Inter-Species Competition of Mono- or Dual Species Biofilms- of MDR-Staphylococcus aureus and Pseudomonas aeruginosa Promotes the Killing Efficacy of Phage or Phage Cocktail
Abstract
1. Introduction
2. Methods and Materials
2.1. Bacterial Strains, Bacteriophages, and Growth Conditions
2.2. Determination of Biofilm Biomass of Single or Dual Species
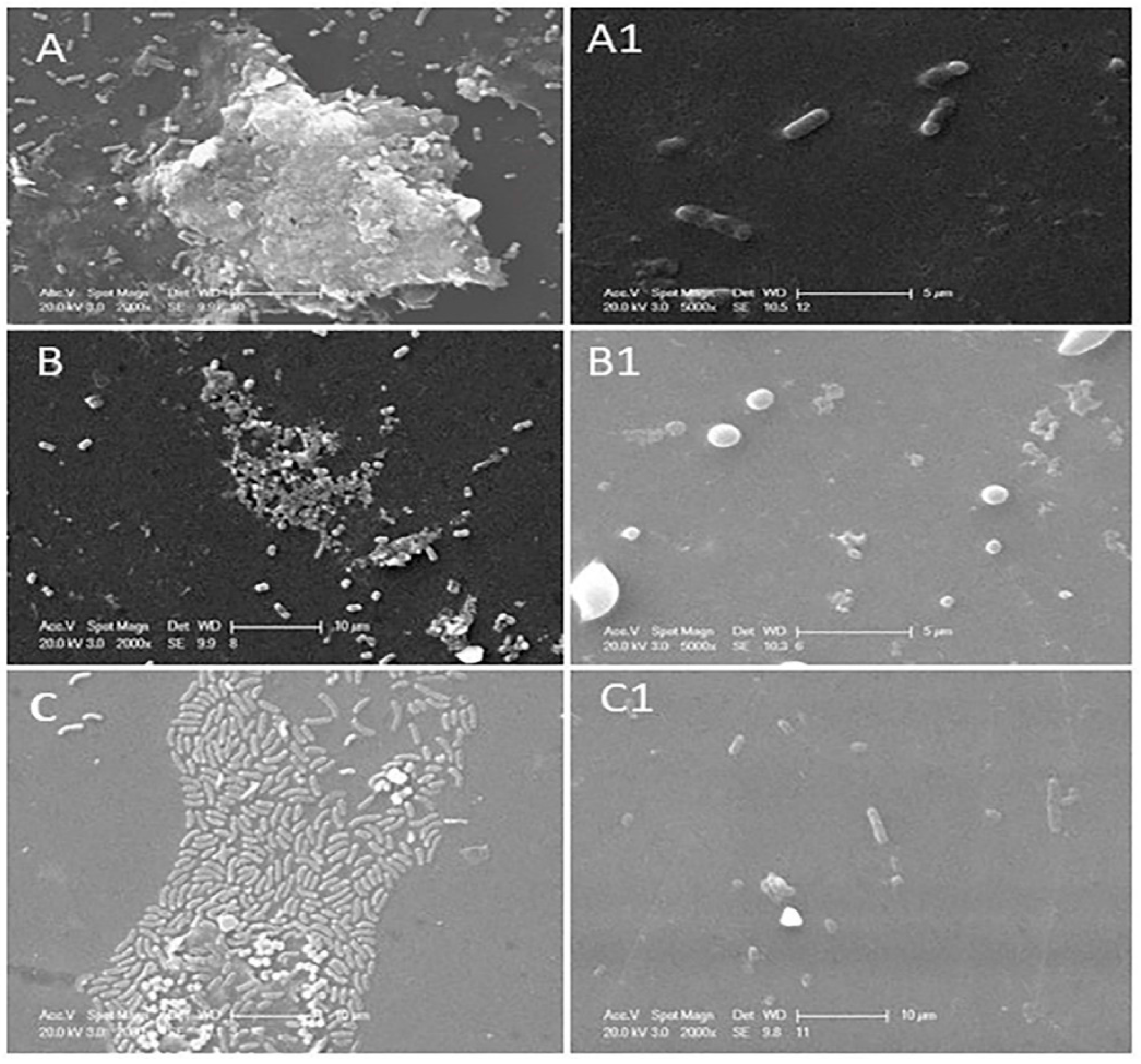
2.3. Scanning Electron Microscopy
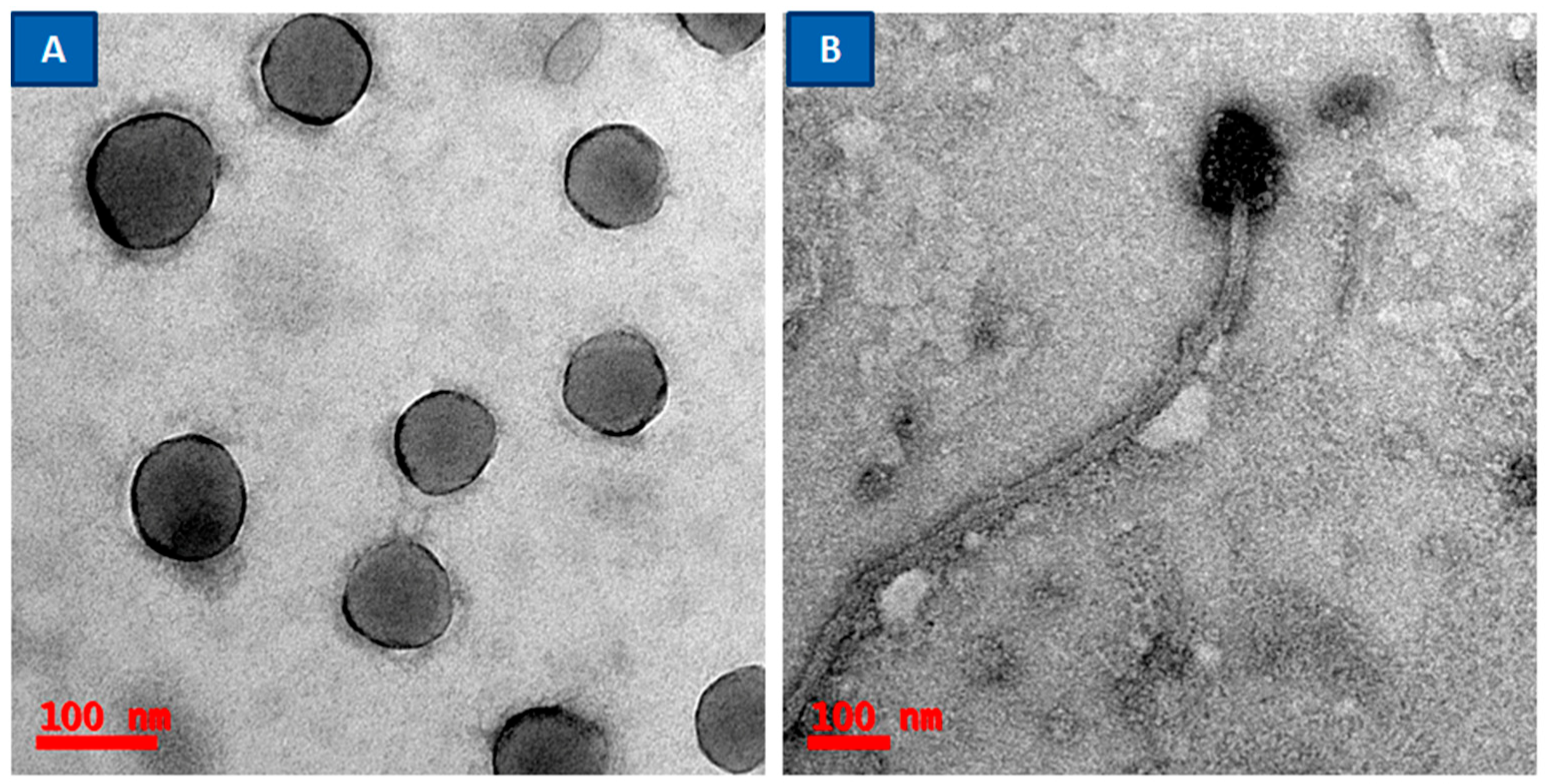
2.4. Confocal Laser Scanning Microscopy
3. Results
3.1. Phage or Phage Cocktail Action on Biofilm Biomass
3.2. Determination of Phage or Phage Cocktail Lytic Action on Single- or Dual-Species Biofilm by Using SEM
3.3. Phage or Phage Cocktail Lytic Action on Single- or Dual-Species Biofilm by Using CLSM
4. Discussion
5. Conclusions
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Tian, F.; Li, J.; Nazir, A.; Tong, Y. Bacteriophage—A promising alternative measure for bacterial biofilm control. Infect. Drug Resist. 2021, 14, 205–217. [Google Scholar] [CrossRef] [PubMed]
- Awuor, S.O.; Omwenga, E.O.; Mariita, R.M.; Musila, J.M.; Musyoki, S. Monitoring the battleground: Exploring antimicrobial resistance and virulence factors in wound bacterial isolates. Access Microbiol. 2023, 5, 000613.v6. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Baral, P.; Afnan, N.; Ahmad Zahra, M.; Akter, B.; Rabia Prapti, S.; Muazzam Hossan, M.; Haque, F.K.M. Bacteriological analysis and antibiotic resistance in patients with diabetic foot ulcers in Dhaka. PLoS ONE 2024, 19, e0301767. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Pallavali, R.R.; Avula, S.; Degati, V.L.; Penubala, M.; Damu, A.G.; Durbaka, V.R.P. Data of antibacterial activity of plant leaves crude extract on bacterial isolates of wound infections. Data Brief 2019, 24, 103896. [Google Scholar] [CrossRef]
- Pallavali, R.R.; Degati, V.L.; Lomada, D.; Reddy, M.C.; Durbaka, V.R.P. Isolation and in vitro evaluation of bacteriophages against MDR-bacterial isolates from septic wound infections. PLoS ONE 2017, 12, e0179245. [Google Scholar] [CrossRef] [PubMed]
