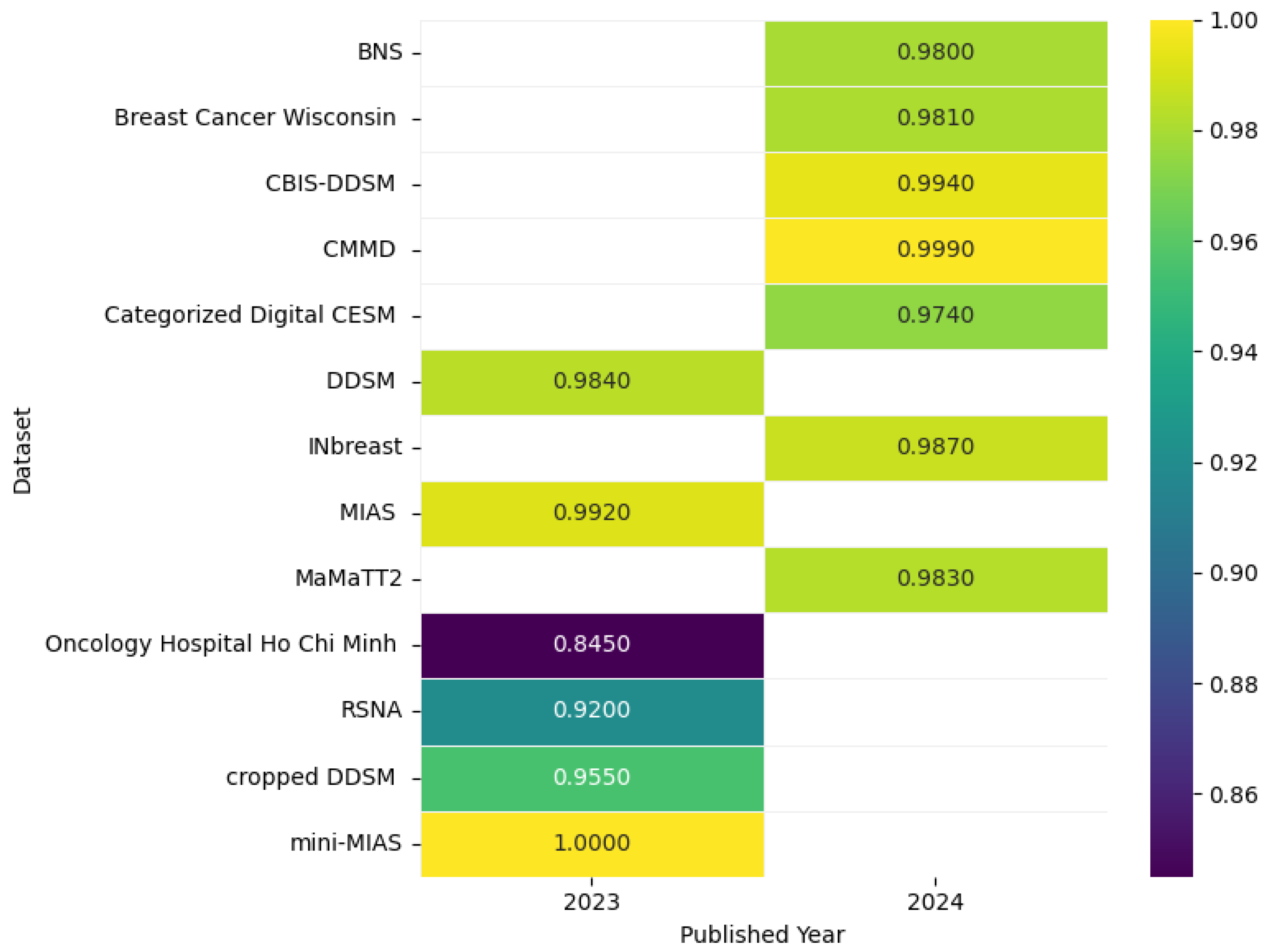
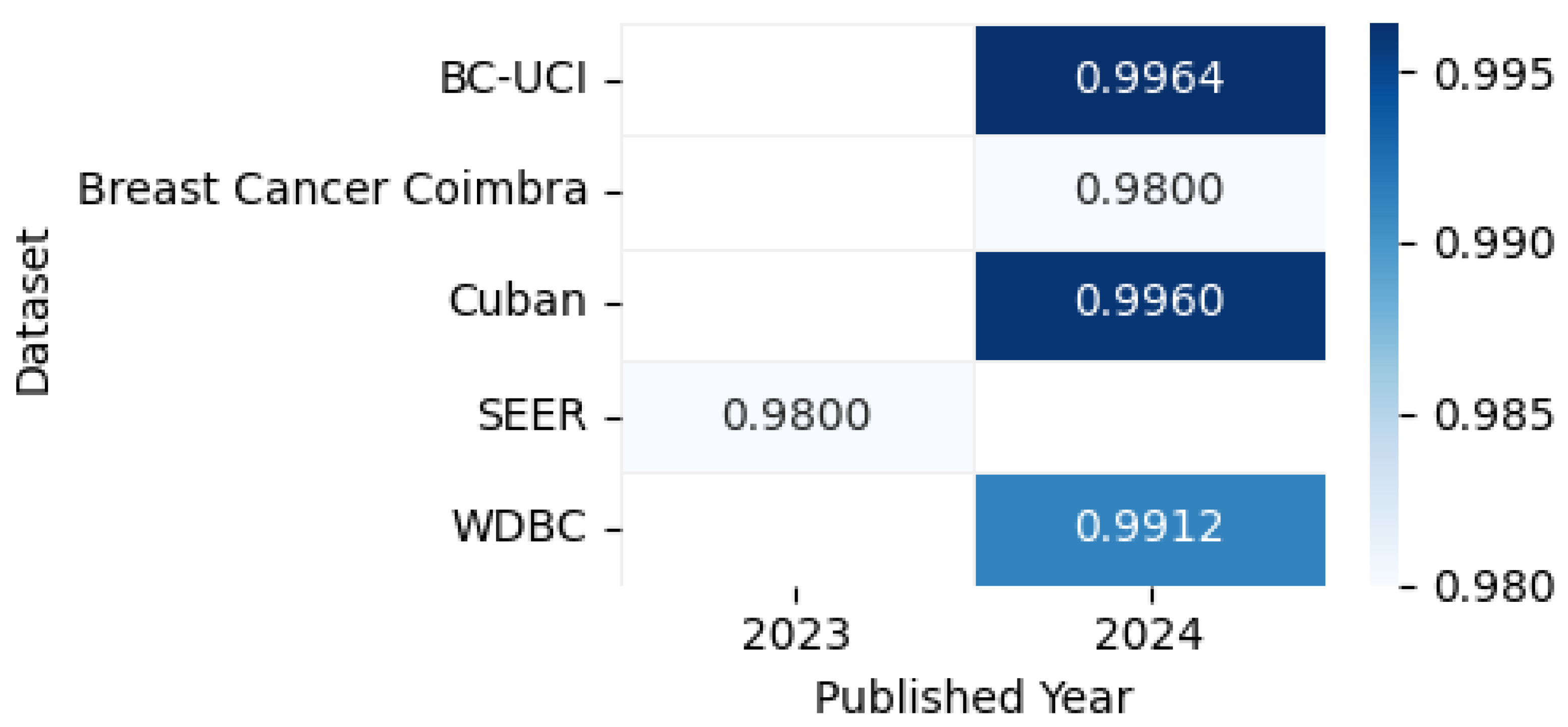
Figure 14 shows the highest accuracies obtained by different models applied to mammogram datasets in 2023 and 2024, including CBIS-DDSM [80], CMMD [80], DDSM [81], INbreast [82], MIAS [81], mini-MIAS [76], Breast Cancer Wisconsin [83], Categorized Digital CESM [84], Oncology Hospital Ho Chi Minh [85], Cropped DDSM [86], MaMaTT2 [78], RSNA [87], and BNS [88], as reported in the articles, while the accompanying correlation figure uses color intensity to represent accuracy levels.
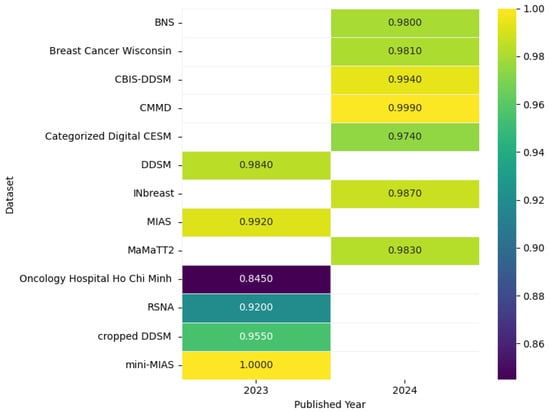
Figure 14.
Highest detection accuracy across multiple mammogram datasets for breast cancer.
8.1.1. Model Performance on Mammogram Image: Pros, Cons, and Future Directions
Table 6 provides an overview of the key benefits, limitations, and future research directions for leading mammogram-based breast cancer detection models, enabling easy comparison of their capabilities and areas for improvement.

Table 6.
Comprehensive overview of top-performing models on mammogram breast cancer datasets.
8.1.2. Summary
Several publicly available datasets, such as CBIS-DDSM, DDSM, MIAS, and INbreast, have been widely used for breast cancer (BC) detection from mammogram images. Recent studies have employed advanced machine learning and deep learning techniques, including DenseNet, VGG16, AlexNet, and InceptionV3, to achieve high accuracy and specificity in BC detection. Preprocessing methods like contrast limited adaptive histogram equalization (CLAHE) and unsharp masking (USM) have been utilized for image enhancement, while ensemble models and hybrid systems combining deep learning with traditional image processing techniques have also shown promising results. The highest reported accuracy reached 99.96% on the DDSM dataset. Table 7 presents a summary of key findings in mammogram-based breast cancer detection.

Table 7.
Summary of key findings in mammogram-based BC detection.
8.1.3. Analysis
The application of deep learning models, particularly CNNs and ensemble models, has significantly improved BC detection performance. Preprocessing techniques like CLAHE and USM, combined with classifiers such as SVM and ANN, have enabled researchers to achieve near-perfect metrics. Advanced architectures like DenseNet and EfficientNet have demonstrated robust performance across multiple datasets, including DDSM, MIAS, and CBIS-DDSM. Hybrid systems, such as those combining deep learning with traditional image processing techniques (e.g., Log Ratio, Gabor Filter, and FCM), have also shown remarkable accuracy. Table 8 provides an analysis of techniques and their impact based on mammogram-based BC detection.

Table 8.
Analysis of techniques and their impact based on mammogram-based BC detection.
8.1.4. Critique
While current models demonstrate impressive performance, several limitations need addressing. First, the interpretability of these models remains a challenge, which is critical for clinical acceptance. Second, the generalization ability of models across diverse datasets and imaging conditions is often untested, raising concerns about their real-world applicability. Additionally, the lack of diversity in datasets (e.g., limited representation of different populations and imaging devices) may lead to overfitting and reduced robustness. Table 9 presents a critique of current limitations.

Table 9.
Critique of current limitations based on mammogram-based BC detection.
8.1.5. Comparison
The results of various studies highlight differing approaches and outcomes. For instance, Yaqub et al. (2024) [77] achieved 89.13% accuracy on the MIAS dataset using a two-stage system, while Nemade et al. (2024) [75] reported 98.10% accuracy with an ensemble model. Avci et al. (2023) [76] demonstrated the importance of preprocessing, achieving 100% accuracy with CLAHE, USM, SVM, and ANN. EfficientNet, as used by Dada et al. (2024) [78], outperformed other models with 98.29% accuracy on the MaMaTT2 dataset. These comparisons underscore the importance of dataset quality, preprocessing, and algorithm choice in achieving high detection accuracy. Table 10 provides a comparison of key studies.

Table 10.
Comparison of key studies based on mammogram-based BC detection.
The advancements in mammogram-based BC detection are promising, with deep learning models and preprocessing techniques driving significant improvements in accuracy and performance. However, challenges related to interpretability, generalization, and dataset diversity must be addressed to ensure these models are clinically viable and applicable across diverse populations and imaging conditions. Future research should focus on developing more interpretable models, expanding dataset diversity, and testing models in real-world clinical settings.
While mammography models achieve >99% accuracy on curated datasets (Table 5), two critical gaps persist: (1) performance drops significantly on dense breast tissue, (2) most studies lack real-world testing with radiologist workflows.
8.2. Ultrasound Image-Based Breast Cancer Detection
Ultrasound images are used to visualize internal structures through high-frequency sound waves. There are several publicly available datasets for ultrasound image-based BC detection, including the Breast Ultrasound Images (BUSI) dataset [34], BUS-BRA [89], BUS-UCLM [90], and Breast-Lesions-USG [91]. Figure 15 presents a sample ultrasound image with an array-marked region, highlighting a potential abnormality for further examination.
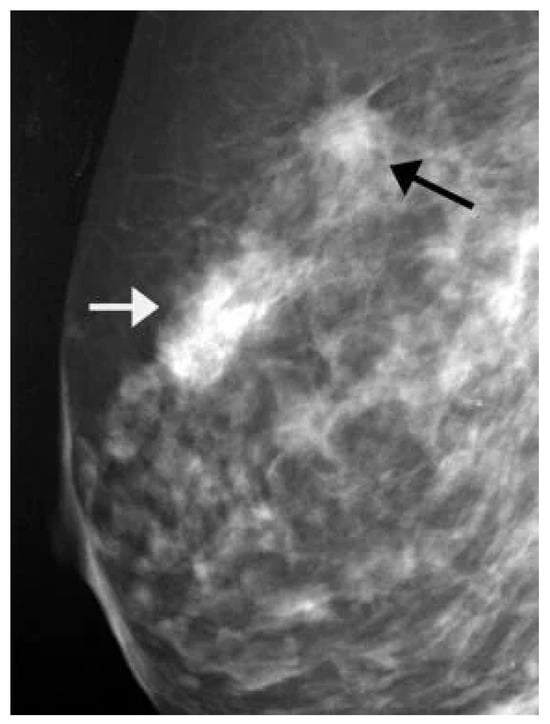
Figure 15.
A sample ultrasound image [92].
The BUSI dataset was created in 2018 and includes 780 images from 600 female patients aged 25–75 categorized as normal, benign, or malignant, with PNG images averaging pixels and ground truth masks. The BUS-BRA dataset features 1875 anonymized images from 1064 patients in Brazil, including biopsy-proven cases (722 benign and 342 malignant), BI-RADS assessments, manual segmentations, and cross-validation partitions to standardize CAD system evaluations. The BUS-UCLM dataset, collected between 2022 and 2023, contains 683 images with RGB segmentation masks for normal, benign, and malignant cases, enabling the development of machine learning models for the detection and diagnosis of breast lesions. Lastly, the Breast-Lesions-USG dataset offers 256 scans with annotated tumors, BIRADS classifications, and histopathological diagnoses, serving as an external testing set and providing support for AI development in BC research.
Mehedi et al. introduced a DL strategy for BC detection using ultrasound images, based on a publicly available breast ultrasound image dataset from Rodrigues [93] that includes 250 images (100 benign and 150 malignant cases). They incorporated Grad-CAM and occlusion mapping to evaluate feature extraction and developed a custom CNN model with fewer parameters for improved efficiency. For classification, they applied DenseNet201, ResNet50, and VGG16 models, achieving 100% accuracy after fine-tuning using this dataset. Specifically, DenseNet201 and ResNet50 performed best with the Adam and RMSprop optimizers, while VGG16 reached 100% accuracy using Stochastic Gradient Descent. The original images with varying sizes (57 × 75 to 161 × 199) were resized to 224 × 224, 227 × 227, and 299 × 299 for different pre-trained models [94].
The DeepBreastCancerNet model consists of 24 layers, with six convolutional layers, nine inception modules, and a fully connected layer. The model achieved 99.35% accuracy on the initial ultrasound dataset [34] and 99.63% accuracy on a second publicly available dataset [93], demonstrating its robustness. By applying transfer learning, a method that utilizes pre-trained models to avoid overfitting, the researchers improved detection accuracy, surpassing state-of-the-art models in BC classification. The model incorporates clipped ReLU and leaky ReLU activation functions, along with batch and cross-channel normalization, that enhanced its performance on both datasets [95].
A study of 603 patients from three institutions (2018–2021) trained and validated four DCNNs on ultrasound images (420 training, 183 validation) to predict the responses to NAC. ResNet50, the best model using only images, achieved an AUC of 0.879 and 82.5% accuracy. When combined with clinical–pathologic variables, the integrated DLR model outperformed both the image-based and clinical models, achieving AUCs of 0.962 and 0.939 in training and validation, surpassing radiologists’ predictions and improving their accuracy as a support tool [96]. The study introduces a novel ROI-free system for BC diagnosis using ultrasound images that incorporates prior anatomical knowledge of the spatial relationships between malignant and benign tumors, captured through the HoVer-Transformer. This model, which extracts inter- and intra-layer spatial information both horizontally and vertically, was evaluated on the GDPH&SYSUCC dataset and compared to four CNN-based and three vision transformer models. The proposed model achieved state-of-the-art results, with an AUC of 0.924, an accuracy of 0.893, a specificity of 0.836, and a sensitivity of 0.926, surpassing two senior sonographers in BC diagnosis [97].
Madhusudan et al. presented a method for BC detection using ultrasound images (USIs) [98], combining transfer learning (TL) models (MobileNetV2, ResNet50, VGG16) with LSTM for feature extraction and SMOTETomek for feature balancing. The method with VGG16 achieved an F1 score of 99.0%, MCC and kappa coefficients of 98.9%, and an AUC of 1.0. Using K-fold cross-validation, the same method achieved an average F1-score of 96%. Grad-CAM and LIME were applied for visualization and interpretability, and confidence intervals were calculated via NAI (LCI 96.50%, UCI 99.75%, MCI 98.13%) and bootstrapping (LCI 93.81%, UCI 96.00%, MCI 94.90%) [99]. Rao et al. proposed Inception V3 + Stacking, a model that combined transfer learning with ensemble stacking of ML models (including MLP and SVM with RBF and polynomial kernels) and applied the model to the ultrasound breast cancer (USBC) image dataset. Their model achieved excellent results, with an AUC of 0.947 and an accuracy of 0.858, outperforming existing diagnostic systems [100].
Appiah et al. explored the use of DL for classifying BC from ultrasound images, focusing on low- and middle-income countries (LMIC) with limited healthcare access. Their aim was to deploy a simple classifier on a mobile device using affordable handheld ultrasound systems to identify cases needing medical attention. Their model achieved 64% accuracy when trained from scratch and 78% accuracy after pre-training, highlighting DL’s potential to aid early BC detection with minimal image data [101]. Table 11 provides a summary of recent research on ultrasound image-based studies.

