Characterization and Proteomic Profiling of Hepatocyte-like Cells Derived from Human Wharton’s Jelly Mesenchymal Stromal Cells: De Novo Expression of Liver-Specific Enzymes
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Cellular Isolation Protocol of WJ-MSCs
2.2. Cell Culturing and Passaging
2.3. Flow Cytometry (FC) Analysis
2.4. Immunocytochemistry (ICC) Analysis
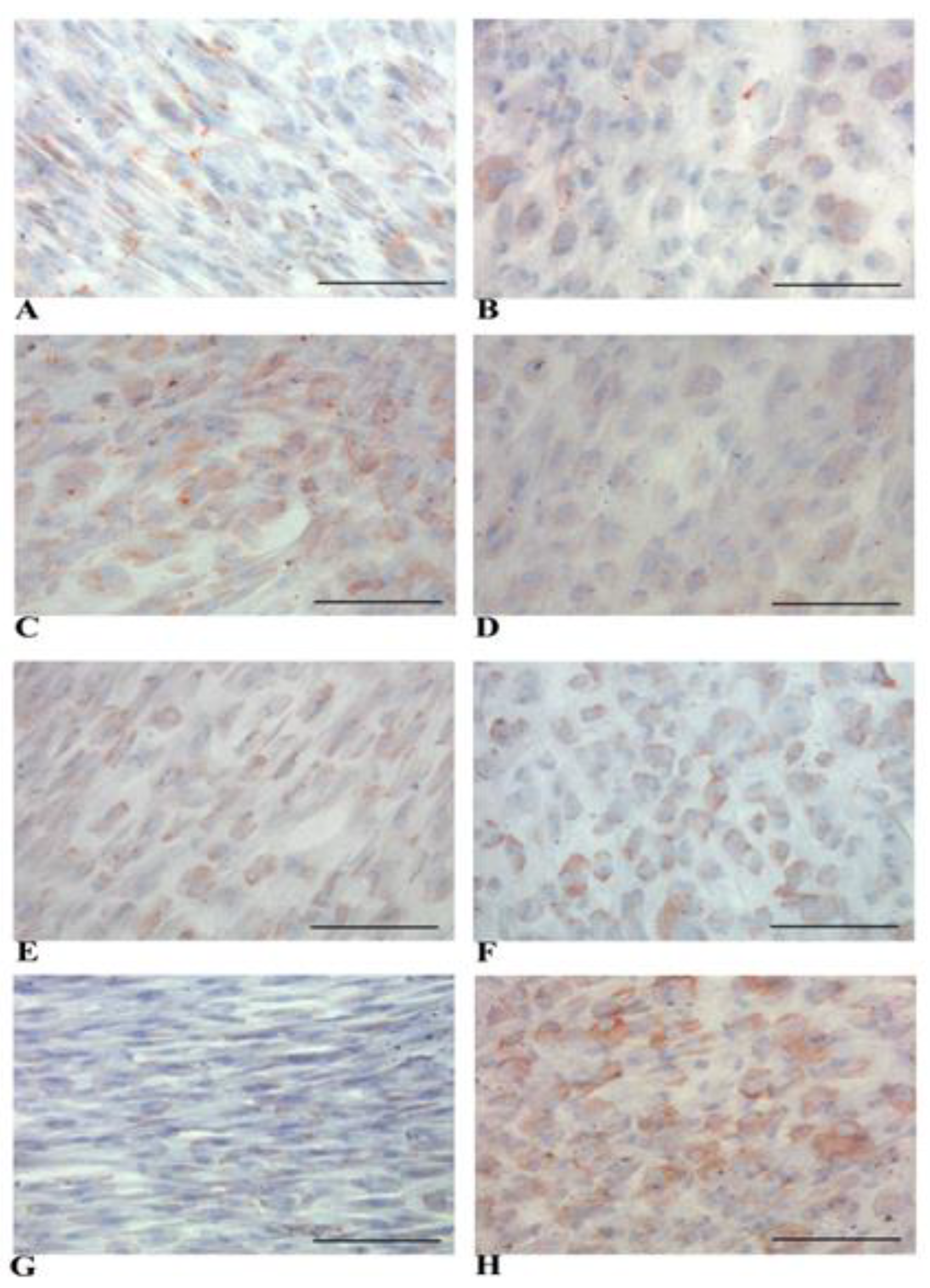
2.5. Immunohistochemistry (IHC) Analysis
2.6. Hepatocyte-Like Cells Differentiation Protocol