- Pallavali, R.R.; Degati, V.L.; Narala, V.R.; Velpula, K.K.; Yenugu, S.; Durbaka, V.R.P. Lytic Bacteriophages Against Bacterial Biofilms Formed by Multidrug-Resistant Pseudomonas aeruginosa, Escherichia coli, Klebsiella pneumoniae, and Staphylococcus aureus Isolated from Burn Wounds. Phage 2021, 2, 120–130. [Google Scholar] [CrossRef]
- Pallavali, R.; Shin, D.; Choi, J. Phage-based Biocontrol of antibiotic-resistant bacterium isolated from Livestock Wastewater Treatment Plant. Water 2023, 15, 1616. [Google Scholar] [CrossRef]
- Sagar, S.S.; Kumar, R.; Kaistha, S.D. Efficacy of Phage and Ciprofloxacin Co-therapy on the Formation and Eradication of Pseudomonas aeruginosa Biofilms. Arab. J. Sci. Eng. 2017, 42, 95–103. [Google Scholar] [CrossRef]
- Liu, S.; Lu, H.; Zhang, S.; Shi, Y.; Chen, Q. Phages against Pathogenic Bacterial Biofilms and Biofilm-Based Infections: A Review. Pharmaceutics 2022, 14, 427. [Google Scholar] [CrossRef]
- Zhu, J.; Miller, M.B.; Vance, R.E.; Dziejman, M.; Bassler, B.L.; Mekalanos, J.J. Quorum-sensing regulators control virulence gene expression in Vibrio cholerae. Proc. Natl. Acad. Sci. USA 2002, 99, 3129–3134. [Google Scholar] [CrossRef]
- Gutiérrez, D.; Fernández, L.; Rodríguez, A.; García, P. Are phage lytic proteins the secret weapon to kill staphylococcus aureus? mBio 2018, 9, e01923-17. [Google Scholar] [CrossRef]
- Fernández, L.; González, S.; Campelo, A.B.; Martínez, B.; Rodríguez, A.; García, P. Downregulation of autolysin-encoding genes by phage-derived lytic proteins inhibits biofilm formation in Staphylococcus aureus. Antimicrob. Agents Chemother. 2017, 61, 1–13. [Google Scholar] [CrossRef] [PubMed]
- Schulze, A.; Mitterer, F.; Pombo, J.P.; Schild, S. Biofilms by bacterial human pathogens: Clinical relevance—Development, composition and regulation—Therapeutical strategies. Microb. Cell 2021, 8, 28–56. [Google Scholar] [CrossRef]
- Vestby, L.K.; Grønseth, T.; Simm, R.; Nesse, L.L. Bacterial biofilm and its role in the pathogenesis of disease. Antibiotics 2020, 9, 59. [Google Scholar] [CrossRef]