Table 11.
Comparison of recent ultrasound-based breast cancer detection techniques.
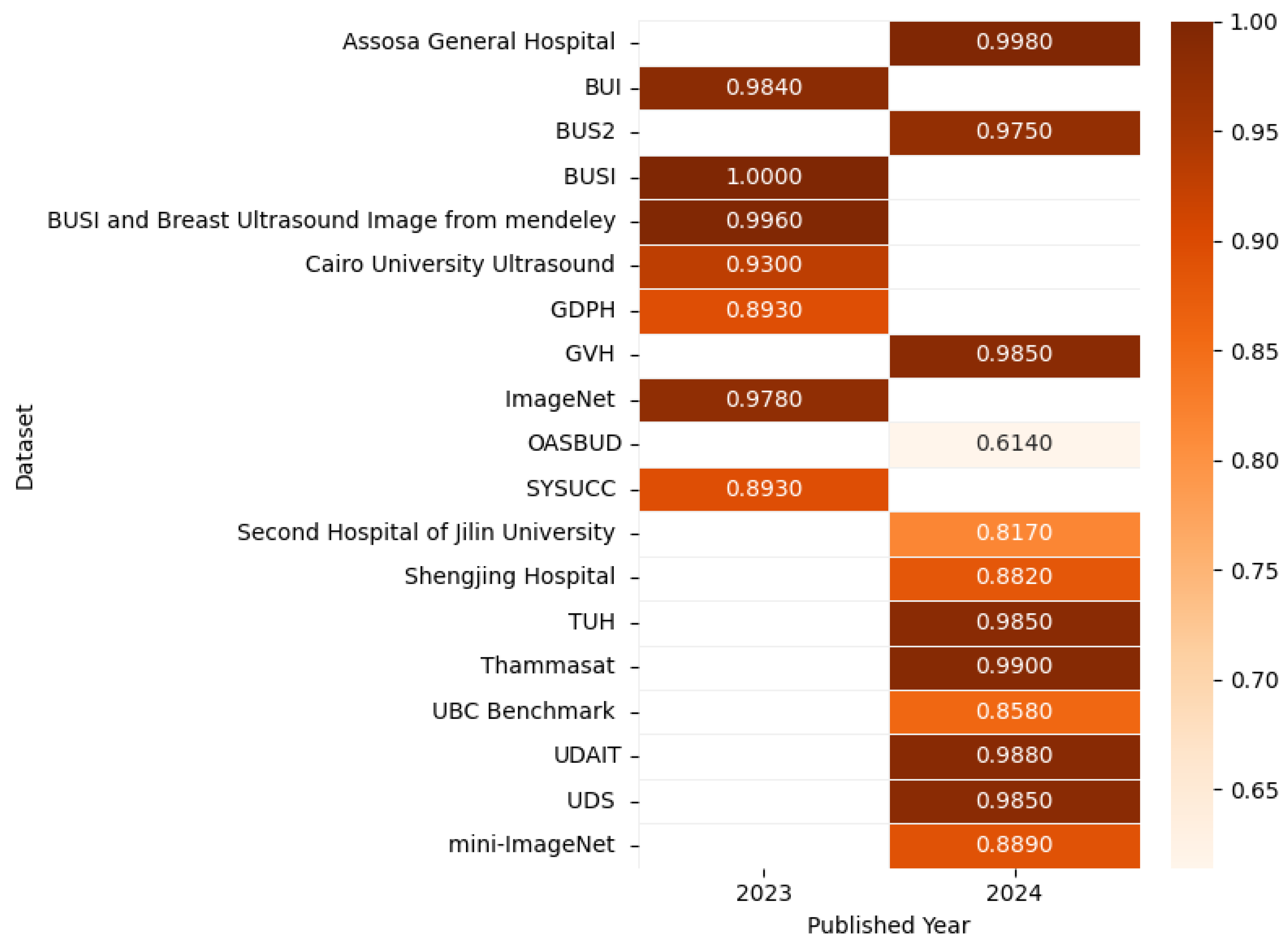
Figure 16 presents the highest accuracy rates achieved by various models on ultrasound datasets in 2023 and 2024, including UBC Benchmark [100], BUSI [102], BUS2 [103], UDAIT [104], multiple hospital datasets, and others [95,104,105,106,107,108,109,110,111,112,113,114], as outlined in the referenced studies, with the corresponding correlation figure using color intensity to depict accuracy levels.
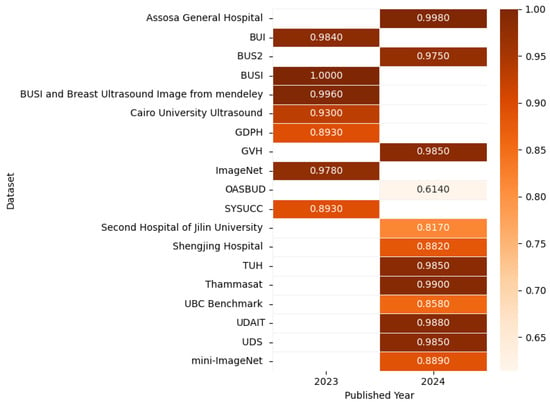
Figure 16.
Highest detection accuracy across multiple ultrasound datasets for breast cancer.
8.2.1. Model Performance on Ultrasound Image: Pros, Cons, and Future Directions
Table 12 summarizes the advantages, disadvantages, and future work recommendations for top-performing models in ultrasound-based breast cancer detection, facilitating direct comparison of their strengths and development needs.

Table 12.
Comprehensive overview of top-performing models on ultrasound breast cancer datasets.
8.2.2. Summary
Ultrasound imaging is a widely used modality for breast cancer (BC) detection, supported by publicly available datasets such as BUSI, BUS-BRA, BUS-UCLM, and Breast-Lesions-USG. Recent studies have employed advanced deep learning (DL) models, including CNNs, transfer learning, and hybrid approaches, to achieve high accuracy and AUC scores. Notable models like DeepBreastCancerNet, HoVer-Transformer, and VGG16-LSTM have demonstrated exceptional performance. Additionally, interpretability techniques such as Grad-CAM and LIME have been integrated to enhance clinical applicability. Despite these advancements, challenges remain in standardizing image preprocessing, improving dataset diversity, and enabling real-world deployment. Table 13 presents a summary of key findings in ultrasound-based breast cancer detection.

Table 13.
Summary of key findings in ultrasound-based BC detection.
8.2.3. Analysis
The studies reviewed demonstrate the effectiveness of DL models, particularly those leveraging transfer learning and ensemble techniques, in classifying BC from ultrasound images. Models like ResNet50, DenseNet201, and VGG16 achieve near-perfect accuracy when fine-tuned, with some studies reporting 100% accuracy. The integration of clinical–pathologic variables further enhances predictive performance, as seen in the integrated DLR model, which achieved an AUC of 0.962. However, dataset limitations, such as small sample sizes and inconsistent imaging protocols, hinder model generalizability. Additionally, the focus on real-time deployment in resource-limited settings highlights the need for computationally efficient models. Table 14 provides an analysis of techniques and their impact based on ultrasound-based breast cancer detection.

Table 14.
Analysis of techniques and their impact in ultrasound-based BC detection.
8.2.4. Critique
While the reported accuracies are impressive, several limitations warrant attention. First, the lack of cross-dataset validation raises concerns about model overfitting and generalizability. Many studies rely on a single dataset, which may not represent diverse populations or imaging conditions. Second, although interpretability techniques like Grad-CAM provide insights into model decisions, they are often insufficient for full clinical trust. Third, models achieving near-100% accuracy may indicate potential biases or dataset-specific overfitting, necessitating independent validation. Finally, the absence of standardized imaging protocols and the limited diversity of datasets hinder the development of robust models. Table 15 presents a critique of current limitations in ultrasound-based breast cancer detection.

Table 15.
Critique of current limitations in ultrasound-based breast cancer detection.
8.2.5. Comparison
The reviewed studies demonstrate varying approaches and outcomes, as summarized in Table 16. DeepBreastCancerNet [95] achieved the highest accuracy (99.63%) on a secondary dataset, followed by HoVer-Transformer (AUC 0.924) and VGG16-LSTM (AUC 1.0, F1-score 99%). The integrated DLR model outperformed image-only approaches by incorporating clinical features, achieving an AUC of 0.962. In contrast, simpler models designed for mobile devices, such as the one proposed by Appiah et al., achieved lower accuracy (78%), highlighting the trade-off between computational efficiency and predictive performance [100]. These comparisons underscore the importance of dataset quality, preprocessing, and algorithm choice in achieving high detection accuracy.

Table 16.
Comparison of key studies in ultrasound-based breast cancer detection.
Ultrasound-based BC detection has seen significant advancements through the application of deep learning models, transfer learning, and hybrid approaches. While models like DeepBreastCancerNet and HoVer-Transformer demonstrate exceptional performance, challenges related to dataset diversity, interpretability, and real-world deployment remain. Future research should focus on cross-dataset validation, standardized imaging protocols, and the development of computationally efficient models for resource-limited settings. Addressing these challenges will enhance the clinical applicability and robustness of ultrasound-based BC detection systems.
Current ultrasound DL models exhibit two key limitations: (1) overreliance on small, single-center datasets (Table 11) leading to poor generalization, (2) lack of standardized evaluation metrics for clinically crucial tasks like tumor boundary delineation.
8.3. Breast Cancer Detection Based on Magnetic Resonance Imaging (MRI) Images
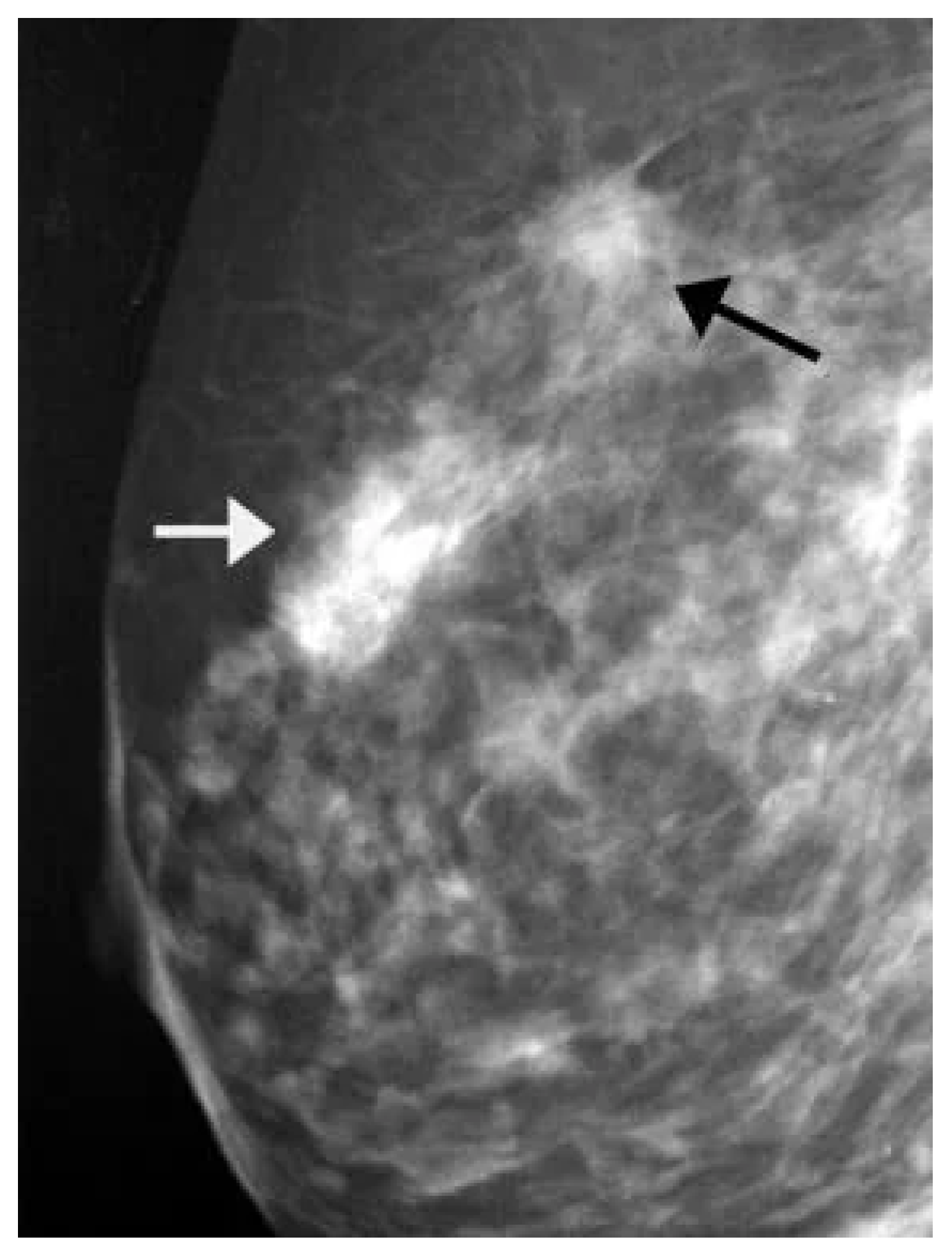
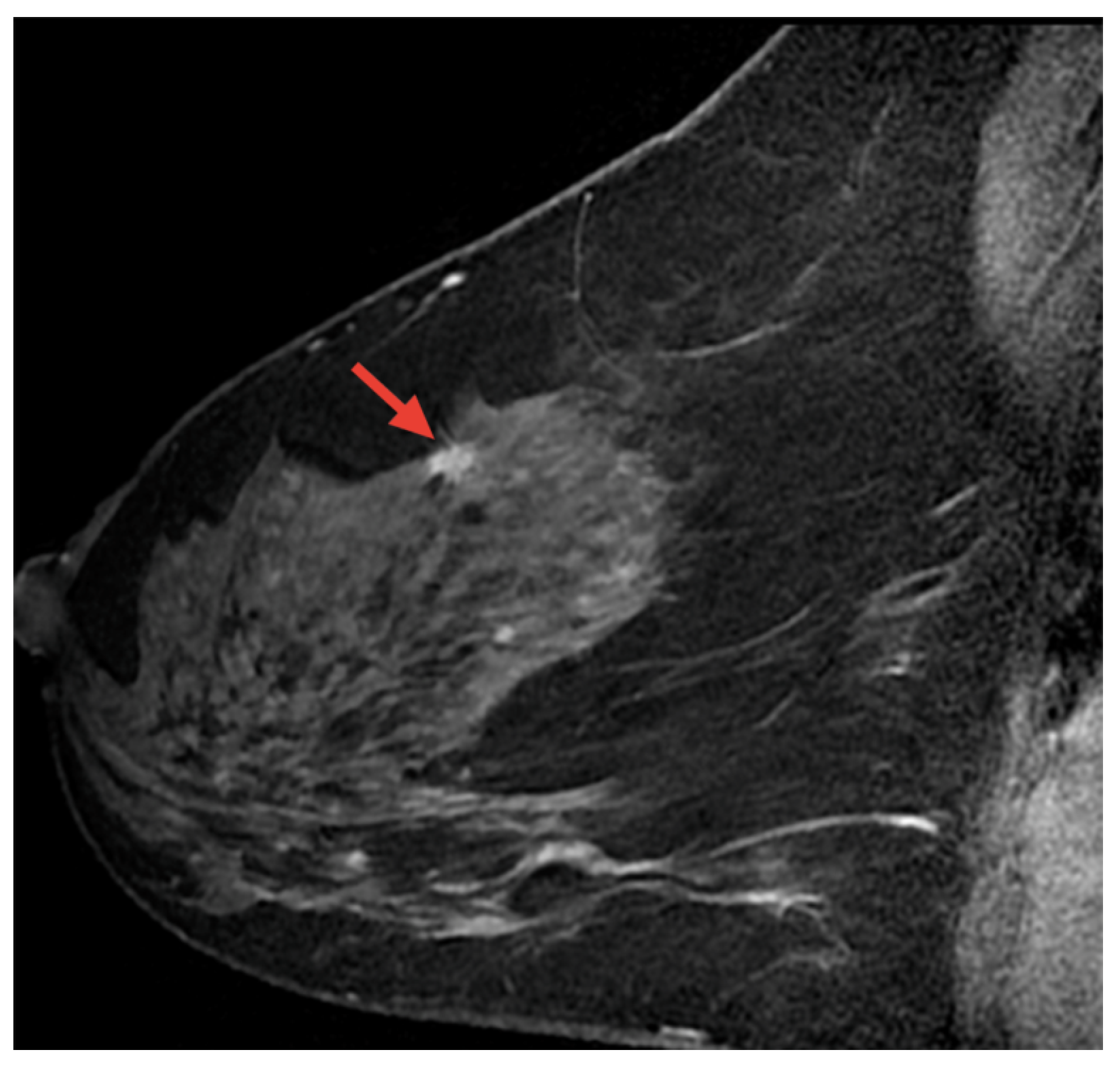
Magnetic resonance imaging (MRI) is a safe, non-invasive technique that uses magnetic fields and radio waves to produce detailed internal body images. MRI is employed in the diagnosis and monitoring of conditions such as soft tissue abnormalities, tumors, and brain and spinal disorders. This section highlights AI-based methods for BC detection using MRI images [115]. Figure 17 displays a sample MRI image with a highlighted area, marked by the red arrow, indicating a potential abnormality for further examination.
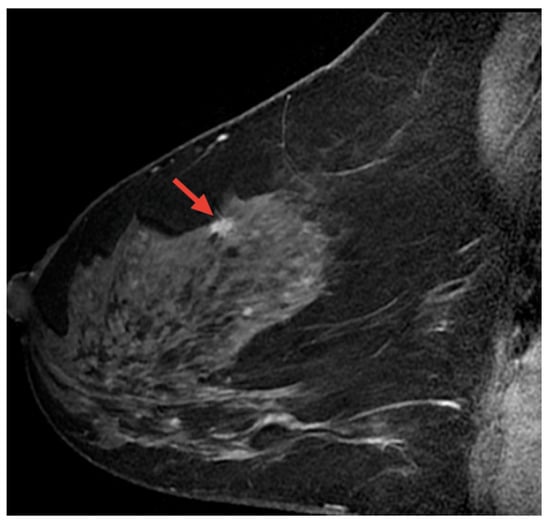
Figure 17.
A sample MRI image [92,116].
Zhang et al. proposed an approach that involved creating radiomics models for diagnosing BC using various MRI modalities (T1WI, T2WI, DWI, ADC, and DCE), with radiomic features extracted from plain, enhanced, and diffuse MRI scans and analyzed through DL for automatic extraction and classification. The diagnostic accuracy was significantly boosted by combining these modalities, with the highest diagnostic performance reflected in an AUC of 0.927 from the combined use of plain scan, enhanced, and diffuse sequences [117]. Liu et al. conducted an IRB-approved study utilizing a weakly supervised ResNet-101-based network for classifying malignant and benign images from a dataset of 278,685 image slices from 438 patients. The slices were grouped into 92,895 three-channel images, with 85% used for training and validation. The Adam optimizer with a SoftMax score threshold of 0.5 was used to train the model. The model achieved an AUC of 0.92 (±0.03), an accuracy of 94.2% (±3.4), a sensitivity of 74.4% (±8.5), and a specificity of 95.3% (±3.3). These results highlight the model’s ability to distinguish among various cancer types [118].
Yunan et al. developed a CNN-based computer-aided diagnostic tool for BC using DCE-MRI images, incorporating tumor geometric and pharmacokinetic features for classification. In a study of 130 patients, 71 were malignant cases and 59 were benign cases. The model achieved an accuracy, precision, sensitivity, and AUC of 87.7%, 91.2%, 86.1%, and 91.2% (±4.0%), respectively, across five-fold testing. Confidence in classification was reinforced by prediction probability, feature heatmaps, and dynamic scan time points, minimizing misclassification [119]. In a study by Yue et al., 516 BC patients were analyzed using a 3D UNet-based automatic segmentation model to extract 1316 radiomics features from regions of interest. Eighteen radiomics methods combining feature selection and classifiers were employed for model selection, and the model achieved an average Dice similarity coefficient of 0.89. The radiomics models effectively predicted four molecular subtypes, with the best performance showing an AUC of 0.8623, an accuracy of 0.6596, a sensitivity of 0.6383, and a specificity of 0.8775. For specific subtypes, the AUC values ranged from 0.8676 to 0.9335, with specificity reaching 0.9865 [120]. Qian et al. used CIBERSORT to analyze immune cell infiltration in BC (BRCA) patients from the TCGA database, applying univariate and multivariate Cox regression to identify M2 macrophages as an independent prognostic factor (HR = 32.288) while also obtaining imaging data from the TCIA database to construct an MRI-based radiomics model by selecting key features via LASSO. The models developed included intratumoral, peritumoral, and combined types, with the peritumoral model showing the highest performance, achieving an accuracy of 0.773, a sensitivity of 0.727, and a specificity of 0.818 [121].
Guo et al. proposed an MRI-based deep learning radiomics (DLR) model to predict HER2-low-positive status in BC patients and evaluate its prognostic value. The model utilized features from traditional radiomics and deep semantic segmentation and achieved AUCs of 0.868 and 0.763 for distinguishing HER2-negative from HER2-overexpressing patients, and 0.855 and 0.750 for differentiating HER2-low-positive from HER2-zero patients in the training and validation cohorts, respectively [122]. The study by Chao et al. used 569 local cases and 125 external cases for lesion classification, employing T1-weighted, DCE-MRI, T2WI, and diffusion-weighted imaging. A CNN and LSTM cascaded network achieved AUCs of 0.98/0.91 and sensitivities of 0.96/0.83 in internal/external cohorts. The DL model outperformed radiologists without DCE-MRI (AUC 0.96 vs. 0.90), and lesion localization had sensitivities of 0.97/0.93 with DCE-MRI/T2WI [123]. Liang et al. proposed a DL model (MLP-radiomic) using MRI radiomics and clinical radiological features, achieving a high AUC of 0.896 in predicting lymphovascular invasion in BC patients and outperforming ML models [124]. Table 17 provides an overview of studies conducted using MRI image-based BC detection.