2.7. Glycogen Staining with Periodic Acid Schiff (PAS)
2.8. Glucose-6-Phosphatase (G-6-Pase) Activity Assay
2.9. CYP450 3A4 Metabolic Activity Assay
2.10. Sample Preparation for Proteome Analyses
2.11. In-Solution Digestion
2.12. High-Performance Liquid Chromatography (HPLC) and Electrospray MS
2.13. Proteomic Data Analysis
2.14. In Silico Analysis
2.15. Label-Free Quantification and Data Analysis
2.16. Statistical Analyses
3. Results
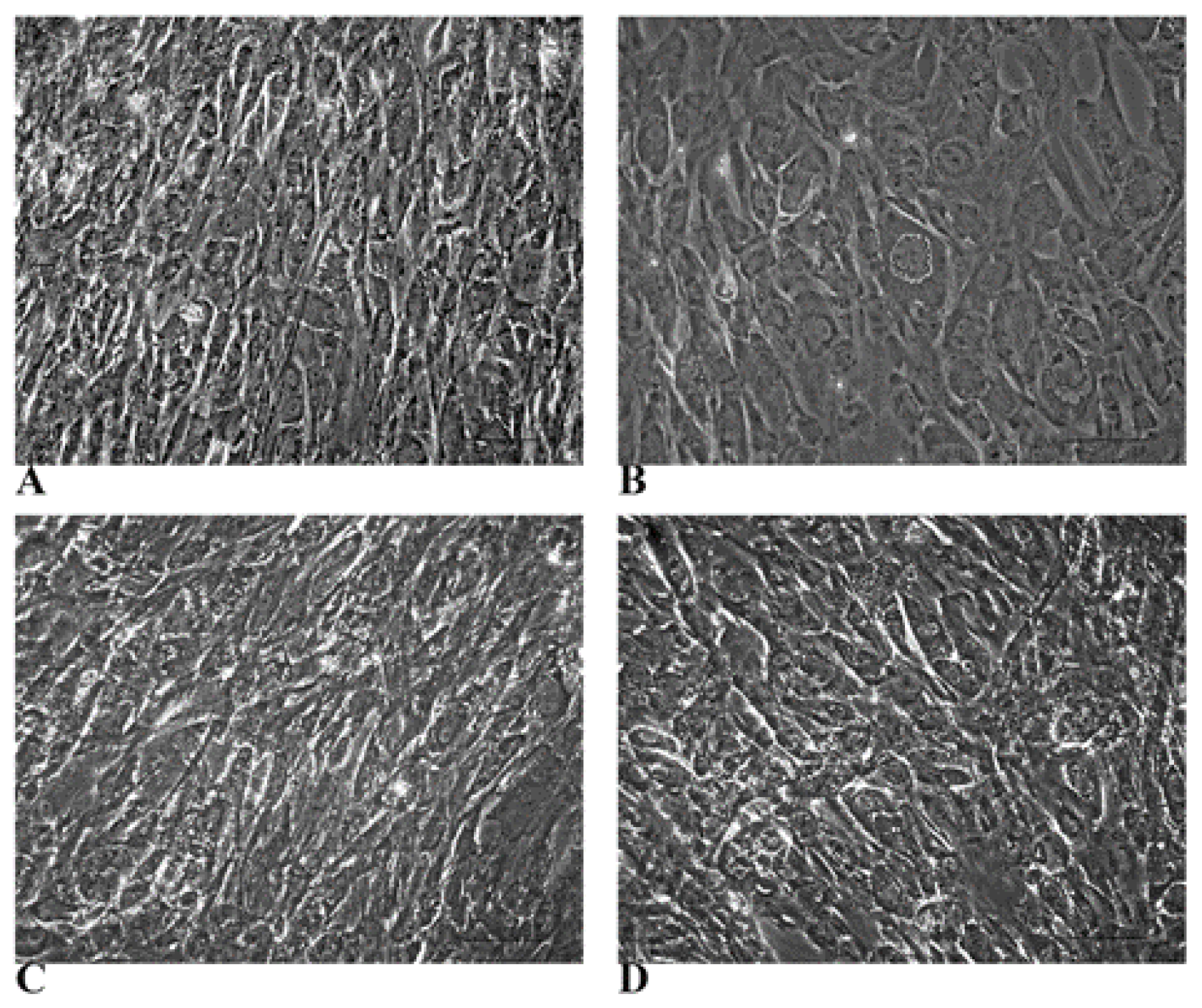
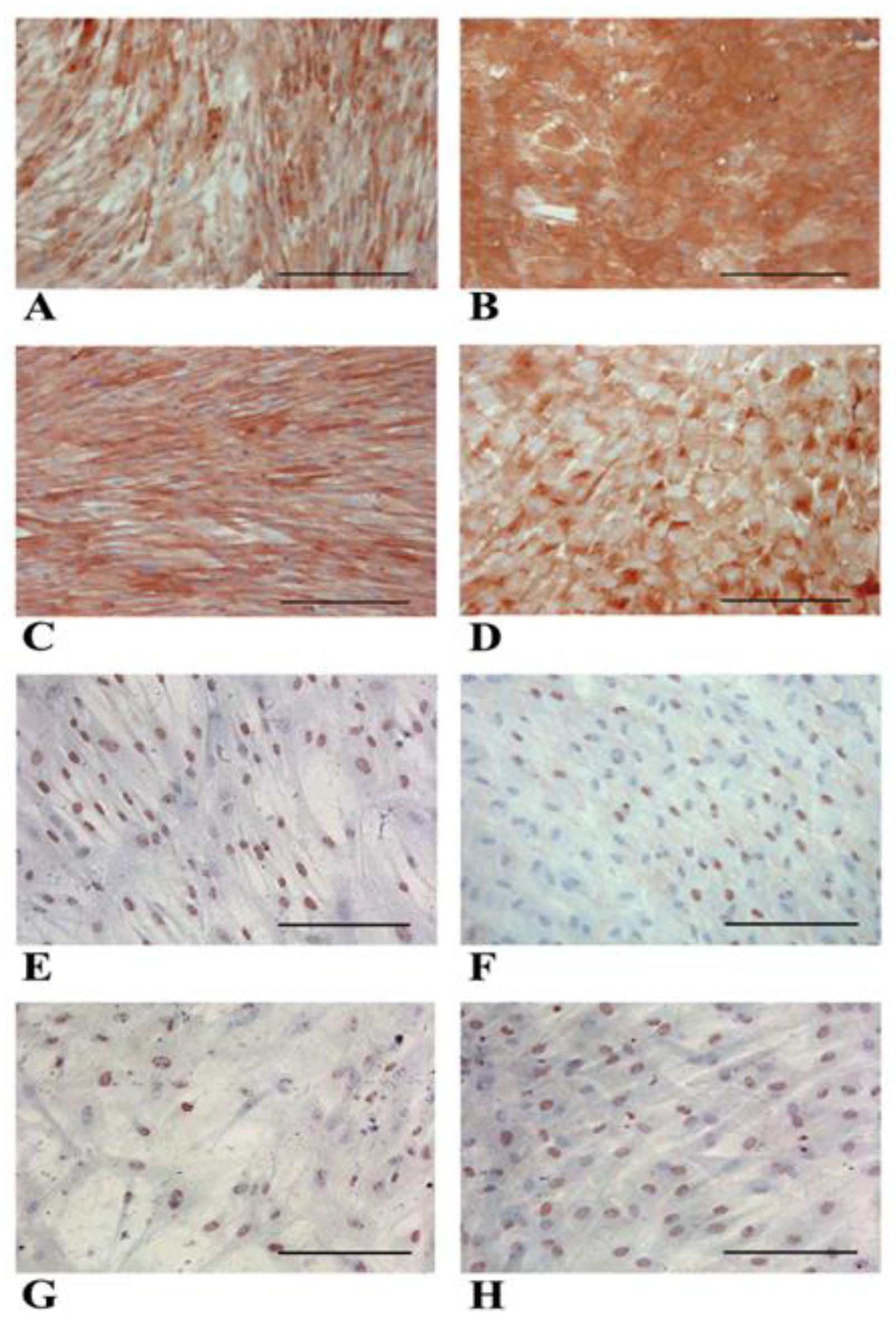