- Hall, C.W.; Mah, T.F. Molecular mechanisms of biofilm-based antibiotic resistance and tolerance in pathogenic bacteria. FEMS Microbiol. Rev. 2017, 41, 276–301. [Google Scholar] [CrossRef] [PubMed]
- Duarte, A.C.; Fernández, L.; de Maesschalck, V.; Gutiérrez, D.; Campelo, A.B.; Briers, Y.; Lavigne, R.; Rodríguez, A.; García, P. Synergistic action of phage phiIPLA-RODI and lytic protein CHAPSH3b: A combination strategy to target Staphylococcus aureus biofilms. NPJ Biofilms Microbiomes 2021, 7, 39. [Google Scholar] [CrossRef]
- Pinto, A.M.; Cerqueira, M.A.; Bañobre-Lópes, M.; Pastrana, L.M.; Sillankorva, S. Bacteriophages for chronic wound treatment: From traditional to novel delivery systems. Viruses 2020, 12, 235. [Google Scholar] [CrossRef]
- Gutiérrez, D.; Rodríguez-Rubio, L.; Martínez, B.; Rodríguez, A.; García, P. Bacteriophages as weapons against bacterial biofilms in the food industry. Front. Microbiol. 2016, 7, 825. [Google Scholar] [CrossRef]
- Chen, L.; Fan, J.; Yan, T.; Liu, Q.; Zhang, S.Y.H.; Yang, J.; Deng, D.; Huang, S.; Ma, Y. Isolation and Characterization of Specific Phages to Prepare a Cocktail Preventing Vibrio sp. Va-F3 Infections in Shrimp (Litopenaeus vannamei). Front. Microbiol. 2019, 10, 2337. [Google Scholar] [CrossRef]
- Motlagh, A.M.; Bhattacharjee, A.S.; Goel, R. Biofilm control with natural and genetically modified phages. World J. Microbiol. Biotechnol. 2016, 32, 67. [Google Scholar] [CrossRef] [PubMed]
- Al-Razem, F.; Al-Aloul, H.; Ishnaiwer, M.; Qadi, R. Isolation and partial characterization of Salmonella Gallinarum bacteriophage. Saudi J. Biol. Sci. 2022, 29, 3308–3312. [Google Scholar] [CrossRef]
- Bragg, R.R.; Boucher, C.E.; van der Westhuizen, W.A.; Lee, J.-Y. The Potential Use of Bacteriophage Therapy as a Treatment Option in a Post-Antibiotic Era. Antibiot. Resist. Mech. New Antimicrob. Approaches 2016, 15, 309–328. [Google Scholar]