Table 17.
Comparison of recent MRI image-based techniques for breast cancer detection.
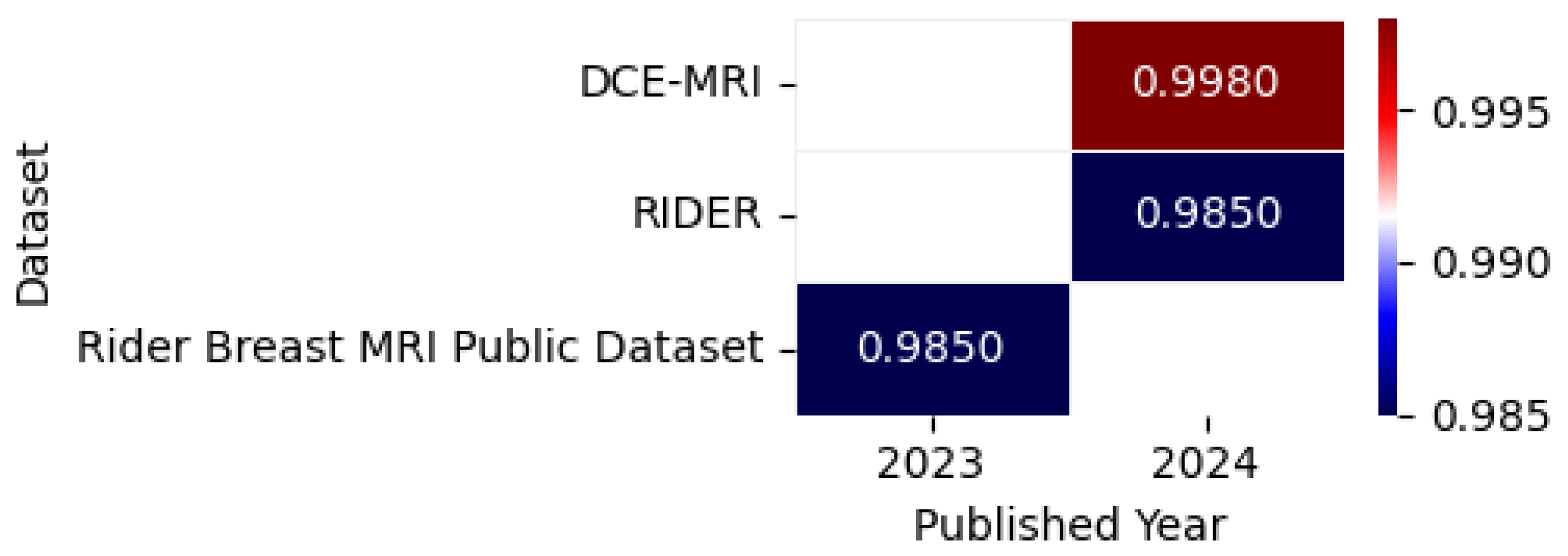
Figure 18 illustrates the highest accuracy rates obtained by various models on MRI datasets in 2023 and 2024, including datasets such as RIDER [125], DCE-MRI [126], and Rider Breast MRI Public Dataset [127], as detailed in the referenced studies, accompanied by a correlation figure that represents accuracy levels through color intensity.

Figure 18.
Highest detection accuracy across multiple MRI datasets for breast cancer.
8.3.1. Model Performance on MRI Image: Pros, Cons, and Future Directions
Table 18 provides an overview of the key benefits, limitations, and future research directions for leading MRI-based breast cancer detection models, enabling easy comparison of their capabilities and areas for improvement.

Table 18.
Comprehensive overview of top-performing models on MRI breast cancer datasets.
8.3.2. Summary
Magnetic resonance imaging (MRI) has emerged as a powerful tool for breast cancer (BC) detection, particularly in cases involving dense breast tissue where other imaging modalities may be less effective. Recent advancements in machine learning (ML) and deep learning (DL) have significantly enhanced the accuracy of MRI-based BC detection. Studies have utilized various MRI modalities, including T1-weighted, T2-weighted, diffusion-weighted imaging (DWI), apparent diffusion coefficient (ADC), and dynamic contrast-enhanced (DCE) MRI, to extract relevant features and detect cancerous lesions. These approaches have achieved impressive results, with models evaluated based on metrics such as accuracy, area under the curve (AUC), and Dice similarity coefficient. However, challenges related to dataset variability, model generalizability, and computational complexity remain. Table 19 presents a summary of key findings in MRI-based breast cancer detection.

Table 19.
Summary of key findings in MRI-based breast cancer detection.
8.3.3. Analysis
The integration of multimodal MRI approaches with deep learning techniques has significantly improved BC detection accuracy. Studies like Zhang et al. (2024) [112] demonstrate the advantage of combining multiple MRI modalities (T1WI, T2WI, DWI, ADC, and DCE) to capture complementary features, achieving an AUC of 0.927. Weakly supervised learning, as employed by Liu et al. (2022) [118], has shown promise in scenarios with limited annotated data, achieving an AUC of 0.92. CNN-based architectures, such as ResNet-101 and 3D UNet, have been widely used for classification and segmentation tasks, with notable performance improvements. Additionally, advanced models incorporating immune composition data, such as Qian et al. (2024) [121], have opened new avenues for predicting patient prognosis and cancer status. Table 20 provides an analysis of techniques and their impact on MRI-based breast cancer detection.

Table 20.
Analysis of techniques and their impact in MRI-based breast cancer detection.
8.3.4. Critique
Despite the impressive results, several limitations need to be addressed. First, the variability in performance across different datasets and cohorts raises concerns about model generalizability. Many models perform well on internal datasets but struggle with external validation, as seen in Chao et al. (2024) [123]. Second, the reliance on large annotated datasets remains a significant challenge, as acquiring such data is costly and time-consuming. Third, the computational complexity of advanced models, such as CNN-LSTM cascaded networks, may limit their feasibility in real-world clinical settings. Finally, while models achieve high-performance metrics, their lack of interpretability hinders clinical trust and adoption. Table 21 presents a critique of current limitations in MRI-based breast cancer detection.

Table 21.
Critique of current limitations in MRI-based breast cancer detection.
8.3.5. Comparison
The reviewed studies demonstrate varying approaches and outcomes, as summarized in Table 22. Chao et al. (2024) [123] achieved the highest AUC of 0.98 using a CNN-LSTM cascaded network, outperforming radiologists in lesion classification. Liu et al. (2022) [118] and Yue et al. (2023) [120] also reported strong performance, with AUC values of 0.92 and 0.86, respectively. The integration of radiomics features, as seen in Qian et al. (2024) [121], further enhanced predictive accuracy, achieving an AUC of 0.927. However, performance discrepancies in external validation datasets highlight the need for further research in model generalization. The table also highlights variations in model complexity, with simpler models like MLP-radiomics offering a balance between performance and computational efficiency.

Table 22.
Comparison of key studies in MRI-based breast cancer detection.
MRI-based breast cancer detection has seen significant advancements through the integration of deep learning and radiomics techniques. While models like CNN-LSTM and ResNet-101 demonstrate exceptional performance, challenges related to generalizability, dataset dependency, and computational complexity remain. Future research should focus on improving model interpretability, expanding dataset diversity, and developing computationally efficient models for real-world clinical deployment. Addressing these challenges will enhance the clinical applicability and robustness of MRI-based BC detection systems.
MRI-based models face unresolved challenges: (1) multi-institutional data variability degrades performance (Table 17), (2) most studies ignore computational costs prohibitive for clinical deployment, (3) temporal dynamics in DCE-MRI remain underutilized.
8.4. Breast Cancer Detection Using Computed Tomography (CT) Scan Images
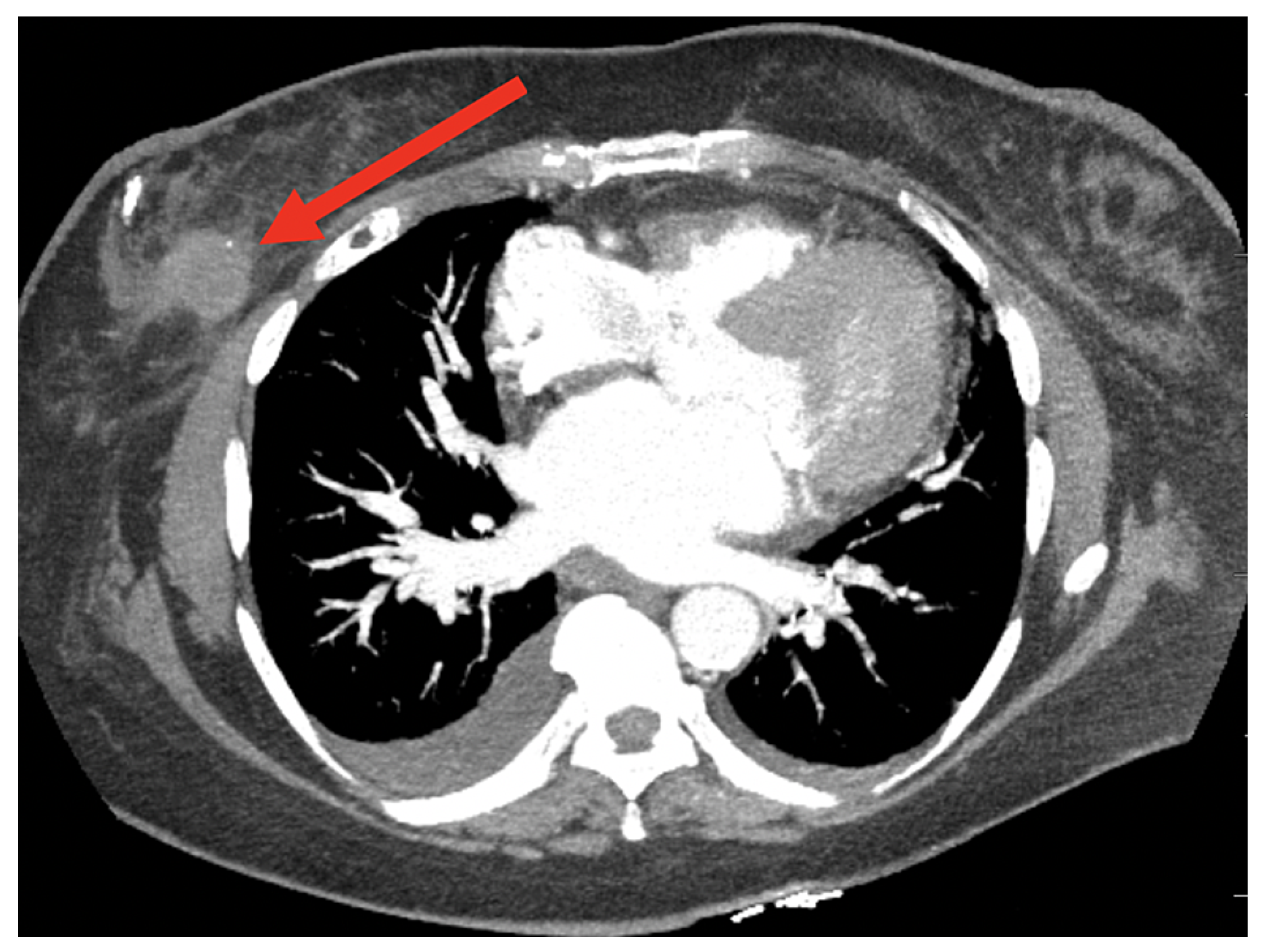
Computed tomography (CT) imaging, a diagnostic method using X-ray technology to produce detailed cross-sectional body images, facilitates the detection and monitoring of abnormalities such as tumors; this section examines AI-driven techniques for BC detection using CT images, with Figure 19 highlighting a region of interest for further analysis.
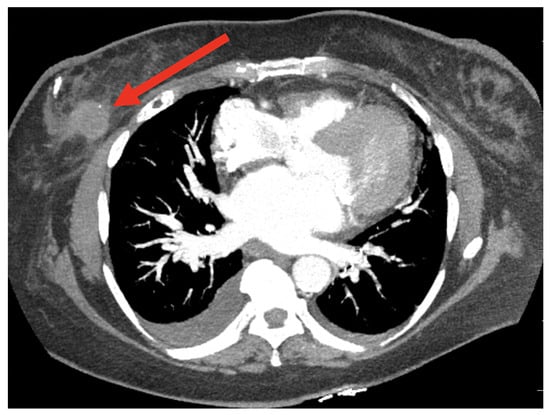
Figure 19.
A sample CT image [116].
Bargalló et al. [128] proposed a comprehensive CT imaging pipeline for BC that employs a trained CNN-Unet model to auto-contour standard tissue structures, thereby enabling accurate classification of patients by laterality and surgical procedures, such as lumpectomy or mastectomy, with a Random Forest classifier achieving 93.75% accuracy and an AUC of 0.993 in a secondary sample of 16 patients. While this method reduced target delineation errors and highlighted the need to consider anatomical and procedural differences, there remains scope for enhancing workflow efficiency. Koh et al. developed a DL model for detecting BC from chest CT scans using augmented axial images with bounding boxes and the RetinaNet algorithm. The model achieved 96.5% sensitivity in the internal test set and 96.1% in the external test set, with sensitivities ranging from 88.5% to 93.0% depending on the candidate probability threshold [129].
Yasak et al. conducted a study on BC detection using DL models applied to contrast-enhanced chest CT images. They used a dataset of 201 training, 26 validation, and 30 test cases and achieved an AUC of 0.976, thus improving the performance of radiologists through model assistance [130]. Table 23 presents a summary of research focused on BC detection using computed tomography (CT) images.

Table 23.
A comparison of recent techniques for breast cancer detection using computed tomography.
Figure 20 displays the highest accuracy rates achieved by various models on computed tomography datasets in 2023 and 2024, including the Breast Wellness Centre [131], Bone Metastases: Breast vs. Non-Breast Cancer [132], GE Healthcare [133], and datasets from hospitals in Pakistan [134], as outlined in the referenced studies, along with a correlation figure illustrating accuracy levels using varying color intensities.
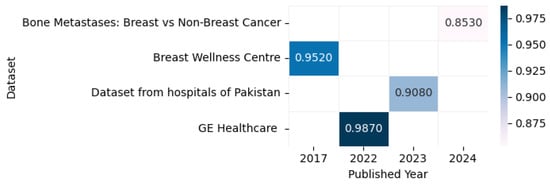
Figure 20.
Highest detection accuracy across multiple computed tomography datasets for breast cancer.
8.4.1. Model Performance on CT Image: Pros, Cons, and Future Directions
The comparative analysis in Table 24 highlights the main strengths, weaknesses, and research opportunities for current CT imaging approaches in breast cancer detection.

Table 24.
Comprehensive overview of top-performing models on CT breast cancer datasets.
8.4.2. Summary
Computed tomography (CT) imaging is a widely used diagnostic tool for breast cancer (BC) detection, offering detailed cross-sectional images of the body. Recent advancements in artificial intelligence (AI) and deep learning (DL) have significantly enhanced the accuracy of BC detection using CT images. Studies have employed various DL models, such as CNN-Unet and RetinaNet, to segment and classify breast cancer lesions with high accuracy. For example, Bargalló et al. (2023) [128] achieved 93.75% accuracy using a CNN-Unet model combined with a Random Forest classifier, while Koh et al. (2022) [129] reported 96.5% sensitivity using RetinaNet on augmented CT images. Despite these advancements, challenges such as dataset dependency, computational complexity, and generalizability remain. Table 25 presents a summary of key findings in CT-based breast cancer detection.

Table 25.
Summary of key findings in CT-based breast cancer detection.
8.4.3. Analysis
The integration of deep learning models with CT imaging has significantly improved BC detection accuracy. Bargalló et al. (2023) [128] proposed a comprehensive CT imaging pipeline using CNN-Unet for automatic segmentation and Random Forest for classification, achieving 93.75% accuracy and an AUC of 0.993. This approach reduces target delineation errors and highlights the importance of considering anatomical and procedural differences. Koh et al. (2022) [129] employed RetinaNet with augmented axial images, achieving 96.5% sensitivity in internal test sets and 96.1% in external test sets, demonstrating robust performance across different datasets. Yasak et al. (2024) [130] utilized deep learning on contrast-enhanced CT images, achieving an AUC of 0.976, which improved radiologists’ performance. These studies underscore the effectiveness of combining advanced CT imaging techniques with deep learning models for accurate BC detection. Table 26 provides an analysis of techniques and their impact in CT-based breast cancer detection.

Table 26.
Analysis of techniques and their impact in CT-based breast cancer detection.
8.4.4. Critique
Despite the promising results, several limitations need to be addressed. First, the reliance on high-quality, annotated CT image datasets limits scalability, as such datasets are not always available in clinical settings. Second, the performance of deep learning models can vary when applied to real-world data, as seen in Koh et al. (2022) [129], where sensitivity varied across test sets. Third, the computational complexity of advanced models, such as CNN-Unet and RetinaNet, may hinder their practical implementation in resource-limited environments. Finally, while automatic segmentation and procedural classification reduce human error, they require further refinement to improve workflow efficiency. The lack of generalizability across diverse datasets and clinical practices remains a significant challenge. Table 27 presents a critique of current limitations in CT-based breast cancer detection.

Table 27.
Critique of current limitations in CT-based breast cancer detection.
8.4.5. Comparison
The reviewed studies demonstrate varying approaches and outcomes, as summarized in Table 28. Bargalló et al. (2023) [128] achieved 93.75% accuracy using CNN-Unet and Random Forest, while Koh et al. (2022) [129] reported 96.5% sensitivity using RetinaNet. Yasak et al. (2024) [130] utilized deep learning on contrast-enhanced CT images, achieving an AUC of 0.976. These studies highlight the diversity of approaches in CT-based BC detection, with each method offering unique advantages. For instance, CNN-Unet excels in automatic segmentation, while RetinaNet performs well in lesion detection. However, performance discrepancies across datasets underscore the need for further research in model generalization.