- Moghadam, M.T.; Amirmozafari, N.; Shariati, A.; Hallajzadeh, M.; Mirkalantari, S.; Khoshbayan, A.; Jazi, F.M. How phages overcome the challenges of drug resistant bacteria in clinical infections. Infect. Drug Resist. 2020, 13, 45–61. [Google Scholar] [CrossRef]
- Viertel, T.M.; Ritter, K.; Horz, H.P. Viruses versus bacteria-novel approaches to phage therapy as a tool against multidrug-resistant pathogens. J. Antimicrob. Chemother. 2014, 69, 2326–2336. [Google Scholar] [CrossRef]
- Abedon, S.T.; Danis-Wlodarczyk, K.M.; Wozniak, D.J.; Sullivan, M.B. Improving phage-biofilm in vitro experimentation. Viruses 2021, 13, 1175. [Google Scholar] [CrossRef] [PubMed]
- Wagner, E.M.; Fischel, K.; Rammer, N.; Beer, C.; Palmetzhofer, A.L.; Conrady, B.; Roch, F.-F.; Hanson, B.T.; Wagner, M.; Rychli, K. Bacteria of eleven different species isolated from biofilms in a meat processing environment have diverse biofilm forming abilities. Int. J. Food Microbiol. 2021, 349, 109232. [Google Scholar] [CrossRef]
- Muhammad, M.H.; Idris, A.L.; Fan, X.; Guo, Y.; Yu, Y.; Jin, X.; Qiu, J.; Guan, X.; Huang, T. Beyond Risk: Bacterial Biofilms and Their Regulating Approaches. Front. Microbiol. 2020, 11, 928. [Google Scholar] [CrossRef] [PubMed]
- O’Neill, E.; Pozzi, C.; Houston, P.; Humphreys, H.; Robinson, D.A.; Loughman, A.; Foster, T.J.; O’Gara, J.P. A novel Staphylococcus aureus biofilm phenotype mediated by the fibronectin-binding proteins, FnBPA and FnBPB. J. Bacteriol. 2008, 190, 3835–3850. [Google Scholar] [CrossRef]
- González, S.; Fernández, L.; Campelo, A.B.; Gutiérrez, D.; Martínez, B.; Rodríguez, A.; García, P. The behavior of Staphylococcus aureus dual-species biofilms treated with bacteriophage phiIPLA-RODI depends on the accompanying microorganism. Appl. Environ. Microbiol. 2017, 83, e02821-16. [Google Scholar] [CrossRef]
- Drago, L.; Agrappi, S.; Bortolin, M.; Toscano, M.; Romanò, C.L.; de Vecchi, E. How to study biofilms after microbial colonization of materials used in orthopaedic implants. Int. J. Mol. Sci. 2016, 17, 293. [Google Scholar] [CrossRef]
- Ayyaru, S.; Choi, J.; Ahn, Y.H. Biofouling reduction in a MBR by the application of a lytic phage on a modified nanocomposite membrane. Environ. Sci. 2018, 4, 1624–1638. [Google Scholar] [CrossRef]
- Baudin, M.; Cinquin, B.; Sclavi, B.; Pareau, D.; Lopes, F. Understanding the fundamental mechanisms of biofilms development and dispersal: BIAM (Biofilm Intensity and Architecture Measurement), a new tool for studying biofilms as a function of their architecture and fluorescence intensity. J. Microbiol. Methods 2017, 140, 47–57. [Google Scholar] [CrossRef]
- Liu, J.; Gao, S.; Dong, Y.; Lu, C.; Liu, Y. Isolation and characterization of bacteriophages against virulent Aeromonas hydrophila. BMC Microbiol. 2020, 20, 141. [Google Scholar] [CrossRef] [PubMed]
- Akturk, E.; Oliveira, H.; Santos, S.B.; Costa, S.; Kuyumcu, S.; Melo, L.D.R.; Azeredo, J. Synergistic action of phage and antibiotics: Parameters to enhance the killing efficacy against mono and dual-species biofilms. Antibiotics 2019, 8, 103. [Google Scholar] [CrossRef] [PubMed]
- Radlinski, L.; Rowe, S.E.; Kartchner, L.B.; Maile, R.; Cairns, B.A.; Vitko, N.P.; Gode, C.J.; Lachiewicz, A.M.; Wolfgang, M.C.; Conlon, B.P. Pseudomonas aeruginosa exoproducts determine antibiotic efficacy against Staphylococcus aureus. PLoS Biol. 2017, 15, e2003981. [Google Scholar] [CrossRef] [PubMed]
- Smith, A.C.; Rice, A.; Sutton, B.; Gabrilska, R.; Wessel, A.K.; Whiteley, M.; Rumbaugh, K.P. Albumin inhibits Pseudomonas aeruginosa quorum sensing and alters polymicrobial interactions. Infect. Immun. 2017, 85, e00116-17. [Google Scholar]
- Tkhilaishvili, T.; Wang, L.; Perka, C.; Trampuz, A.; Moreno, M.G. Using Bacteriophages as a Trojan Horse to the Killing of Dual-Species Biofilm Formed by Pseudomonas aeruginosa and Methicillin Resistant Staphylococcus aureus. Front. Microbiol. 2020, 11, 695. [Google Scholar]
- Szymczak, M.; Pankowski, K.A.; Kwiatek, A.; Grygorcewicz, B.; Karczewska-Golec, J.; Sadowska, K.; Golec, P. An effective antibiofilm strategy based on bacteriophages armed with silver nanoparticles. Sci. Rep. 2024, 14, 9088. [Google Scholar] [CrossRef]