Table 28.
Comparison of key studies in CT-based breast cancer detection.
CT-based breast cancer detection has seen significant advancements through the integration of deep learning models and advanced imaging techniques. While models like CNN-Unet and RetinaNet demonstrate exceptional performance, challenges related to dataset dependency, computational complexity, and generalizability remain. Future research should focus on improving model interpretability, expanding dataset diversity, and developing computationally efficient models for real-world clinical deployment. Addressing these challenges will enhance the clinical applicability and robustness of CT-based BC detection systems.
While CT-based models show promise (Table 28), two fundamental gaps limit clinical translation: (1) Radiation concerns—Most studies use standard-dose CT [133], ignoring the trade-offs between detection accuracy and patient safety in low-dose protocols; (2) Anatomical specificity—Models like CNN-Unet [128] focus on lesion detection but fail to incorporate breast density patterns that critically impact diagnostic interpretation.
8.5. Histopathological Image-Based Breast Cancer Detection
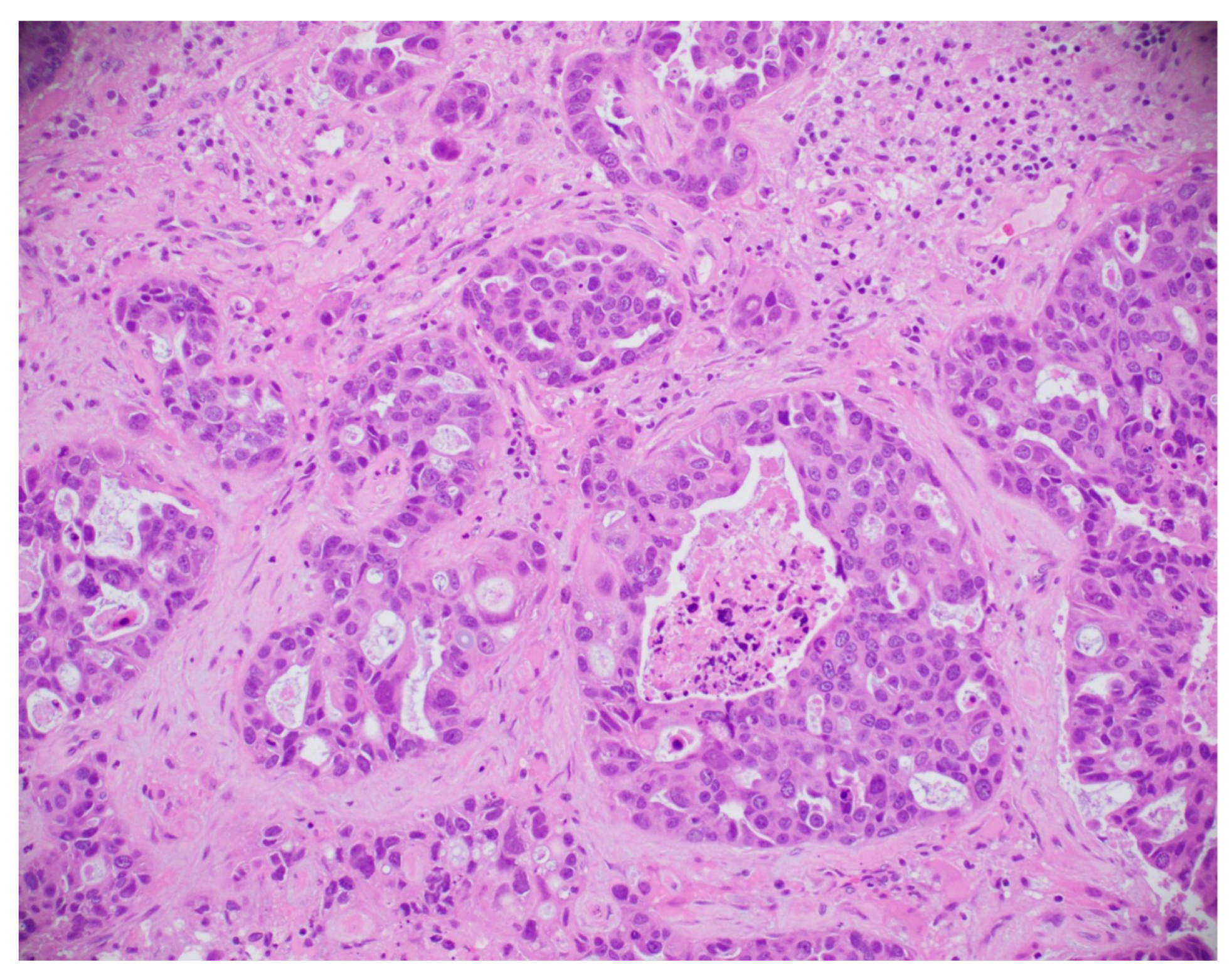
This subsection explores the application of histopathological and microscopic imaging, key diagnostic tools for BC detection that involve examining tissue samples at the cellular level to identify malignancies and support accurate diagnosis and treatment planning. A simple histopathological image is shown in Figure 21.
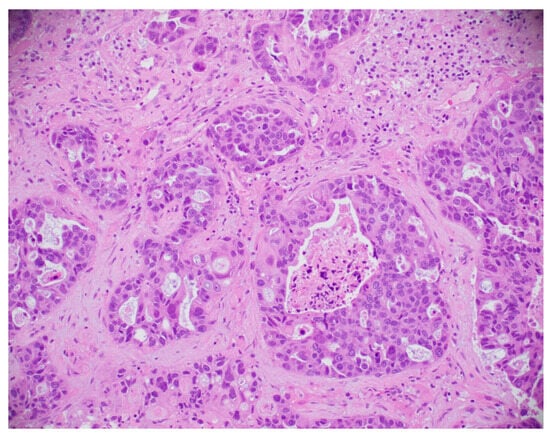
Figure 21.
A sample histopathological image [135].
Muntean and Chowkkar compared CNN and DenseNet121 models for BC detection using 7909 histopathological images. The CNN achieved 90.9% accuracy with 100× magnification, while DenseNet121 reached 86.6% accuracy, with a 16.4% accuracy improvement through transfer learning. Both models demonstrated effective performance [136]. Bhowal et al. [137] proposed a BC histology classification method that combined multiple DL models—VGG16, VGG19, Xception, Inception V3, and InceptionResnet V2—using Choquet Integral, Coalition Game Theory, and Information Theory for synthesis. The method incorporated image preprocessing, feature extraction, and classification, achieving improved accuracy, precision, and recall over traditional techniques. The dataset used for evaluation was from the BACH dataset, with an accuracy of 96% for the two-class problem and 95% for the four-class problem, outperforming state-of-the-art methods.
Yang et al. proposed a multimodal DL model to predict the prognosis of HER2-positive BC patients by integrating whole slide H&E images and clinical data. The model utilized a deep convolutional neural network (CNN) to extract features from 512 × 512-pixel H&E images that were combined with clinical information to assess the risks of relapse and metastasis. The model achieved an AUC of 0.76 in two-fold cross-validation and 0.72 in an independent TCGA testing set, demonstrating its effectiveness in predicting the prognosis of HER2-positive BC patients across different datasets [138].
Maleki et al. proposed a method for histopathological image classification using DL and transfer learning. The approach utilized pre-trained networks for feature extraction and then applied the XGBoost classifier. Evaluated on the BreakHis dataset, the proposed method achieved accuracies of 93.6%, 91.3%, 93.8%, and 89.1% for magnifications of 40×, 100×, 200×, and 400×, respectively. The DenseNet201 model was used for feature extraction, and XGBoost was employed as the final classifier [139]. Majumdar et al. proposed a rank-based ensemble method for BC detection from histopathological images, utilizing three CNN models: GoogleNet, VGG11, and MobileNetV3_Small. The decision scores from these models were combined using the Gamma function for a two-class classification problem. Their approach was evaluated on the BreakHis and ICIAR-2018 datasets, achieving classification accuracies of 99.16%, 98.24%, 98.67%, and 96.16% for 40×, 100×, 200×, and 400× magnification levels on the BreakHis dataset and 96.95% on the ICIAR-2018 dataset [140]. Ray et al. proposed the use of pre-trained deep transfer learning models, including ResNet50, ResNet101, VGG16, and VGG19, for the detection of BC using histopathological images. The study utilized a dataset of 2453 images, divided into invasive ductal carcinoma (IDC) and non-IDC categories. Among the models analyzed, ResNet50 showed superior performance, achieving an accuracy of 92.2%, an AUC of 91.0%, and a recall of 95.7%, with a minimal loss of 3.5% [141].
Rajkumar et al. proposed a method for BC detection using the Darknet-53 convolutional neural network (CNN) that enhanced the accuracy of image classification. They utilized the contrast-limited adaptive histogram equalization (CLAHE) technique for image preprocessing and the Haralick grey-level co-occurrence matrix (HGLCM) for feature extraction. The model achieved an accuracy of 95.6% [142]. Alhassan et al. proposed a comprehensive classification framework for differentiating types of lung, colon, and breast cancers by analyzing histopathological images using AI, machine learning, and digital image processing techniques. They utilized the BreakHis and LC25000 datasets and applied various preprocessing steps, including quadruple clipped adaptive histogram equalization for noise reduction and stain color adaptive normalization (SCAN) to enhance image contrast. The VGG16 model was used for feature extraction, followed by an ensemble max-voting classifier for classification. The results demonstrated strong performance, with 89.03% accuracy, 88.56% precision, 88.09% sensitivity, and an F1-score of 88.7%, highlighting the effectiveness of their approach for faster and more accurate cancer diagnoses [143]. The study used the BreaKHis and BACH datasets for histopathological BC image classification. The image sizes varied according to the magnification level, with images from 40×, 100×, 200×, and 400× magnifications used for evaluation. The proposed model was BCHI-CovNet, a lightweight AI model employing multiscale depth-wise separable convolution, an additional pooling module, and a multi-head self-attention mechanism to reduce computational complexity while maintaining high accuracy. The model achieved impressive results, with accuracies of 99.15% at 40×, 99.08% at 100×, 99.22% at 200×, and 98.87% at 400× magnification on the BreaKHis dataset, and 99.38% on the BACH dataset [144]. A summary of BC classification using histopathological images is provided in Table 29.

Table 29.
A review of recent approaches for detecting breast cancer through histopathological images.
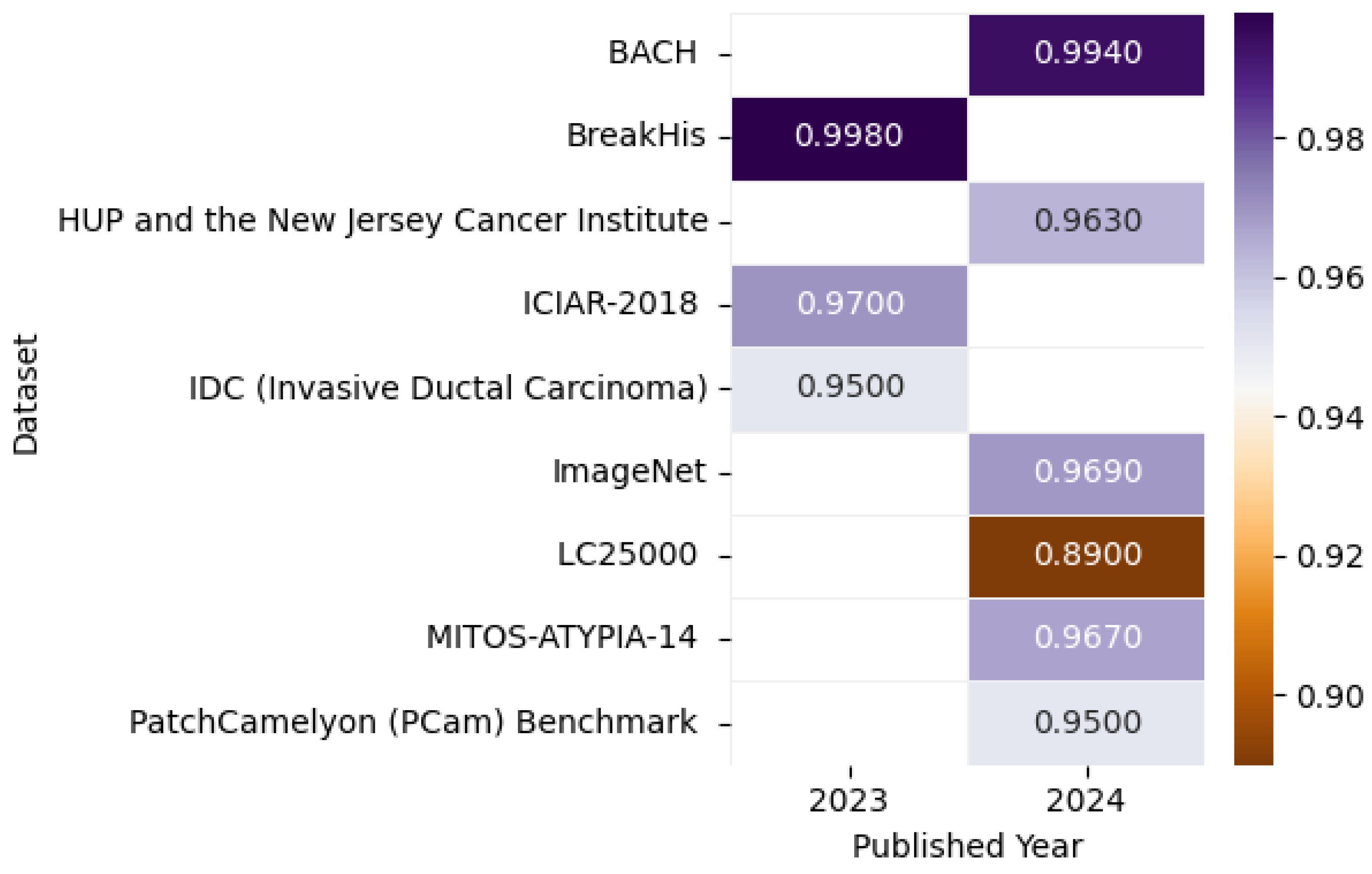
Figure 22 presents the highest accuracy rates achieved by various models on Histopathological datasets in 2023 and 2024, including the ImageNet [145], BreakHis [146], Pennsylvania University Hospital (HUP) and the New Jersey Cancer Institute [147], BACH [144], LC25000 [143], MITOS-ATYPIA-14 [148], PatchCamelyon (PCam) Benchmark [149], IDC (Invasive Ductal Carcinoma) [150], and ICIAR-2018 [140] datasets, as detailed in the referenced studies, paired with a correlation figure that conveys accuracy levels through color intensity variation.
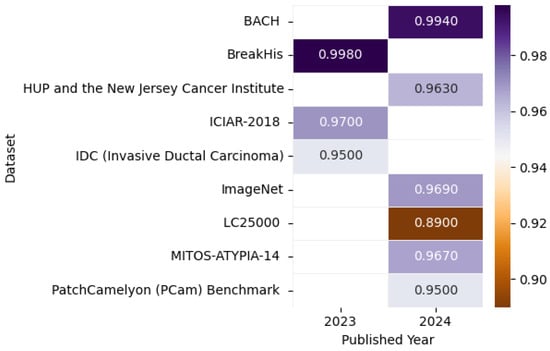
Figure 22.
Highest detection accuracy across multiple histopathological datasets for breast cancer.
8.5.1. Model Performance on Histopathological Image: Pros, Cons, and Future Directions
The comparative analysis in Table 30 highlights the main strengths, weaknesses, and research opportunities for current histopathological imaging approaches in breast cancer detection.

Table 30.
Comprehensive overview of top-performing models on histopathological breast cancer datasets.
8.5.2. Summary
Histopathological imaging is a critical tool for breast cancer (BC) detection, enabling the examination of tissue samples at the cellular level to identify malignancies. Recent advancements in deep learning (DL) have significantly improved the accuracy of BC detection using histopathological images. Studies have employed various DL models, such as CNNs, DenseNet, ResNet, and ensemble methods, to achieve high accuracy and AUC scores. For example, Muntean and Chowkkar (2022) [136] achieved 90.9% accuracy using a CNN, while Bhowal et al. (2022) [137] reported 96.0% accuracy using a fusion model. Despite these advancements, challenges such as dataset dependency, computational complexity, and generalizability remain. Table 31 presents a summary of key findings in histopathology-based breast cancer detection.

Table 31.
Summary of key findings in histopathological-based breast cancer detection.
8.5.3. Analysis
The integration of deep learning models with histopathological imaging has significantly improved BC detection accuracy. Muntean and Chowkkar (2022) [136] demonstrated the effectiveness of CNNs and DenseNet121, achieving 90.9% and 86.6% accuracy, respectively, with transfer learning further improving performance. Bhowal et al. [137] proposed a fusion model combining VGG16, Xception, and InceptionResNet V2, achieving 96.0% accuracy on the ICIAR 2018 dataset. Yang et al. (2022) [138] integrated whole slide H&E images with clinical data, achieving an AUC of 0.76, showcasing the potential of multimodal approaches. Ensemble methods, such as those proposed by Majumdar et al. (2023) [140], further enhance robustness, achieving 99.16% accuracy on the BreakHis dataset. Table 32 provides an analysis of techniques and their impact on histopathological-based breast cancer detection.

Table 32.
Analysis of techniques and their impact in histopathological-based breast cancer detection.
8.5.4. Critique
Despite the promising results, several limitations need to be addressed. First, the reliance on high-quality, annotated histopathological image datasets limits scalability, as such datasets are not always available in clinical settings. Second, the performance of deep learning models can vary when applied to real-world data, as seen in Yang et al. (2022) [138], where the AUC dropped to 0.76 in external validation. Third, the computational complexity of advanced models, such as ensemble methods and multimodal approaches, may hinder their practical implementation in resource-limited environments. Finally, while models achieve high-performance metrics, their lack of interpretability hinders clinical trust and adoption. Table 33 presents a critique of current limitations in histopathological-based breast cancer detection.

Table 33.
Critique of current limitations in histopathological-based breast cancer detection.
8.5.5. Comparison
The reviewed studies demonstrate varying approaches and outcomes, as summarized in Table 34. Muntean and Chowkkar (2022) [136] achieved 90.9% accuracy using a CNN, while Bhowal et al. (2022) [137] reported 95.6% accuracy using a fusion model. Yang et al. (2022) [138] integrated clinical data with imaging, achieving an AUC of 0.76. Majumdar et al. (2023) [140] employed an ensemble method, achieving 99.16% accuracy on the BreakHis dataset. Addo et al. (2024) [144] proposed BCHI-CovNet, achieving 99.38% accuracy on the BACH dataset, showcasing the effectiveness of lightweight models. These comparisons highlight the diversity of approaches in histopathology-based BC detection, with each method offering unique advantages.