- Melo, L.D.R.; Pinto, G.; Oliveira, F.; Vilas-Boas, D.; Almeida, C.; Sillankorva, S.; Cerca, N.; Azeredo, J. The Protective Effect of Staphylococcus epidermidis Biofilm Matrix against Phage Predation. Viruses 2020, 12, 1076. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]

| S.No. | Bacteria | Biofilm at 24 h | Phage with 4 h |
|---|---|---|---|
| 1. | P. aeruginosa | 0.761 ± 0.031 | 0.18 ± 0.016 |
| 2. | S. aureus | 0.856 ± 0.055 | 0.205 ± 0.018 |
| 3. | P. aeruginosa + S. aureus | 0.67 ± 0.020 | 0.16 ± 0.020 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
RojaRani, P.; Reddy, G.D.; Vijayalakshmi, D.; Prasad, D.V.R.; Choi, J.D. Inter-Species Competition of Mono- or Dual Species Biofilms- of MDR-Staphylococcus aureus and Pseudomonas aeruginosa Promotes the Killing Efficacy of Phage or Phage Cocktail. Appl. Microbiol. 2024, 4, 1247-1256. https://doi.org/10.3390/applmicrobiol4030085
RojaRani P, Reddy GD, Vijayalakshmi D, Prasad DVR, Choi JD. Inter-Species Competition of Mono- or Dual Species Biofilms- of MDR-Staphylococcus aureus and Pseudomonas aeruginosa Promotes the Killing Efficacy of Phage or Phage Cocktail. Applied Microbiology. 2024; 4(3):1247-1256. https://doi.org/10.3390/applmicrobiol4030085
Chicago/Turabian StyleRojaRani, Pallavali, Guda Dinneswara Reddy, Degati Vijayalakshmi, Durbaka Vijaya Raghava Prasad, and Jeong Dong Choi. 2024. "Inter-Species Competition of Mono- or Dual Species Biofilms- of MDR-Staphylococcus aureus and Pseudomonas aeruginosa Promotes the Killing Efficacy of Phage or Phage Cocktail" Applied Microbiology 4, no. 3: 1247-1256. https://doi.org/10.3390/applmicrobiol4030085
APA StyleRojaRani, P., Reddy, G. D., Vijayalakshmi, D., Prasad, D. V. R., & Choi, J. D. (2024). Inter-Species Competition of Mono- or Dual Species Biofilms- of MDR-Staphylococcus aureus and Pseudomonas aeruginosa Promotes the Killing Efficacy of Phage or Phage Cocktail. Applied Microbiology, 4(3), 1247-1256. https://doi.org/10.3390/applmicrobiol4030085