Table 34.
Comparison of key studies in histopathological-based breast cancer detection.
Histopathological imaging combined with deep learning models has significantly advanced breast cancer detection, achieving high accuracy and AUC scores. While models like CNNs, DenseNet, and ensemble methods demonstrate exceptional performance, challenges related to dataset dependency, computational complexity, and generalizability remain. Future research should focus on improving model interpretability, expanding dataset diversity, and developing computationally efficient models for real-world clinical deployment. Addressing these challenges will enhance the clinical applicability and robustness of histopathological-based BC detection systems.
Four fundamental gaps emerge in histopathology analysis: (1) stain variation robustness is rarely tested, (2) whole-slide analysis remains computationally expensive, (3) few models incorporate pathologist feedback loops, (4) clinical significance of extracted features is often unverified.
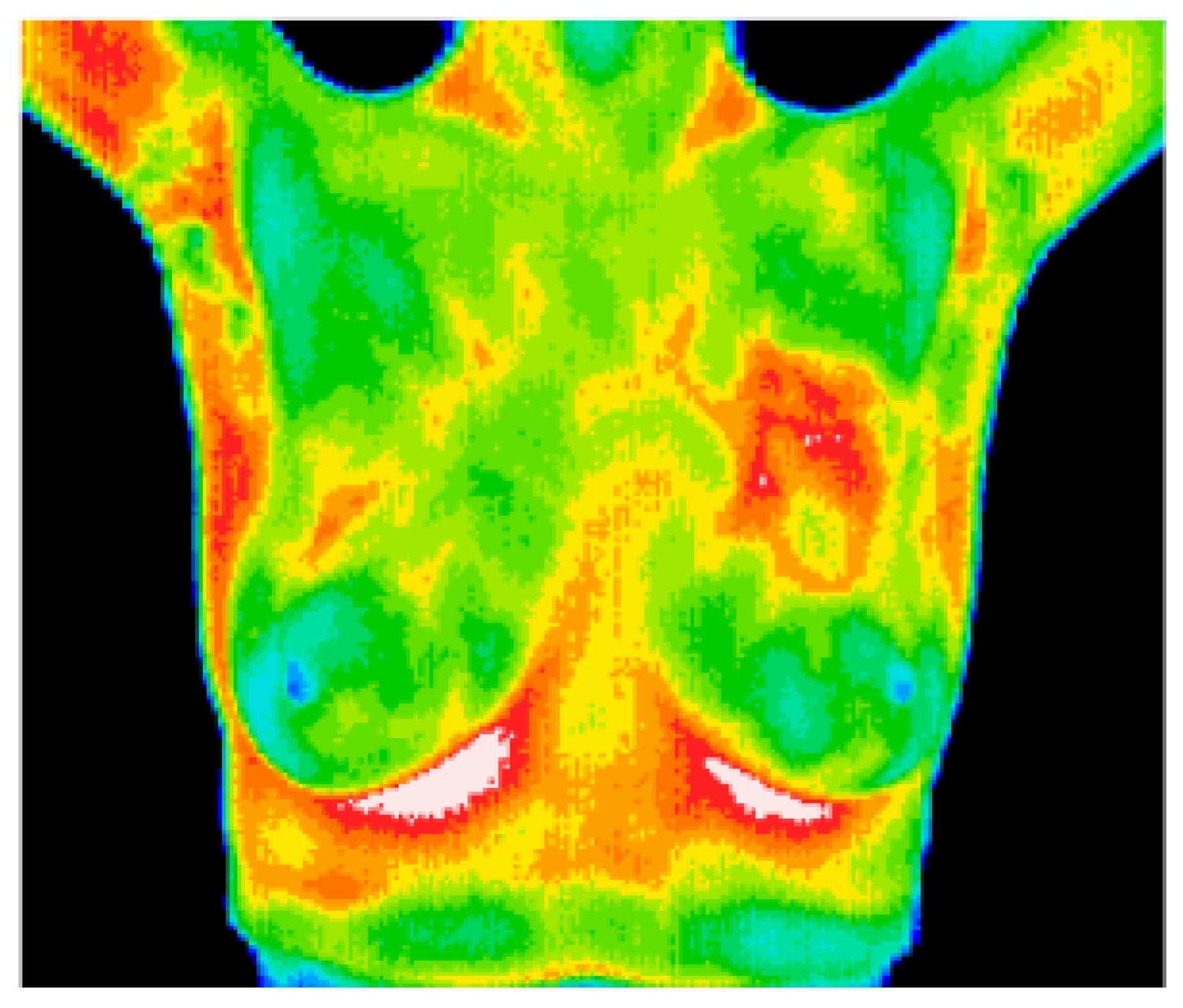
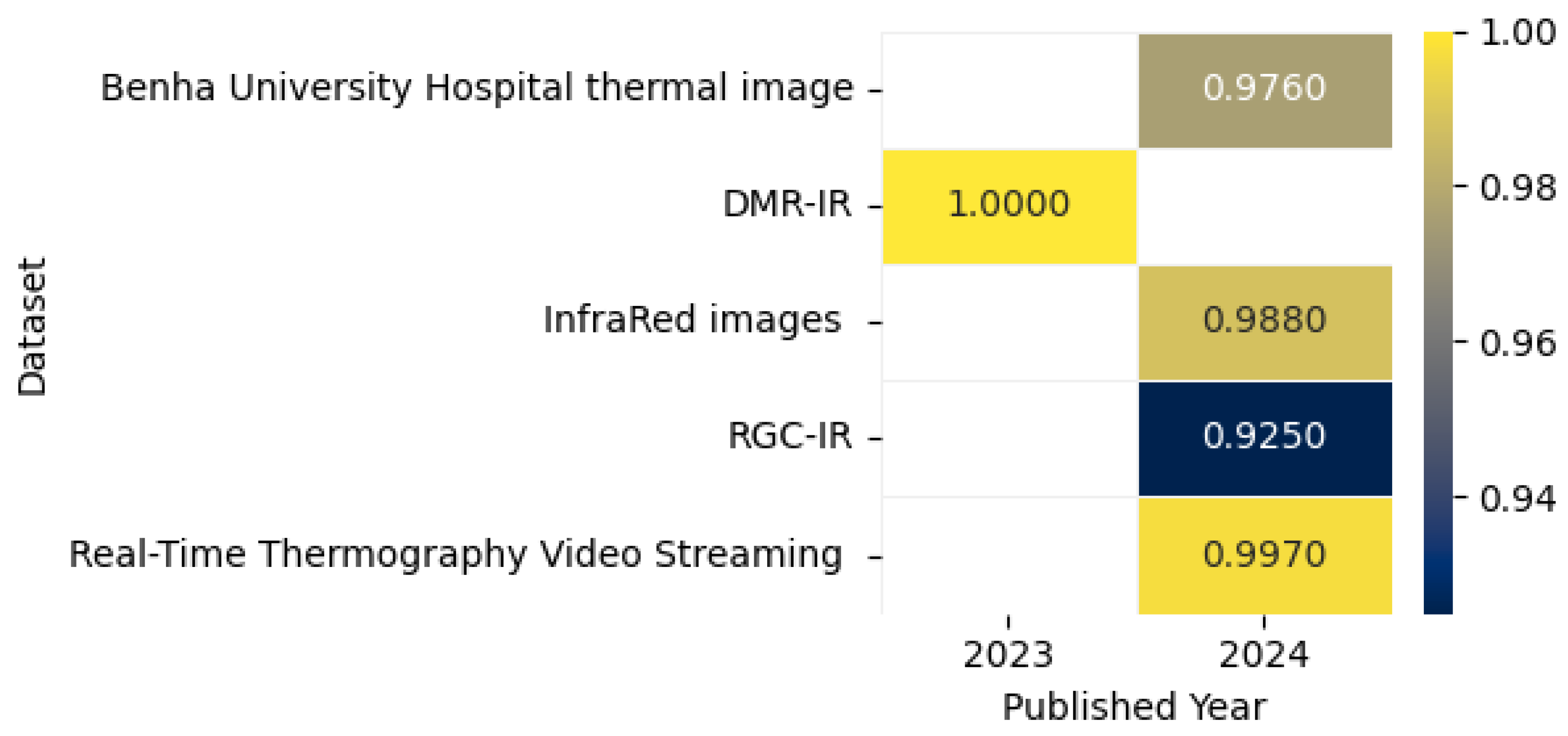
8.6. Breast Cancer Detection Using Thermal Imaging
The thermal image shown in Figure 23 captures the heat emitted by objects, visualizing patterns of temperature variation. This study presents a CADx system for BC detection using DL, integrating multi-view thermograms (frontal and lateral) with patient clinical data. The system employs transfer learning with pre-trained models and focuses on regions of interest (ROIs) for targeted analysis. By addressing the limitations of single-view thermograms and incorporating critical clinical data, the proposed approach achieved 90.48% accuracy, 93.33% sensitivity, and an AUROC of 0.94, offering a cost-effective, less hazardous screening option [151]. Chatterjee et al. proposed a two-stage model for BC detection using thermographic images. The model utilizes VGG16 for feature extraction and a memory-based version of the Dragonfly Algorithm (DA) enhanced by the Grunwald–Letnikov (GL) method for optimal feature selection. Evaluated on the DMR-IR dataset, the framework achieved 100% diagnostic accuracy while reducing features by 82%, demonstrating its efficiency and potential for accurate early detection [152].
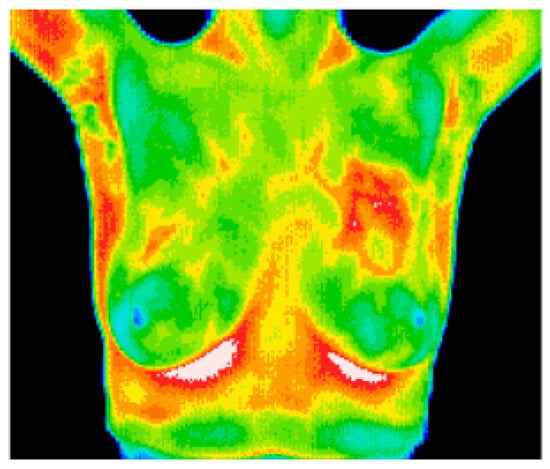
Figure 23.
A sample thermal image [153].
Civilibal et al. proposed a Mask R-CNN model with ResNet-50 and ResNet-101 backbones for breast tumor detection, segmentation, and classification using thermal images. The ResNet-50 model pre-trained on COCO achieved superior performance, with 97.1% accuracy, an mAP of 0.921, and an overlap score of 0.868, outperforming models used in prior studies. This single DL model effectively delineates tumors and adjacent tissues for accurate diagnosis [154]. The study utilized the DMR database for thermal breast image classification, employing a custom CNN model trained on augmented, standardized, and enhanced images with histogram of oriented gradients (HOG) feature extraction. The model achieved impressive accuracy rates ranging from 95.7% to 98.5%, outperforming other classifiers and demonstrating a significant potential for real-time BC diagnosis [155]. Mahoro et al. proposed a BC detection system using DL techniques. They employed TransUNet for breast region segmentation and four models—ResNet-50, EfficientNet-B7, VGG-16, and DenseNet-201—for classifying images into healthy, sick, or unknown categories. The best performance was achieved with ResNet-50 and demonstrated an accuracy of 97.26%, a sensitivity of 97.26%, and a specificity of 100%, 96.94%, and 99.72% for healthy, sick, and unknown classes, respectively [156].
Husaini et al. investigated the impact of various types of noise on DL models for early BC detection using thermal images. They evaluated the performance of several models, particularly Inception MV4, under different noise conditions, showing that noise significantly reduced classification accuracy without proper preprocessing. The proposed method achieved impressive results, with 99.748% accuracy, 0.996 sensitivity, 1.0 specificity, and 0.998 AUC. Their findings highlight the importance of noise reduction strategies and preprocessing techniques to improve the reliability and accuracy of thermal imaging for BC diagnosis [157]. Table 35 provides a summary of the results of BC detection using thermal imaging.

Table 35.
A review of recent methods for breast cancer detection using thermal imaging.
Figure 24 illustrates the highest accuracy rates achieved by various models on thermograph datasets in 2023 and 2024, including the Real-Time Thermography Video Streaming [158], thermal images from Benha University Hospital [159], RGC-IR [42], DMR-IR [160], and infrared images [161], as outlined in the referenced studies, accompanied by a correlation figure where accuracy levels are represented by different color intensities.
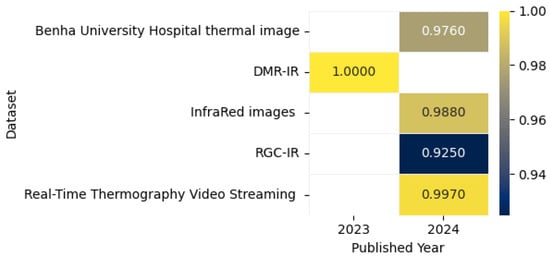
Figure 24.
Highest detection accuracy across multiple thermograph datasets for breast cancer.
8.6.1. Model Performance on Thermal Image: Pros, Cons, and Future Directions
Table 36 presents a comparative analysis of the top performing models, outlining their key strengths, limitations, and potential avenues for future research in thermal imaging-based breast cancer detection.

Table 36.
Comprehensive overview of top-performing models on thermal breast cancer datasets.
8.6.2. Summary
Thermal imaging has emerged as a promising, cost-effective, and radiation-free modality for breast cancer (BC) detection. Recent advancements in deep learning (DL) have significantly improved the accuracy of BC detection using thermal images. Studies have employed techniques such as multi-view thermograms, transfer learning, feature selection, and deep segmentation models. Notable results include Chatterjee et al.’s (2022) [152] 100% accuracy using a two-stage model with VGG16 and the Dragonfly Algorithm, Civilibal et al.’s (2023) 97.1% accuracy with Mask R-CNN and ResNet [154], and Mahoro et al.’s (2024) 97.26% accuracy using TransUNet [156]. Additionally, Husaini et al. (2024) [157] achieved 99.82% accuracy with noise mitigation strategies. These advancements demonstrate the potential of DL-based thermal imaging for accurate and non-invasive BC screening. Table 37 presents a summary of key findings in thermal imaging-based breast cancer detection.

Table 37.
Summary of key findings in thermal imaging-based breast cancer detection.
8.6.3. Analysis
The integration of multi-view thermograms and clinical data, along with the use of transfer learning and advanced feature extraction techniques, has led to improved accuracy rates. Segmentation models, like TransUNet, and feature selection algorithms, such as the Dragonfly Algorithm, enhance model performance. Noise reduction techniques have proven essential, highlighting the importance of preprocessing in thermal imaging. Despite these advancements, challenges in dataset variability, model architectures, and preprocessing techniques remain, hindering standardization. Table 38 provides an analysis of techniques and their impact on thermal imaging-based breast cancer detection.

Table 38.
Analysis of techniques and their impact on thermal imaging-based breast cancer detection.
8.6.4. Critique
While accuracy rates are impressive, several limitations need to be addressed. First, the reliance on small or proprietary datasets limits generalizability. Second, some claims, such as 100% accuracy, may indicate overfitting. Third, the lack of standardized datasets and preprocessing techniques complicates comparisons across studies. Fourth, models like EfficientNet-B7 and TransUNet have high computational demands, limiting their real-time application. Finally, more comprehensive studies are needed on noise and thermal image quality to ensure robust and reliable BC detection. Table 39 presents a critique of current limitations in thermal imaging-based breast cancer detection.

Table 39.
Critique of current limitations in thermal imaging-based breast cancer detection.
8.6.5. Comparison
The reviewed studies demonstrate varying approaches and outcomes, as summarized in Table 40. Chatterjee et al. (2022) [152] achieved 100% accuracy using a two-stage model with VGG16 and the Dragonfly Algorithm, significantly reducing feature dimensionality. Civilibal et al. (2023) [154] employed Mask R-CNN with ResNet-50, achieving 97.1% accuracy and precise tumor localization. Mahoro et al. (2024) [156] utilized TransUNet for segmentation, achieving 97.26% accuracy. Husaini et al. (2024) [157] emphasized noise mitigation, achieving 99.82% accuracy with Inception MV4. These comparisons highlight the diversity of approaches in thermal imaging-based BC detection, with each method offering unique advantages.

Table 40.
Comparison of key studies in thermal imaging-based breast cancer detection.
Thermal imaging combined with deep learning models has significantly advanced breast cancer detection, achieving high accuracy and precision. While models like VGG16, Mask R-CNN, and TransUNet demonstrate exceptional performance, challenges related to dataset dependency, computational complexity, and noise reduction remain. Future research should focus on improving model interpretability, expanding dataset diversity, and developing computationally efficient models for real-world clinical deployment. Addressing these challenges will enhance the clinical applicability and robustness of thermal imaging-based BC detection systems.
Two critical gaps persist in thermal imaging-based breast cancer detection: (1) generalizability remains limited due to reliance on small or proprietary datasets, (2) real-time deployment is hindered by the high computational costs of models like TransUNet and EfficientNet.
8.7. CSV Dataset-Based Breast Cancer Detection
The proposed method using the WDBC dataset applies preprocessing and employs 13 significant features with a gradient boosting regressor and Bonferroni correction. The eXtreme Gradient Boosting model achieved 99.12% accuracy, 0.9767 precision, 1.0 recall, 0.9861 specificity, and a 0.9882 F1-score, demonstrating superior performance and reliability [162]. Manikandan et al. utilized the SEER dataset to classify BC patients’ survival status using a two-step feature selection method (Variance Threshold and PCA) and various classifiers, including Decision Tree, AdaBoosting, XGBoosting, Gradient Boosting, and Naive Bayes. The Decision Tree model achieved 98% accuracy when evaluated with train–test split and k-fold cross-validation, outperforming other approaches [163]. Albadr et al. proposed the Online Sequential Extreme Learning Machine (OSELM) approach to enhance diagnostic accuracy. This method was applied to the WDBC (Wisconsin Diagnostic Breast Cancer) dataset and achieved impressive results, with a precision of 94.09%, a recall of 95.57%, an accuracy of 96.13%, a G-Mean of 94.82%, an F-Measure of 94.80%, a specificity of 96.51%, and an MCC of 91.76%. The results indicate that OSELM is a reliable and efficient technique for BC diagnosis [164]. Table 41 provides a summary of BC detection methods using CSV-based data.

Table 41.
An overview of contemporary approaches for BC detection utilizing CSV datasets.
Figure 25 displays the highest accuracy rates attained by various models on CSV datasets in 2023 and 2024, including WDBC [162], SEER [163], BC-UCI [165], Breast Cancer Coimbra [165], and Cuban [166], as detailed in the referenced studies, along with a correlation figure that visualizes accuracy levels through changes in color intensity.
Figure 25.
Highest detection accuracy across multiple CSV datasets for breast cancer.
8.7.1. Model Performance on Tabular Data: Pros, Cons, and Future Directions
Table 42 provides a comparison of the top-performing models, highlighting their primary advantages, drawbacks, and prospective research directions in breast cancer detection using tabular data.

Table 42.
Detailed review of high-performing models applied to tabular breast cancer datasets.
8.7.2. Summary
Recent advancements in breast cancer (BC) detection using CSV datasets have demonstrated high accuracy and reliability. The eXtreme Gradient Boosting (XGBoost) model applied to the WDBC dataset achieved 99.12% accuracy, with excellent precision, recall, specificity, and F1-score [162]. Manikandan et al. (2023) [163] utilized the SEER dataset and a two-step feature selection method, achieving 98% accuracy with a Decision Tree model. Albadr et al. (2024) [164] proposed the Online Sequential Extreme Learning Machine (OSELM) approach, achieving 96.13% accuracy on the WDBC dataset. These results highlight the effectiveness of feature selection, preprocessing, and advanced machine learning models in improving BC detection accuracy. Table 43 presents a summary of key findings in CSV-based breast cancer detection.

Table 43.
Summary of key findings in CSV-based breast cancer detection.
8.7.3. Analysis
The integration of feature selection and preprocessing techniques has significantly improved the accuracy of BC detection using CSV datasets. Rahman et al. (2024) [162] employed gradient boosting with Bonferroni correction, achieving 99.12% accuracy on the WDBC dataset. Manikandan et al. (2023) [163] utilized a two-step feature selection method (Variance Threshold and PCA) with a Decision Tree model, achieving 98% accuracy on the SEER dataset. Albadr et al. (2024) [164] demonstrated the effectiveness of OSELM, achieving 96.13% accuracy on the WDBC dataset. These approaches underscore the importance of feature engineering and model optimization in achieving high classification accuracy. Table 44 provides an analysis of techniques and their impact on CSV-based breast cancer detection.

Table 44.
Analysis of techniques and their impact on CSV-based Bbeast cancer detection.
8.7.4. Critique
Despite the impressive results, several limitations need to be addressed. First, the reliance on limited datasets, such as WDBC and SEER, may affect the generalizability of the models. Second, the computational complexity of methods like XGBoost and OSELM may hinder their real-time application. Third, the lack of standardized preprocessing techniques complicates comparisons across studies. Finally, more comprehensive validation on larger and more diverse datasets is needed to confirm the robustness of these models across different populations. Table 45 presents a critique of current limitations in CSV-based breast cancer detection.

Table 45.
Critique of current limitations in CSV-based breast cancer detection.
8.7.5. Comparison
The reviewed studies demonstrate varying approaches and outcomes, as summarized in Table 46. Rahman et al. (2024) [162] achieved the highest accuracy (99.12%) using XGBoost on the WDBC dataset. Manikandan et al. (2023) [163] employed a Decision Tree model on the SEER dataset, achieving 98% accuracy. Albadr et al. (2024) [164] proposed OSELM, achieving 96.13% accuracy on the WDBC dataset. While the Decision Tree model has a simpler architecture, XGBoost and OSELM offer better performance, particularly in terms of precision and recall. These comparisons highlight the diversity of approaches in CSV-based BC detection, with each method offering unique advantages.

Table 46.
Comparison of key studies in CSV-based breast cancer detection.
CSV-based breast cancer detection has seen significant advancements through the integration of feature selection, preprocessing, and advanced machine learning models. While models like XGBoost and Decision Tree demonstrate exceptional performance, challenges related to dataset dependency, computational complexity, and generalizability remain. Future research should focus on improving model interpretability, expanding dataset diversity, and developing computationally efficient models for real-world clinical deployment. Addressing these challenges will enhance the clinical applicability and robustness of CSV-based BC detection systems.
Four persistent challenges emerge in CSV-based breast cancer detection: (1) limited generalizability due to small or homogeneous datasets like WDBC and SEER, (2) computational inefficiency of high-performing models (e.g., XGBoost) for real-time clinical use, (3) lack of standardized feature selection and preprocessing pipelines hindering reproducibility, (4) minimal clinical integration, with few models validated against multi-center or prospective data.
9. Imaging Techniques and Data in Breast Cancer Detection: Applications, Strengths, and Weaknesses
Table 47 provides an overview of the various medical imaging modalities used in breast cancer detection, along with corresponding CSV data that highlight their advantages, disadvantages, and applications. Understanding these factors is crucial for effective BC detection, as selecting the appropriate imaging technique and data can significantly impact early diagnosis, treatment planning, and patient outcomes.

Table 47.
Imaging modalities—applications, strengths, and weaknesses.
Model Interpretability in Clinical Practice
Despite achieving high accuracy, deep learning (DL) models often encounter skepticism due to their opaque, black-box nature of decision-making. In imaging modalities such as ultrasound and MRI, models like HoVer-Transformer and DLR have integrated attention mechanisms to generate visual explanations, but still rely heavily on radiologist validation to ensure reliability. Similarly, in histopathological analysis, architectures such as BCHI-CovNet incorporate self-attention to enhance interpretability, though their real-world clinical adoption remains limited. A persistent trade-off exists—simpler models like SVM applied to structured CSV data offer greater interpretability but typically lag behind in accuracy. Moving forward, it is essential to strike a balance between performance and transparency by leveraging explainable AI (XAI) frameworks.
10. Trends in Breast Cancer Detection: ML, Statistical, and DL Techniques Across Journals
Breast cancer detection is currently the leading type of cancer worldwide, garnering significant attention from researchers aiming to develop efficient detection systems. Figure 26 illustrates the number of papers published from 2022 to 2024 across different journals, with a focus on the keyword “Breast Cancer Detection” in the article title. As shown in the figure, the IEEE journal published the highest number of articles with this keyword. The year 2023 saw a peak, with 138 publications. Moreover, research in BC detection has increased over the years, with 187 publications in 2022, 284 in 2023, and 303 in 2024, an increase of 38.28% in 2024 compared to 2022 and 6.27% compared to 2023.
Figure 26.
Publications on breast cancer detection in different journals from 2022 to 2024.
11. Conclusions
This review highlights the remarkable advancements in breast cancer (BC) detection achieved through the integration of machine learning (ML) and deep learning (DL) technologies. Recent studies from 2023 to 2024 have demonstrated the transformative impact of DL models, particularly convolutional neural networks (CNNs), in improving diagnostic accuracy across various imaging modalities, including mammography, ultrasound, MRI, CT scans, histopathology, and thermal imaging. These advancements have enabled earlier and more reliable detection, significantly contributing to improved patient outcomes and survival rates.
However, the clinical adoption of these models hinges on overcoming their ’black-box’ nature. While DL achieves high accuracy, interpretability remains a barrier to trust among physicians. Future work must prioritize explainable AI (XAI) techniques—such as attention maps, Grad-CAM, and clinician feedback loops—to align model decisions with medical reasoning and ensure transparent diagnostics.
Beyond technological innovations, this review underscores the critical role of lifestyle interventions and risk factor management in BC prevention and control. Reducing red meat consumption, maintaining a healthy body mass index (BMI), addressing high fasting plasma glucose, limiting alcohol use, increasing physical activity, and avoiding smoking and secondhand smoke exposure are essential strategies for reducing BC risk. A comprehensive understanding of genetic, hormonal, and environmental risk factors further enhances early detection and prevention efforts.
While technological advancements have revolutionized BC detection, a holistic approach that combines cutting-edge diagnostic tools with lifestyle modifications and early detection strategies remains vital for effective BC management. This review provides valuable insights into the current state of BC detection technologies and emphasizes the need for continued research to refine these tools, improve diagnostic accuracy, and reduce the global burden of breast cancer. By integrating technological innovation with preventive healthcare, we can move closer to a future where breast cancer is detected early, treated effectively, and, ultimately, prevented.
12. Future Research Directions
To advance deep learning applications in breast cancer detection and management, future research should focus on the following key areas:
- Exploring advanced data augmentation techniques remains essential to address the challenge of limited medical imaging data in breast cancer detection. While traditional augmentation methods can enhance dataset diversity, they may compromise data integrity, potentially affecting model reliability. Future research should focus on developing and refining augmentation strategies, such as implicit data augmentation and novel generative approaches, to ensure both diversity and authenticity in the augmented data. Additionally, integrating these methods with deep learning models, including CNNs and transfer learning architectures, can further improve diagnostic accuracy and robustness, ultimately aiding in more reliable breast cancer detection [175,176].
- Develop deep learning models that are trained on diverse, high-quality datasets to mitigate biases and improve generalizability across various clinical settings. This will ensure that models perform consistently across different populations and healthcare systems.
- Prioritize XAI techniques (e.g., counterfactual explanations, concept activation vectors) to align model decisions with clinical reasoning. For example, integrating radiologist feedback loops during training could improve trust in CNN-based mammography systems [78].
- Design user-friendly tools and interfaces that seamlessly integrate deep learning models into existing healthcare practices. Adequate training for clinicians on AI-driven tools will be essential to ensure their adoption and practical utility.
- Focus on early breast cancer diagnosis by integrating longitudinal and multi-modal data to predict risk trajectories and enable preventive measures, thereby allowing for timely interventions.
- Foster collaboration among researchers, healthcare institutions, and data scientists with the goal of sharing datasets. Data should be collected from multiple sources to avoid biases and be properly formatted for ease of analysis, ensuring that deep learning models are trained on diverse and high-quality data.
- Develop advanced image preprocessing techniques for noise reduction, contrast enhancement, and image normalization. These techniques will improve image quality, leading to more accurate detection and classification.
- Innovate new segmentation architectures to improve the extraction of the region of interest (ROI) in medical images. Better segmentation will enhance the overall system performance and accuracy in identifying cancerous regions.
- Integrate various medical image modalities such as mammography, ultrasound, MRI, CT scans, histopathology, and thermal imaging to improve the accuracy and reliability of breast cancer detection. The synthesis of these modalities can provide a more comprehensive view of the disease.
- Incorporate non-imaging data such as cancer history, age, and other health issues alongside imaging data. This combination will allow for earlier and more accurate diagnosis, particularly for high-risk individuals.
- Conduct rigorous validation of deep learning models in real-world clinical settings, with the involvement of medical professionals. This ensures that the models are relevant, applicable, and safe for clinical use.
- Future studies should (1) develop fault-tolerant cloud architectures for reliable deployment, (2) integrate blockchain/IoT for secure health data workflows, and (3) validate AI monitoring systems in real-world clinical settings to enhance diagnostic pipelines [177,178,179].
By addressing these key areas, future research will be able to overcome current limitations, ensuring that deep learning can achieve its full potential in transforming breast cancer diagnosis, treatment, and patient care.
Author Contributions
Conceptualization, methodology, software, validation, formal analysis, investigation, data curation, visualization, and project administration, M.A.R.; Writing—original draft preparation, M.A.R. and J.T.P.; Writing—review and editing, Y.W., M.S.H.K., T.T.A., S.R., M.S.H., S.A.A., R.I.T., V.D., A.H. and T.B.; Supervision, T.B. All authors have read and agreed to the published version of the manuscript.
Funding
This research received no external funding.
Data Availability Statement
The authors state that the data supporting this study are included in the review article.
Conflicts of Interest
The authors declare no conflicts of interest.
References
- Chugh, G.; Kumar, S.; Singh, N. TransNet: A Comparative Study on Breast Carcinoma Diagnosis with Classical Machine Learning and Transfer Learning Paradigm. Multimed. Tools Appl. 2024, 83, 33855–33877. [Google Scholar] [CrossRef]
- World Health Organization. Breast Cancer—who.int. 2024. Available online: https://www.who.int/news-room/fact-sheets/detail/breast-cancer (accessed on 23 November 2024).
- World Health Organization International Agency for Research on Cancer. Cancer Today—gco.iarc.who.int. 2022. Available online: https://gco.iarc.who.int/today/en/dataviz (accessed on 13 December 2024).
- Sekine, C.; Horiguchi, J. Current status and prospects of breast cancer imaging-based diagnosis using artificial intelligence. Int. J. Clin. Oncol. 2024, 29, 1641–1647. [Google Scholar] [CrossRef]
- Institute for Health Metrics and Evaluation (IHME). The Breast Cancer Record. Available online: https://vizhub.healthdata.org/gbd-compare/cancer (accessed on 22 November 2024).
- Saroğlu, H.E.; Shayea, I.; Saoud, B.; Azmi, M.H.; El-Saleh, A.A.; Saad, S.A.; Alnakhli, M. Machine learning, IoT and 5G technologies for breast cancer studies: A review. Alex. Eng. J. 2024, 89, 210–223. [Google Scholar] [CrossRef]
- Amgad, M.; Hodge, J.M.; Elsebaie, M.A.; Bodelon, C.; Puvanesarajah, S.; Gutman, D.A.; Siziopikou, K.P.; Goldstein, J.A.; Gaudet, M.M.; Teras, L.R.; et al. A population-level digital histologic biomarker for enhanced prognosis of invasive breast cancer. Nat. Med. 2024, 30, 85–97. [Google Scholar] [CrossRef]
- Wang, R.; Zhu, Y.; Liu, X.; Liao, X.; He, J.; Niu, L. The Clinicopathological features and survival outcomes of patients with different metastatic sites in stage IV breast cancer. BMC Cancer 2019, 19, 1091. [Google Scholar] [CrossRef]
- Shahidi, F.; Daud, S.M.; Abas, H.; Ahmad, N.A.; Maarop, N. Breast cancer classification using deep learning approaches and histopathology image: A comparison study. IEEE Access 2020, 8, 187531–187552. [Google Scholar] [CrossRef]
- Alom, M.Z.; Yakopcic, C.; Nasrin, M.S.; Taha, T.M.; Asari, V.K. Breast cancer classification from histopathological images with inception recurrent residual convolutional neural network. J. Digit. Imaging 2019, 32, 605–617. [Google Scholar] [CrossRef] [PubMed]
- Chattopadhyay, S.; Dey, A.; Singh, P.K.; Sarkar, R. DRDA-Net: Dense residual dual-shuffle attention network for breast cancer classification using histopathological images. Comput. Biol. Med. 2022, 145, 105437. [Google Scholar] [CrossRef] [PubMed]
- Ukwuoma, C.C.; Hossain, M.A.; Jackson, J.K.; Nneji, G.U.; Monday, H.N.; Qin, Z. Multi-classification of breast cancer lesions in histopathological images using DEEP_Pachi: Multiple self-attention head. Diagnostics 2022, 12, 1152. [Google Scholar] [CrossRef] [PubMed]
- Momenimovahed, Z.; Salehiniya, H. Epidemiological characteristics of and risk factors for breast cancer in the world. Breast Cancer Targets Ther. 2019, 11, 151–164. [Google Scholar] [CrossRef]
- Azamjah, N.; Soltan-Zadeh, Y.; Zayeri, F. Global trend of breast cancer mortality rate: A 25-year study. Asian Pac. J. Cancer Prev. APJCP 2019, 20, 2015. [Google Scholar] [CrossRef]
- Rautela, K.; Kumar, D.; Kumar, V. A comprehensive review on computational techniques for breast cancer: Past, present, and future. Multimed. Tools Appl. 2024, 83, 76267–76300. [Google Scholar] [CrossRef]
- WHO. WHO Report on Cancer: Setting Priorities, Investing Wisely and Providing Care for All|Knowledge Action Portal on NCDs—knowledge-action-portal.com. 6 February 2020. Available online: https://www.knowledge-action-portal.com/en/content/who-report-cancer-setting-priorities-investing-wisely-and-providing-care-all (accessed on 22 November 2024).
- Karthikeyan, A.; Priyakumar, U.D. Artificial intelligence: Machine learning for chemical sciences. J. Chem. Sci. 2022, 134, 2. [Google Scholar] [CrossRef] [PubMed]
- Stone, J.P.; Hartley, R.L.; Temple-Oberle, C. Breast cancer in transgender patients: A systematic review. Part 2: Female to Male. Eur. J. Surg. Oncol. 2018, 44, 1463–1468. [Google Scholar] [CrossRef]
- Chokshi, M.; Morgan, O.; Carroll, E.F.; Fraker, J.L.; Holligan, H.; Kling, J.M. Disparities in Study Inclusion and Screening Rates in Breast Cancer Screening Rates among Transgender People: A Systematic Review. J. Am. Coll. Radiol. 2024, 21, 1430–1443. [Google Scholar] [CrossRef]
- Wahlström, E.; Audisio, R.A.; Selvaggi, G. Aspects to consider regarding breast cancer risk in trans men: A systematic review and risk management approach. PLoS ONE 2024, 19, e0299333. [Google Scholar] [CrossRef] [PubMed]
- Abhisheka, B.; Biswas, S.K.; Purkayastha, B. A comprehensive review on breast cancer detection, classification and segmentation using deep learning. Arch. Comput. Methods Eng. 2023, 30, 5023–5052. [Google Scholar] [CrossRef]
- Nasser, M.; Yusof, U.K. Deep learning based methods for breast cancer diagnosis: A systematic review and future direction. Diagnostics 2023, 13, 161. [Google Scholar] [CrossRef] [PubMed]
- Thakur, N.; Kumar, P.; Kumar, A. A systematic review of machine and deep learning techniques for the identification and classification of breast cancer through medical image modalities. Multimed. Tools Appl. 2024, 83, 35849–35942. [Google Scholar] [CrossRef]
- Sharafaddini, A.M.; Esfahani, K.K.; Mansouri, N. Deep learning approaches to detect breast cancer: A comprehensive review. Multimed. Tools Appl. 2024, 84, 24079–24190. [Google Scholar] [CrossRef]
- Sushanki, S.; Bhandari, A.K.; Singh, A.K. A review on computational methods for breast cancer detection in ultrasound images using multi-image modalities. Arch. Comput. Methods Eng. 2024, 31, 1277–1296. [Google Scholar] [CrossRef]
- Cui, C.; Li, L.; Cai, H.; Fan, Z.; Zhang, L.; Dan, T.; Li, J.; Wang, J. The Chinese Mammography Database (CMMD): An online mammography database with biopsy confirmed types for machine diagnosis of breast. Cancer Imaging Arch. 2021. [CrossRef]
- Suckling, J.; Boggis, C.R.M.; Hutt, I.; Astley, S.; Betal, D.; Cerneaz, N.; Dance, D.R.; Kok, S.L.; Parker, J.; Ricketts, I.; et al. The Mammographic Image Analysis Society Digital Mammogram Database. Online, 1994. Exerpta Medica. International Congress Series 1069. pp. 375–378. Available online: http://peipa.essex.ac.uk/info/mias.html (accessed on 20 April 2025).
- Heath, M.; Bowyer, K.; Kopans, D.; Moore, R.; Kegelmeyer, W.P. The Digital Database for Screening Mammography. In Proceedings of the Fifth International Workshop on Digital Mammography; Yaffe, M., Ed.; Medical Physics Publishing: Madison, WI, USA, 2001; pp. 212–218. [Google Scholar]
- Sawyer-Lee, R.; Gimenez, F.; Hoogi, A.; Rubin, D. Curated Breast Imaging Subset of Digital Database for Screening Mammography (CBIS-DDSM). 2016. Available online: https://www.cancerimagingarchive.net/collection/cbis-ddsm/ (accessed on 20 April 2025).
- Moreira, I.C.; Amaral, I.; Domingues, I.; Cardoso, A.; Cardoso, M.J.; Cardoso, J.S. Inbreast: Toward a full-field digital mammographic database. Acad. Radiol. 2012, 19, 236–248. [Google Scholar] [CrossRef] [PubMed]
- Prapavesis, S.; Fornage, B.; Palko, A.; Weismann, C.F.; Zoumpoulis, P. Breast Ultrasound and US-Guided Interventional Techniques: A Multimedia Teaching File, 2003. CD-ROM Multimedia Teaching File. Available online: https://www.auntminnie.com/clinical-news/article/15565901/breast-ultrasound-and-us-guided-interventional-techniques-a-multimedia-teaching-file (accessed on 20 April 2025).
- Yap, M.H.; Pons, G.; Marti, J.; Ganau, S.; Sentis, M.; Zwiggelaar, R.; Davison, A.K.; Marti, R. Automated Breast Ultrasound Lesions Detection Using Convolutional Neural Networks. IEEE J. Biomed. Health Inform. 2018, 22, 1218–1226. [Google Scholar] [CrossRef]
- Dar, R.A.; Rasool, M.; Assad, A. Breast cancer detection using deep learning: Datasets, methods, and challenges ahead. Comput. Biol. Med. 2022, 149, 106073. [Google Scholar] [CrossRef]
- Al-Dhabyani, W.; Gomaa, M.; Khaled, H.; Fahmy, A. Dataset of breast ultrasound images. Data Brief 2020, 28, 104863. [Google Scholar] [CrossRef]
- Islam, M.R.; Rahman, M.M.; Ali, M.S.; Nafi, A.A.N.; Alam, M.S.; Godder, T.K.; Miah, M.S.; Islam, M.K. Enhancing breast cancer segmentation and classification: An Ensemble Deep Convolutional Neural Network and U-net approach on ultrasound images. Mach. Learn. Appl. 2024, 16, 100555. [Google Scholar] [CrossRef]
- Rehman, N.U.; Wang, J.; Weiyan, H.; Ali, I.; Akbar, A.; Assam, M.; Ghadi, Y.Y.; Algarni, A. Edge of discovery: Enhancing breast tumor MRI analysis with boundary-driven deep learning. Biomed. Signal Process. Control 2024, 95, 106291. [Google Scholar] [CrossRef]
- The Cancer Genome Atlas Research Network. The Cancer Genome Atlas Program. Available online: https://www.cancer.gov/ccg/research/genome-sequencing/tcga (accessed on 12 April 2025).
- Li, X.; Abramson, R.G.; Arlinghaus, L.R.; Chakravarthy, A.B.; Abramson, V.G.; Sanders, M.; Yankeelov, T.E. Data From QIN-BREAST (Version 2). 2016. Available online: https://www.cancerimagingarchive.net/collection/qin-breast/ (accessed on 21 April 2025).
- Aresta, G.; Araújo, T.; Campilho, A.; Eloy, C.; Polónia, A.; Aguiar, P. BACH: Grand Challenge on Breast Cancer Histology Images. Med. Image Anal. 2019, 56, 122–139. [Google Scholar] [CrossRef]
- Spanhol, F.A.; Oliveira, L.S.; Petitjean, C.; Heutte, L. A Dataset for Breast Cancer Histopathological Image Classification. IEEE Trans. Biomed. Eng. 2016, 63, 1455–1462. [Google Scholar] [CrossRef]
- Bhowmik, M.K.; Gogoi, U.R.; Majumdar, G.; Datta, D.; Ghosh, A.K.; Bhattacharjee, D. DBT-TU-JU Breast Thermogram Dataset. 2018. Available online: https://www.mkbhowmik.in/dbtTu.aspx (accessed on 21 April 2025).
- Gupta, T.; Agrawal, R.; Sangal, R.; Rao, S.A. Performance Evaluation of Thermography-Based Computer-Aided Diagnostic Systems for Detecting Breast Cancer: An Empirical Study. ACM Trans. Comput. Healthc. 2024, 5, 1–30. [Google Scholar] [CrossRef]
- Street, W.; Wolberg, W.; Mangasarian, O. Nuclear feature extraction for breast tumor diagnosis. IS T/SPIE 1993 Int. Symp. Electron. Imaging Sci. Technol. 1993, 1905, 861–870. [Google Scholar]
- Teng, J. SEER Breast Cancer Data, 2019. Dataset Derived from the November 2017 Update of the SEER Program of the National Cancer Institute, Encompassing 4024 Female Patients Diagnosed with Infiltrating Duct and Lobular Carcinoma Breast Cancer Between 2006 and 2010. Available online: https://ieee-dataport.org/open-access/seer-breast-cancer-data (accessed on 21 April 2025).
- Curtis, C.; Shah, S.P.; Chin, S.F.; Turashvili, G.; Rueda, O.M.; Dunning, M.J.; Speed, D.; Lynch, A.G.; Samarajiwa, S.A.; Yuan, Y.; et al. The genomic and transcriptomic architecture of 2000 breast tumours reveals novel subgroups. Nature 2012, 486, 346–352. [Google Scholar] [CrossRef] [PubMed]
- Elter, M.; Horsch, A.; Nauck, D.; Spies, M. Mammographic Mass Dataset. UCI Machine Learning Repository. 2007. Available online: https://archive.ics.uci.edu/dataset/161/mammographic+mass (accessed on 22 April 2025).
- Alnuaimi, A.F.; Albaldawi, T.H. An overview of machine learning classification techniques. In Proceedings of the BIO Web of Conferences. EDP Sciences, Copenhagen, Denmark, 25–30 August 2024; Volume 97, p. 00133. [Google Scholar]
- Mendonça, M.O.; Netto, S.L.; Diniz, P.S.; Theodoridis, S. Machine learning: Review and trends. In Signal Processing and Machine Learning Theory; Elsevier: Amsterdam, The Netherlands, 2024; pp. 869–959. [Google Scholar]
- Kronberger, G.; Burlacu, B.; Kommenda, M.; Winkler, S.M.; Affenzeller, M. Symbolic Regression; CRC Press: Boca Raton, FL, USA, 2024. [Google Scholar]
- Shmueli, G.; Polak, J. Practical Time Series Forecasting with r: A Hands-On Guide; Axelrod Schnall Publishers: Green Cove Springs, FL, USA, 2024. [Google Scholar]
- Zhou, Z.H. Ensemble Methods: Foundations and Algorithms; CRC Press: Boca Raton, FL, USA, 2025. [Google Scholar]
- Veeranjaneyulu, K.; Lakshmi, M.; Janakiraman, S. Swarm Intelligent Metaheuristic Optimization Algorithms-Based Artificial Neural Network Models for Breast Cancer Diagnosis: Emerging Trends, Challenges and Future Research Directions. Arch. Comput. Methods Eng. 2025, 32, 381–398. [Google Scholar] [CrossRef]
- Patil, P.; Fulkar, B.; Sharma, M.; Rewatkar, R. Detecting Breast Cancer: A Comparative Study of Various Machine Learning Models. In Proceedings of the 2024 Parul International Conference on Engineering and Technology (PICET), Vadodara, India, 3–4 May 2024; pp. 1–5. [Google Scholar]
- Barnils, N.P.; Schüz, B. Identifying intersectional groups at risk for missing breast cancer screening: Comparing regression-and decision tree-based approaches. Ssm-Popul. Health 2025, 29, 101736. [Google Scholar] [CrossRef] [PubMed]
- Win, N.S.S.; Li, G.; Lin, L. Revolutionizing early breast cancer screening: Advanced multi-spectral transmission imaging classification with improved Otsu’s method and K-means clustering. Comput. Biol. Med. 2025, 184, 109373. [Google Scholar] [CrossRef] [PubMed]
- Zhang, L.; Wang, L.; Liang, R.; He, X.; Wang, D.; Sun, L.; Yu, S.; Su, W.; Zhang, W.; Zhou, Q.; et al. An Effective Ultrasound Features-Based Diagnostic Model via Principal Component Analysis Facilitated Differentiating Subtypes of Mucinous Breast Cancer From Fibroadenomas. Clin. Breast Cancer 2024, 24, e583–e592. [Google Scholar] [CrossRef]
- Tan, Y.N.; Lam, P.D.; Tinh, V.P.; Le, D.D.; Nam, N.H.; Khoa, T.A. Joint Federated Learning Using Deep Segmentation and the Gaussian Mixture Model for Breast Cancer Tumors. IEEE Access 2024, 12, 94231–94249. [Google Scholar] [CrossRef]
- Dimri, S.C.; Indu, R.; Negi, H.S.; Panwar, N.; Sarda, M. Hidden Markov Model-Applications, Strengths, and Weaknesses. In Proceedings of the 2024 2nd International Conference on Device Intelligence, Computing and Communication Technologies (DICCT), Dehradun, India, 15–16 March 2024; pp. 300–305. [Google Scholar]
- Costanzo, S.; Flores, A. Reinforcement Learning to Enhanced Microwave Imaging for Accurate Tumor Detection in Breast Images. In Proceedings of the 2024 IEEE International Conference on Metrology for eXtended Reality, Artificial Intelligence and Neural Engineering (MetroXRAINE), St Albans, UK, 21–23 October 2024; pp. 101–106. [Google Scholar]
- Jayakrishnan, R.; Meera, S. Multiclass Classification of Leukemia Cancer Subtypes using Gene Expression Data and Optimized Dueling Double Deep Q-Network. Chemom. Intell. Lab. Syst. 2025, 262, 105402. [Google Scholar] [CrossRef]
- Wang, Q.; Chang, C. Automating the optimization of proton PBS treatment planning for head and neck cancers using policy gradient-based deep reinforcement learning. Med. Phys. 2025, 52, 1997–2014. [Google Scholar] [CrossRef] [PubMed]
- Institute for Health Metrics and Evaluation (IHME). GBD Cancer Compare—vizhub.healthdata.org. Available online: https://vizhub.healthdata.org/gbd-compare/cancer (accessed on 22 November 2024).
- Global Burden of Disease Collaborative Network. Global Burden of Disease Study 2021 (GBD 2021) Results, 2022. Available from the GBD Results Tool. Available online: https://vizhub.healthdata.org/gbd-results/ (accessed on 23 April 2025).
- Ivanova, M.; Pescia, C.; Trapani, D.; Venetis, K.; Frascarelli, C.; Mane, E.; Cursano, G.; Sajjadi, E.; Scatena, C.; Cerbelli, B.; et al. Early Breast Cancer Risk Assessment: Integrating Histopathology with Artificial Intelligence. Cancers 2024, 16, 1981. [Google Scholar] [CrossRef]
- Hussain, S.; Ali, M.; Naseem, U.; Nezhadmoghadam, F.; Jatoi, M.A.; Gulliver, T.A.; Tamez-Peña, J.G. Breast cancer risk prediction using machine learning: A systematic review. Front. Oncol. 2024, 14, 1343627. [Google Scholar] [CrossRef]
- IBM. What Is Machine Learning (ML)?|IBM—ibm.com. 2023. Available online: https://www.ibm.com/cloud/learn/machine-learning (accessed on 23 November 2024).
- Hristov, D.; Stojanov, D. In silico report on five high-risk Protein C pathogenic variants: G403R, P405S, S421N, C238S, and I243T. Mutat. Res.-Fundam. Mol. Mech. Mutagen. 2025, 831, 111907. [Google Scholar] [CrossRef]
- Spek, C.A.; Arruda, V.R. The protein C pathway in cancer metastasis. Thromb. Res. 2012, 129, S80–S84. [Google Scholar] [CrossRef]
- Lee, R.S.; Gimenez, F.; Hoogi, A.; Miyake, K.K.; Gorovoy, M.; Rubin, D.L. A Curated Mammography Data Set for Use in Computer-Aided Detection and Diagnosis Research. Sci. Data 2017, 4, 170177. [Google Scholar] [CrossRef] [PubMed]
- Heath, M.; Bowyer, K.; Kopans, D.; Kegelmeyer, P., Jr.; Moore, R.; Chang, K.; Munishkumaran, S. Current Status of the Digital Database for Screening Mammography. In Digital Mammography; Karssemeijer, N., Thijssen, M., Hendriks, J., van Erning, L., Eds.; Computational Imaging and Vision; Springer: Dordrecht, The Netherlands, 1998; Volume 13. [Google Scholar] [CrossRef]
- Suckling, J.; Parker, J.; Dance, D.; Astley, S.; Hutt, I.; Boggis, C.; Ricketts, I.; Stamatakis, E.; Cerneaz, N.; Kok, S.; et al. Mammographic Image Analysis Society (MIAS) Database v1.21 2015. Available online: https://www.repository.cam.ac.uk/handle/1810/250394 (accessed on 23 April 2025).
- Kiros, H. Doctors Using AI Catch Breast Cancer More Often than Either Does Alone—technologyreview.com. 2022. Available online: https://www.technologyreview.com/2022/07/11/1055677/ai-diagnose-breast-cancer-mammograms/ (accessed on 30 November 2024).
- Qu, J.; Zhao, X.; Chen, P.; Wang, Z.; Liu, Z.; Yang, B.; Li, H. Deep learning on digital mammography for expert-level diagnosis accuracy in breast cancer detection. Multimed. Syst. 2022, 28, 1263–1274. [Google Scholar] [CrossRef]
- Jamil, R.; Dong, M.; Bano, S.; Javed, A.; Abdullah, M. Precancerous Change Detection Technique on Mammography Breast Cancer Images based on Mean Ratio and Log Ratio using Fuzzy c Mean Classification with Gabor Filter. Curr. Med. Imaging 2024, 20, e18749445290351. [Google Scholar] [CrossRef]
- Nemade, V.; Pathak, S.; Dubey, A.K. Deep learning-based ensemble model for classification of breast cancer. Microsyst. Technol. 2024, 30, 513–527. [Google Scholar] [CrossRef]
- Avcı, H.; Karakaya, J. A novel medical image enhancement algorithm for breast cancer detection on mammography images using machine learning. Diagnostics 2023, 13, 348. [Google Scholar] [CrossRef]
- Yaqub, M.; Jinchao, F.; Aijaz, N.; Ahmed, S.; Mehmood, A.; Jiang, H.; He, L. Intelligent breast cancer diagnosis with two-stage using mammogram images. Sci. Rep. 2024, 14, 16672. [Google Scholar] [CrossRef] [PubMed]
- Dada, E.G.; Oyewola, D.O.; Misra, S. Computer-aided diagnosis of breast cancer from mammogram images using deep learning algorithms. J. Electr. Syst. Inf. Technol. 2024, 11, 38. [Google Scholar] [CrossRef]
- Mudeng, V.; Jeong, J.w.; Choe, S.w. Simply Fine-Tuned Deep Learning-Based Classification for Breast Cancer with Mammograms. Comput. Mater. Contin. 2022, 73, 4677. [Google Scholar] [CrossRef]
- Sait, A.R.W.; Nagaraj, R. An Enhanced LightGBM-Based Breast Cancer Detection Technique Using Mammography Images. Diagnostics 2024, 14, 227. [Google Scholar] [CrossRef]
- Raaj, R.S. Breast cancer detection and diagnosis using hybrid deep learning architecture. Biomed. Signal Process. Control 2023, 82, 104558. [Google Scholar] [CrossRef]
- Mohammed, A.D.; Ekmekci, D. Breast Cancer Diagnosis Using YOLO-Based Multiscale Parallel CNN and Flattened Threshold Swish. Appl. Sci. 2024, 14, 2680. [Google Scholar] [CrossRef]
- Islam, R.; Tarique, M. Artificial Intelligence (AI) and Nuclear Features from the Fine Needle Aspirated (FNA) Tissue Samples to Recognize Breast Cancer. J. Imaging 2024, 10, 201. [Google Scholar] [CrossRef]
- Sakaida, M.; Yoshimura, T.; Tang, M.; Ichikawa, S.; Sugimori, H.; Hirata, K.; Kudo, K. The Effectiveness of Semi-Supervised Learning Techniques in Identifying Calcifications in X-ray Mammography and the Impact of Different Classification Probabilities. Appl. Sci. 2024, 14, 5968. [Google Scholar] [CrossRef]
- Trang, N.T.H.; Long, K.Q.; An, P.L.; Dang, T.N. Development of an artificial intelligence-based breast cancer detection model by combining mammograms and medical health records. Diagnostics 2023, 13, 346. [Google Scholar] [CrossRef]
- Dehghan Rouzi, M.; Moshiri, B.; Khoshnevisan, M.; Akhaee, M.A.; Jaryani, F.; Salehi Nasab, S.; Lee, M. Breast cancer detection with an ensemble of deep learning networks using a consensus-adaptive weighting method. J. Imaging 2023, 9, 247. [Google Scholar] [CrossRef]
- Jafari, Z.; Karami, E. Breast cancer detection in mammography images: A CNN-based approach with feature selection. Information 2023, 14, 410. [Google Scholar] [CrossRef]
- Anas, M.; Haq, I.U.; Husnain, G.; Jaffery, S.A.F. Advancing Breast Cancer Detection: Enhancing YOLOv5 Network for Accurate Classification in Mammogram Images. IEEE Access 2024, 12, 16474–16488. [Google Scholar] [CrossRef]
- Gómez-Flores, W.; Gregorio-Calas, M.J.; Coelho de Albuquerque Pereira, W. BUS-BRA: A breast ultrasound dataset for assessing computer-aided diagnosis systems. Med. Phys. 2024, 51, 3110–3123. [Google Scholar] [CrossRef] [PubMed]
- Vallez, N.; Bueno, G.; Deniz, O.; Rienda, M.A.; Pastor, C. BUS-UCLM: Breast Ultrasound Lesion Segmentation Dataset. Mendeley Data, V1, 2024. Available online: https://data.mendeley.com/datasets/7fvgj4jsp7/1 (accessed on 23 April 2025).
- Pawłowska, A.; Ćwierz Pieńkowska, A.; Domalik, A.; Jaguś, D.; Kasprzak, P.; Matkowski, R.; Fura, Ł.; Nowicki, A.; Zolek, N. A Curated Benchmark Dataset for Ultrasound-Based Breast Lesion Analysis. Sci. Data 2024, 11, 148. [Google Scholar] [CrossRef]
- Fisher, P.R. Breast Cancer Ultrasonography: Practice Essentials, Role of Ultrasonography in Screening, Breast Imaging Reporting and Data System—emedicine.medscape.com. 2021. Available online: https://emedicine.medscape.com/article/346725-overview (accessed on 30 November 2024).
- Rodrigues, P.S. Breast Ultrasound Image. Mendeley Data, V1, 2017. Available online: https://data.mendeley.com/datasets/wmy84gzngw/1 (accessed on 24 April 2025).
- Masud, M.; Hossain, M.S.; Alhumyani, H.; Alshamrani, S.S.; Cheikhrouhou, O.; Ibrahim, S.; Muhammad, G.; Rashed, A.E.E.; Gupta, B. Pre-trained convolutional neural networks for breast cancer detection using ultrasound images. ACM Trans. Internet Technol. (TOIT) 2021, 21, 1–17. [Google Scholar] [CrossRef]
- Raza, A.; Ullah, N.; Khan, J.A.; Assam, M.; Guzzo, A.; Aljuaid, H. DeepBreastCancerNet: A novel deep learning model for breast cancer detection using ultrasound images. Appl. Sci. 2023, 13, 2082. [Google Scholar] [CrossRef]
- Yu, F.H.; Miao, S.M.; Li, C.Y.; Hang, J.; Deng, J.; Ye, X.H.; Liu, Y. Pretreatment ultrasound-based deep learning radiomics model for the early prediction of pathologic response to neoadjuvant chemotherapy in breast cancer. Eur. Radiol. 2023, 33, 5634–5644. [Google Scholar] [CrossRef]
- Mo, Y.; Han, C.; Liu, Y.; Liu, M.; Shi, Z.; Lin, J.; Zhao, B.; Huang, C.; Qiu, B.; Cui, Y.; et al. Hover-trans: Anatomy-aware hover-transformer for roi-free breast cancer diagnosis in ultrasound images. IEEE Trans. Med. Imaging 2023, 42, 1696–1706. [Google Scholar] [CrossRef]
- Shah, A. Breast Ultrasound Images Dataset. Kaggle. 2021. Available online: https://www.kaggle.com/datasets/aryashah2k/breast-ultrasound-images-dataset (accessed on 24 April 2025).
- Lanjewar, M.G.; Panchbhai, K.G.; Patle, L.B. Fusion of transfer learning models with LSTM for detection of breast cancer using ultrasound images. Comput. Biol. Med. 2024, 169, 107914. [Google Scholar] [CrossRef]
- Rao, K.S.; Terlapu, P.V.; Jayaram, D.; Raju, K.K.; Kumar, G.K.; Pemula, R.; Gopalachari, V.; Rakesh, S. Intelligent ultrasound imaging for enhanced breast cancer diagnosis: Ensemble transfer learning strategies. IEEE Access 2024, 12, 22243–22263. [Google Scholar] [CrossRef]
- Ellis, J.; Appiah, K.; Amankwaa-Frempong, E.; Kwok, S.C. Classification of 2D Ultrasound Breast Cancer Images with Deep Learning. In Proceedings of the IEEE/CVF Conference on Computer Vision and Pattern Recognition, Seattle, WA, USA, 16–22 June 2024; pp. 5167–5173. [Google Scholar]
- Muduli, D.; Kumar, R.R.; Pradhan, J.; Kumar, A. An empirical evaluation of extreme learning machine uncertainty quantification for automated breast cancer detection. Neural Comput. Appl. 2023, 37, 7909–7924. [Google Scholar] [CrossRef]
- Sahu, A.; Das, P.K.; Meher, S. An efficient deep learning scheme to detect breast cancer using mammogram and ultrasound breast images. Biomed. Signal Process. Control 2024, 87, 105377. [Google Scholar] [CrossRef]
- Himel, M.H.A.M.H.; Chowdhury, P.; Hasan, M.A.M. A robust encoder decoder based weighted segmentation and dual staged feature fusion based meta classification for breast cancer utilizing ultrasound imaging. Intell. Syst. Appl. 2024, 22, 200367. [Google Scholar] [CrossRef]
- Umer, M.J.; Sharif, M.; Raza, M. A Multi-attention Triple Decoder Deep Convolution Network for Breast Cancer Segmentation Using Ultrasound Images. Cogn. Comput. 2024, 16, 581–594. [Google Scholar] [CrossRef]
- Guo, D.; Lu, C.; Chen, D.; Yuan, J.; Duan, Q.; Xue, Z.; Liu, S.; Huang, Y. A multimodal breast cancer diagnosis method based on Knowledge-Augmented Deep Learning. Biomed. Signal Process. Control 2024, 90, 105843. [Google Scholar] [CrossRef]
- Admass, W.S.; Munaye, Y.Y.; Salau, A.O. Integration of feature enhancement technique in Google inception network for breast cancer detection and classification. J. Big Data 2024, 11, 78. [Google Scholar] [CrossRef]
- Ru, J.; Zhu, Z.; Shi, J. Spatial and geometric learning for classification of breast tumors from multi-center ultrasound images: A hybrid learning approach. Bmc Med. Imaging 2024, 24, 133. [Google Scholar] [CrossRef]
- Karunanayake, N.; Makhanov, S.S. When deep learning is not enough: Artificial life as a supplementary tool for segmentation of ultrasound images of breast cancer. Med. Biol. Eng. Comput. 2024, 63, 2497–2520. [Google Scholar] [CrossRef] [PubMed]
- Gupta, S.; Agrawal, S.; Singh, S.K.; Kumar, S. A novel transfer learning-based model for ultrasound breast cancer image classification. In Computational Vision and Bio-Inspired Computing: Proceedings of ICCVBIC 2022; Springer: Berlin/Heidelberg, Germany, 2023; pp. 511–523. [Google Scholar]
- Zakareya, S.; Izadkhah, H.; Karimpour, J. A new deep-learning-based model for breast cancer diagnosis from medical images. Diagnostics 2023, 13, 1944. [Google Scholar] [CrossRef]
- Zhang, B.; Vakanski, A.; Xian, M. BI-RADS-NET-V2: A Composite Multi-Task Neural Network for Computer-Aided Diagnosis of Breast Cancer in Ultrasound Images With Semantic and Quantitative Explanations. IEEE Access 2023, 11, 79480–79494. [Google Scholar] [CrossRef]
- Işık, G.; Paçal, İ. Few-shot classification of ultrasound breast cancer images using meta-learning algorithms. Neural Comput. Appl. 2024, 36, 12047–12059. [Google Scholar] [CrossRef]
- Hossain, S.; Azam, S.; Montaha, S.; Karim, A.; Chowa, S.S.; Mondol, C.; Hasan, M.Z.; Jonkman, M. Automated breast tumor ultrasound image segmentation with hybrid UNet and classification using fine-tuned CNN model. Heliyon 2023, 9, e21369. [Google Scholar] [CrossRef]
- Mohammadi, M.; Mohammadi, S.; Hadizadeh, H.; Olfati, M.; Moradi, F.; Tanzifi, G.; Ghaderi, S. Brain metastases from breast cancer using magnetic resonance imaging: A systematic review. J. Med. Radiat. Sci. 2024, 71, 133–141. [Google Scholar] [CrossRef]
- Berg, W. Breast MRI|DenseBreast-info, Inc.—densebreast-info.org. Available online: https://densebreast-info.org/screening-technologies/breast-mri/ (accessed on 1 December 2024).
- Zhang, Z.; Lan, H.; Zhao, S. Analysis of the Value of Quantitative Features in Multimodal MRI Images to Construct a Radio-Omics Model for Breast Cancer Diagnosis. Breast Cancer Targets Ther. 2024, 16, 305–318. [Google Scholar] [CrossRef]
- Liu, M.Z.; Swintelski, C.; Sun, S.; Siddique, M.; Desperito, E.; Jambawalikar, S.; Ha, R. Weakly supervised deep learning approach to breast MRI assessment. Acad. Radiol. 2022, 29, S166–S172. [Google Scholar] [CrossRef] [PubMed]
- Wu, Y.; Wu, J.; Dou, Y.; Rubert, N.; Wang, Y.; Deng, J. A deep learning fusion model with evidence-based confidence level analysis for differentiation of malignant and benign breast tumors using dynamic contrast enhanced MRI. Biomed. Signal Process. Control 2022, 72, 103319. [Google Scholar] [CrossRef]
- Yue, W.Y.; Zhang, H.T.; Gao, S.; Li, G.; Sun, Z.Y.; Tang, Z.; Cai, J.M.; Tian, N.; Zhou, J.; Dong, J.H.; et al. Predicting breast cancer subtypes using magnetic resonance imaging based radiomics with automatic segmentation. J. Comput. Assist. Tomogr. 2023, 47, 729–737. [Google Scholar] [CrossRef]
- Qian, H.; Ren, X.; Xu, M.; Fang, Z.; Zhang, R.; Bu, Y.; Zhou, C. Magnetic resonance imaging-based radiomics was used to evaluate the level of prognosis-related immune cell infiltration in breast cancer tumor microenvironment. Bmc Med. Imaging 2024, 24, 31. [Google Scholar] [CrossRef]
- Guo, Y.; Xie, X.; Tang, W.; Chen, S.; Wang, M.; Fan, Y.; Lin, C.; Hu, W.; Yang, J.; Xiang, J.; et al. Noninvasive identification of HER2-low-positive status by MRI-based deep learning radiomics predicts the disease-free survival of patients with breast cancer. Eur. Radiol. 2024, 34, 899–913. [Google Scholar] [CrossRef]
- Cong, C.; Li, X.; Zhang, C.; Zhang, J.; Sun, K.; Liu, L.; Ambale-Venkatesh, B.; Chen, X.; Wang, Y. MRI-based breast cancer classification and localization by multiparametric feature extraction and combination using deep learning. J. Magn. Reson. Imaging 2024, 59, 148–161. [Google Scholar] [CrossRef] [PubMed]
- Liang, R.; Li, F.; Yao, J.; Tong, F.; Hua, M.; Liu, J.; Shi, C.; Sui, L.; Lu, H. Predictive value of MRI-based deep learning model for lymphovascular invasion status in node-negative invasive breast cancer. Sci. Rep. 2024, 14, 16204. [Google Scholar] [CrossRef]
- Kumar, A.; Kumar, P.; Mahto, M.; Srivastava, S. Breast Cancer Detection and Localization Using a Novel Multi Modal Approach. IEEE Trans. Instrum. Meas. 2024, 74, 4000413. [Google Scholar]
- Hasan, A.M.; Al-Waely, N.K.; Aljobouri, H.K.; Jalab, H.A.; Ibrahim, R.W.; Meziane, F. Molecular subtypes classification of breast cancer in DCE-MRI using deep features. Expert Syst. Appl. 2024, 236, 121371. [Google Scholar] [CrossRef]
- Wang, W.; Wang, Y. Deep Learning-Based Modified YOLACT Algorithm on Magnetic Resonance Imaging Images for Screening Common and Difficult Samples of Breast Cancer. Diagnostics 2023, 13, 1582. [Google Scholar] [CrossRef]
- Vivancos Bargalló, H.; Stick, L.B.; Korreman, S.S.; Kronborg, C.; Nielsen, M.M.; Borgen, A.C.; Offersen, B.V.; Nørrevang, O.; Kallehauge, J.F. Classification of laterality and mastectomy/lumpectomy for breast cancer patients for improved performance of deep learning auto segmentation. Acta Oncol. 2023, 62, 1546–1550. [Google Scholar] [CrossRef]
- Koh, J.; Yoon, Y.; Kim, S.; Han, K.; Kim, E.K. Deep learning for the detection of breast cancers on chest computed tomography. Clin. Breast Cancer 2022, 22, 26–31. [Google Scholar] [CrossRef]
- Yasaka, K.; Sato, C.; Hirakawa, H.; Fujita, N.; Kurokawa, M.; Watanabe, Y.; Kubo, T.; Abe, O. Impact of deep learning on radiologists and radiology residents in detecting breast cancer on CT: A cross-vendor test study. Clin. Radiol. 2024, 79, e41–e47. [Google Scholar] [CrossRef] [PubMed]
- Jalalian, A.; Mashohor, S.; Mahmud, R.; Karasfi, B.; Iqbal Saripan, M.; Ramli, A.R. Computer-assisted diagnosis system for breast cancer in computed tomography laser mammography (CTLM). J. Digit. Imaging 2017, 30, 796–811. [Google Scholar] [CrossRef] [PubMed]
- Zhao, X.; Dong, Y.h.; Xu, L.Y.; Shen, Y.Y.; Qin, G.; Zhang, Z.B. Deep bone oncology Diagnostics: Computed tomography based Machine learning for detection of bone tumors from breast cancer metastasis. J. Bone Oncol. 2024, 48, 100638. [Google Scholar] [CrossRef]
- Desperito, E.; Schwartz, L.; Capaccione, K.M.; Collins, B.T.; Jamabawalikar, S.; Peng, B.; Patrizio, R.; Salvatore, M.M. Chest CT for breast cancer diagnosis. Life 2022, 12, 1699. [Google Scholar] [CrossRef]
- Shehzad, I.; Zafar, A. Breast Cancer CT-Scan Image Classification Using Transfer Learning. Sn Comput. Sci. 2023, 4, 789. [Google Scholar] [CrossRef]
- Rakha, E.; Tozbikian, G. Invasive Breast Carcinoma of No Special Type and Variants Invasive Breast Cancer of No Special Type (NST). 2024. Available online: https://www.pathologyoutlines.com/topic/breastmalignantductalnos.html (accessed on 9 December 2024).
- Hava Muntean, C.; Chowkkar, M. Breast cancer detection from histopathological images using deep learning and transfer learning. In Proceedings of the 2022 7th International Conference on Machine Learning Technologies, Rome, Italy, 11–13 March 2022; pp. 164–169. [Google Scholar]
- Bhowal, P.; Sen, S.; Velasquez, J.D.; Sarkar, R. Fuzzy ensemble of deep learning models using choquet fuzzy integral, coalition game and information theory for breast cancer histology classification. Expert Syst. Appl. 2022, 190, 116167. [Google Scholar] [CrossRef]
- Yang, J.; Ju, J.; Guo, L.; Ji, B.; Shi, S.; Yang, Z.; Gao, S.; Yuan, X.; Tian, G.; Liang, Y.; et al. Prediction of HER2-positive breast cancer recurrence and metastasis risk from histopathological images and clinical information via multimodal deep learning. Comput. Struct. Biotechnol. J. 2022, 20, 333–342. [Google Scholar] [CrossRef]
- Maleki, A.; Raahemi, M.; Nasiri, H. Breast cancer diagnosis from histopathology images using deep neural network and XGBoost. Biomed. Signal Process. Control 2023, 86, 105152. [Google Scholar] [CrossRef]
- Majumdar, S.; Pramanik, P.; Sarkar, R. Gamma function based ensemble of CNN models for breast cancer detection in histopathology images. Expert Syst. Appl. 2023, 213, 119022. [Google Scholar] [CrossRef]
- Ray, R.K.; Linkon, A.A.; Bhuiyan, M.S.; Jewel, R.M.; Anjum, N.; Ghosh, B.P.; Mia, M.T.; Badruddowza; Sarker, M.S.U.; Shaima, M. Transforming Breast Cancer Identification: An In-Depth Examination of Advanced Machine Learning Models Applied to Histopathological Images. J. Comput. Sci. Technol. Stud. 2024, 6, 155–161. [Google Scholar] [CrossRef]
- Rajkumar, R.; Gopalakrishnan, S.; Praveena, K.; Venkatesan, M.; Ramamoorthy, K.; Hephzipah, J.J. Darknet-53 convolutional neural network-based image processing for breast cancer detection. Mesop. J. Artif. Intell. Healthc. 2024, 2024, 59–68. [Google Scholar] [CrossRef] [PubMed]
- Alhassan, A.M. Enhanced pre-processing technique for histopathological image stain normalization and cancer detection. Multimed. Tools Appl. 2024, 84, 29733–29761. [Google Scholar] [CrossRef]
- Addo, D.; Zhou, S.; Sarpong, K.; Nartey, O.T.; Abdullah, M.A.; Ukwuoma, C.C.; Al-antari, M.A. A hybrid lightweight breast cancer classification framework using the histopathological images. Biocybern. Biomed. Eng. 2024, 44, 31–54. [Google Scholar] [CrossRef]
- Sampath, N.; Srinath, N. Breast cancer detection from histopathological image dataset using hybrid convolution neural network. Int. J. Model. Simul. Sci. Comput. 2024, 15, 2441003. [Google Scholar] [CrossRef]
- Yu, D.; Lin, J.; Cao, T.; Chen, Y.; Li, M.; Zhang, X. SECS: An effective CNN joint construction strategy for breast cancer histopathological image classification. J. King Saud Univ.-Comput. Inf. Sci. 2023, 35, 810–820. [Google Scholar] [CrossRef]
- Zarif, S.; Abdulkader, H.; Elaraby, I.; Alharbi, A.; Elkilani, W.S.; Pławiak, P. Using hybrid pre-trained models for breast cancer detection. PLoS ONE 2024, 19, e0296912. [Google Scholar] [CrossRef]
- Maheshwari, N.U.; SatheesKumaran, S. Automatic Mitosis and Nuclear Atypia Detection for Breast Cancer Grading in Histopathological Images using Hybrid Machine Learning Technique. Multimed. Tools Appl. 2024, 83, 90105–90132. [Google Scholar] [CrossRef]
- Naz, A.; Khan, H.; Din, I.U.; Ali, A.; Husain, M. An Efficient Optimization System for Early Breast Cancer Diagnosis based on Internet of Medical Things and Deep Learning. Eng. Technol. Appl. Sci. Res. 2024, 14, 15957–15962. [Google Scholar] [CrossRef]
- Sharmin, S.; Ahammad, T.; Talukder, M.A.; Ghose, P. A hybrid dependable deep feature extraction and ensemble-based machine learning approach for breast cancer detection. IEEE Access 2023, 11, 87694–87708. [Google Scholar] [CrossRef]
- Tsietso, D.; Yahya, A.; Samikannu, R.; Tariq, M.U.; Babar, M.; Qureshi, B.; Koubaa, A. Multi-input deep learning approach for breast cancer screening using thermal infrared imaging and clinical data. IEEE Access 2023, 11, 52101–52116. [Google Scholar] [CrossRef]
- Chatterjee, S.; Biswas, S.; Majee, A.; Sen, S.; Oliva, D.; Sarkar, R. Breast cancer detection from thermal images using a Grunwald-Letnikov-aided Dragonfly algorithm-based deep feature selection method. Comput. Biol. Med. 2022, 141, 105027. [Google Scholar] [CrossRef] [PubMed]
- Roubidoux, M.A.; Sabel, M.S.; Bailey, J.E.; Kleer, C.G.; Klein, K.A.; Helvie, M.A. Breast Scans—Dynamic Thermal Imaging—nydti.com. Available online: https://www.nydti.com/services/breast-scans/ (accessed on 9 December 2024).
- Civilibal, S.; Cevik, K.K.; Bozkurt, A. A deep learning approach for automatic detection, segmentation and classification of breast lesions from thermal images. Expert Syst. Appl. 2023, 212, 118774. [Google Scholar] [CrossRef]
- Ikechukwu, A.V.; Bhimshetty, S.; Deepu, R.; Mala, M. Advances in Thermal Imaging: A Convolutional Neural Network Approach for Improved Breast Cancer Diagnosis. In Proceedings of the 2024 International Conference on Distributed Computing and Optimization Techniques (ICDCOT), Bengaluru, India, 15–16 March 2024; pp. 1–7. [Google Scholar]
- Mahoro, E.; Akhloufi, M.A. Breast cancer classification on thermograms using deep CNN and transformers. Quant. Infrared Thermogr. J. 2024, 21, 30–49. [Google Scholar] [CrossRef]
- Al Husaini, M.A.; Habaebi, M.H.; Elsheikh, E.A.; Islam, M.R.; Suliman, F.; Husaini, Y.N.A. Evaluating the Effect of Noisy Thermal Images On the Detection of Early Breast Cancer Using Deep Learning. Res. Sq. 2024, 5, 3923–3953. [Google Scholar]
- Al Husaini, M.A.S.; Habaebi, M.H.; Islam, M.R. Real-time thermography for breast cancer detection with deep learning. Discov. Artif. Intell. 2024, 4, 57. [Google Scholar] [CrossRef]
- Ahmed, K.S.; Sherif, F.F.; Abdallah, M.S.; Cho, Y.I.; ElMetwally, S.M. An Innovative Thermal Imaging Prototype for Precise Breast Cancer Detection: Integrating Compression Techniques and Classification Methods. Bioengineering 2024, 11, 764. [Google Scholar] [CrossRef]
- Pramanik, R.; Pramanik, P.; Sarkar, R. Breast cancer detection in thermograms using a hybrid of GA and GWO based deep feature selection method. Expert Syst. Appl. 2023, 219, 119643. [Google Scholar] [CrossRef]
- Moayedi, S.M.Z.; Rezai, A.; Hamidpour, S.S.F. Toward Effective Breast Cancer Detection in Thermal Images Using Efficient Feature Selection Algorithm and Feature Extraction Methods. Biomed. Eng. Appl. Basis Commun. 2024, 36, 2450007. [Google Scholar] [CrossRef]
- Rahman, M.A.; Hamada, M.; Sharmin, S.; Rimi, T.A.; Talukder, A.S.; Imran, N.; Kobra, K.; Ahmed, M.R.; Rabbi, M.; Matin, M.M.H.; et al. Enhancing Early Breast Cancer Detection through Advanced Data Analysis. IEEE Access 2024, 12, 161941–161953. [Google Scholar] [CrossRef]
- Manikandan, P.; Durga, U.; Ponnuraja, C. An integrative machine learning framework for classifying SEER breast cancer. Sci. Rep. 2023, 13, 5362. [Google Scholar] [CrossRef]
- Albadr, M.A.A.; AL-Dhief, F.T.; Man, L.; Arram, A.; Abbas, A.H.; Homod, R.Z. Online sequential extreme learning machine approach for breast cancer diagnosis. Neural Comput. Appl. 2024, 36, 10413–10429. [Google Scholar] [CrossRef]
- Islam, M.R.; Islam, M.S.; Majumder, S. Breast Cancer Prediction: A Fusion of Genetic Algorithm, Chemical Reaction Optimization, and Machine Learning Techniques. Appl. Comput. Intell. Soft Comput. 2024, 2024, 7221343. [Google Scholar] [CrossRef]
- Valencia-Moreno, J.M.; Gonzalez-Fraga, J.A.; Gutierrez-Lopez, E.; Estrada-Senti, V.; Cantero-Ronquillo, H.A.; Kober, V. Breast cancer risk estimation with intelligent algorithms and risk factors for Cuban women. Comput. Biol. Med. 2024, 179, 108818. [Google Scholar] [CrossRef]
- Heidelbaugh, J.J. Breast Cancer: A Multidisciplinary Approach; Elsevier: Amsterdam, The Netherlands, 2024. [Google Scholar]
- Jochelson, M. Advanced imaging techniques for the detection of breast cancer. Am. Soc. Clin. Oncol. Educ. Book 2012, 32, 65–69. [Google Scholar] [CrossRef]
- Basurto-Hurtado, J.A.; Cruz-Albarran, I.A.; Toledano-Ayala, M.; Ibarra-Manzano, M.A.; Morales-Hernandez, L.A.; Perez-Ramirez, C.A. Diagnostic Strategies for Breast Cancer Detection: From Image Generation to Classification Strategies Using Artificial Intelligence Algorithms. Cancers 2022, 14, 3442. [Google Scholar] [CrossRef]
- Iranmakani, S.; Mortezazadeh, T.; Sajadian, F.; Ghaziani, M.F.; Ghafari, A.; Khezerloo, D.; Musa, A.E. A review of various modalities in breast imaging: Technical aspects and clinical outcomes. Egypt. J. Radiol. Nucl. Med. 2020, 51, 57. [Google Scholar] [CrossRef]
- Caballo, M.; Pangallo, D.R.; Mann, R.M.; Sechopoulos, I. Deep learning-based segmentation of breast masses in dedicated breast CT imaging: Radiomic feature stability between radiologists and artificial intelligence. Comput. Biol. Med. 2020, 118, 103629. [Google Scholar] [CrossRef] [PubMed]
- Roslidar, R.; Rahman, A.; Muharar, R.; Syahputra, M.R.; Arnia, F.; Syukri, M.; Pradhan, B.; Munadi, K. A review on recent progress in thermal imaging and deep learning approaches for breast cancer detection. IEEE Access 2020, 8, 116176–116194. [Google Scholar] [CrossRef]
- Rasheed, M.E.H.; Youseffi, M. Breast Cancer and Medical Imaging; IOP Publishing: Bristol, UK, 2024. [Google Scholar] [CrossRef]
- Al Jarroudi, O.; El Bairi, K.; Curigliano, G. Breast Cancer Research and Treatment: Innovative Concepts; Springer Nature: Berlin, Germany, 2024; Volume 188. [Google Scholar]
- Darakh, A.; Shah, A.; Oza, P. Exploring the Benefits of Data Augmentation for Breast Cancer Classification using Transfer Learning. In Proceedings of the World Conference on Information Systems for Business Management, Bangkok, Thailand, 7–8 September; Springer: Berlin/Heidelberg, Germany, 2023; pp. 509–520. [Google Scholar]
- Adama, S.; dite Soukoura, D.F.; Ismaël, K.; Lahsen, B. Reliable Medical Data Augmentation for Deep Learning: A Case Study on Breast Cancer Prediction. In Proceedings of the 2024 Sixth International Conference on Intelligent Computing in Data Sciences (ICDS), Marrakech, Morocco, 23–24 October 2024; pp. 1–10. [Google Scholar]
- Talwar, B.; Arora, A.; Bharany, S. An energy efficient agent aware proactive fault tolerance for preventing deterioration of virtual machines within cloud environment. In Proceedings of the 2021 9th International Conference on Reliability, Infocom Technologies and Optimization (Trends and Future Directions) (ICRITO), Noida, India, 3–4 September 2021; pp. 1–7. [Google Scholar]
- Emmanuel, A.A.; Awokola, J.A.; Alam, S.; Bharany, S.; Agboola, P.; Shuaib, M.; Ahmed, R. A hybrid framework of blockchain and IoT technology in the pharmaceutical industry: A comprehensive study. Mob. Inf. Syst. 2023, 2023, 3265310. [Google Scholar] [CrossRef]
- Sundas, A.; Badotra, S.; Shahi, G.S.; Verma, A.; Bharany, S.; Ibrahim, A.O.; Abulfaraj, A.W.; Binzagr, F. Smart patient monitoring and recommendation (SPMR) using cloud analytics and deep learning. IEEE Access 2024, 12, 54238–54255. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).