3.1. WJ-MSCs Show Mesenchymal Phenotype and Intrinsic Hepatogenic Potential
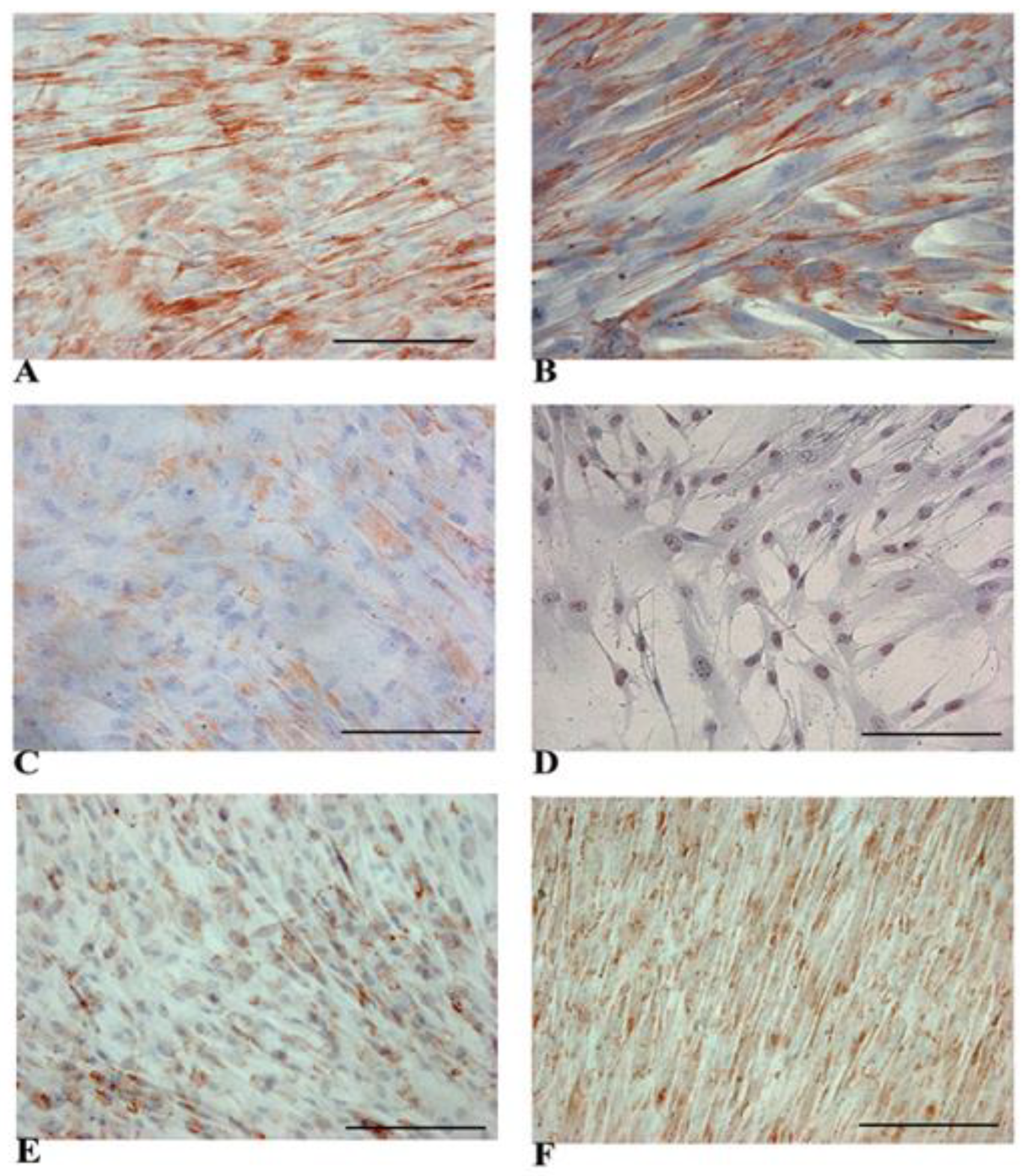
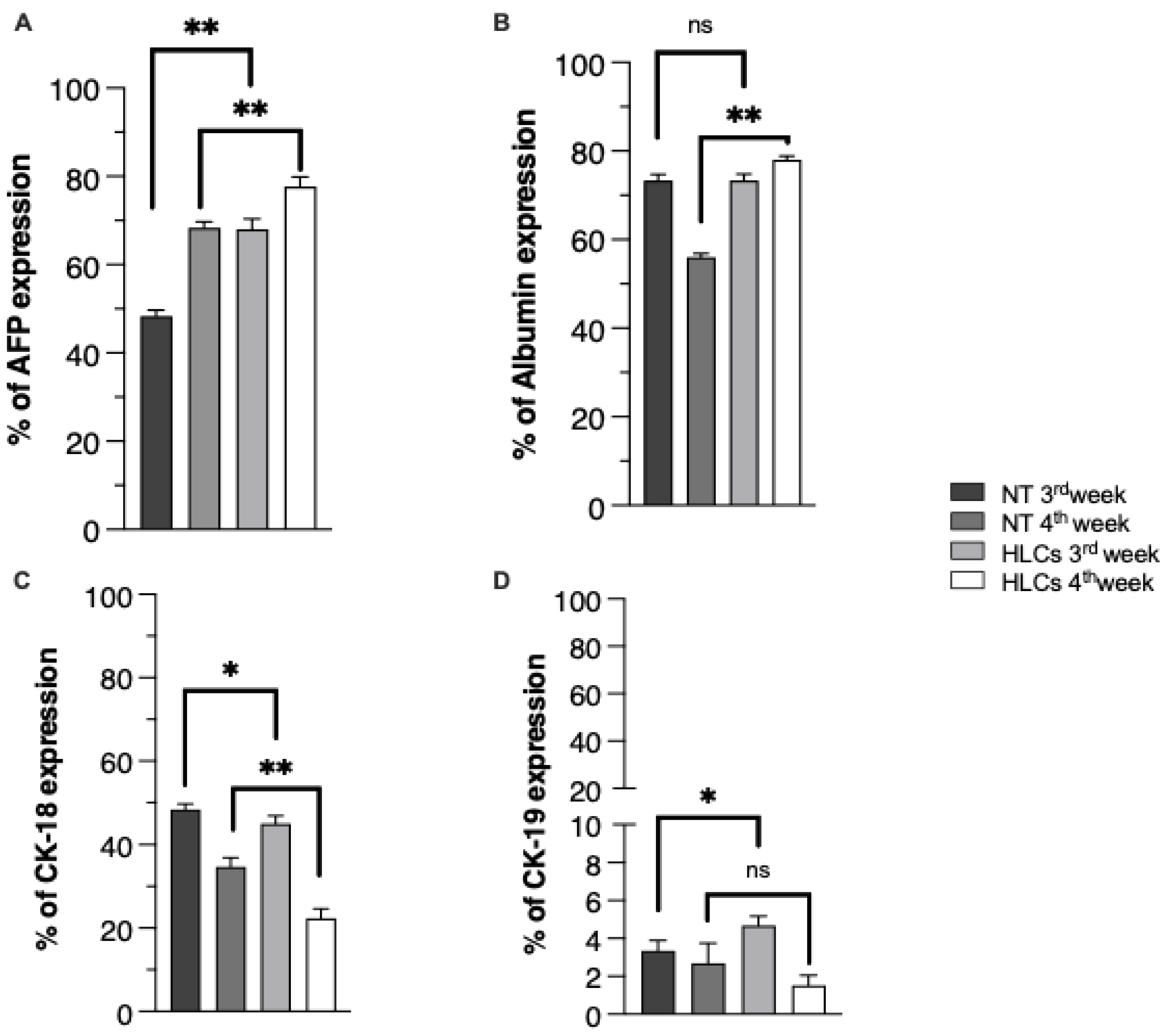
3.2. Evidence of Hepatic Phenotype Acquisition in WJ-MSCs After Hepatogenic Differentiation Protocol
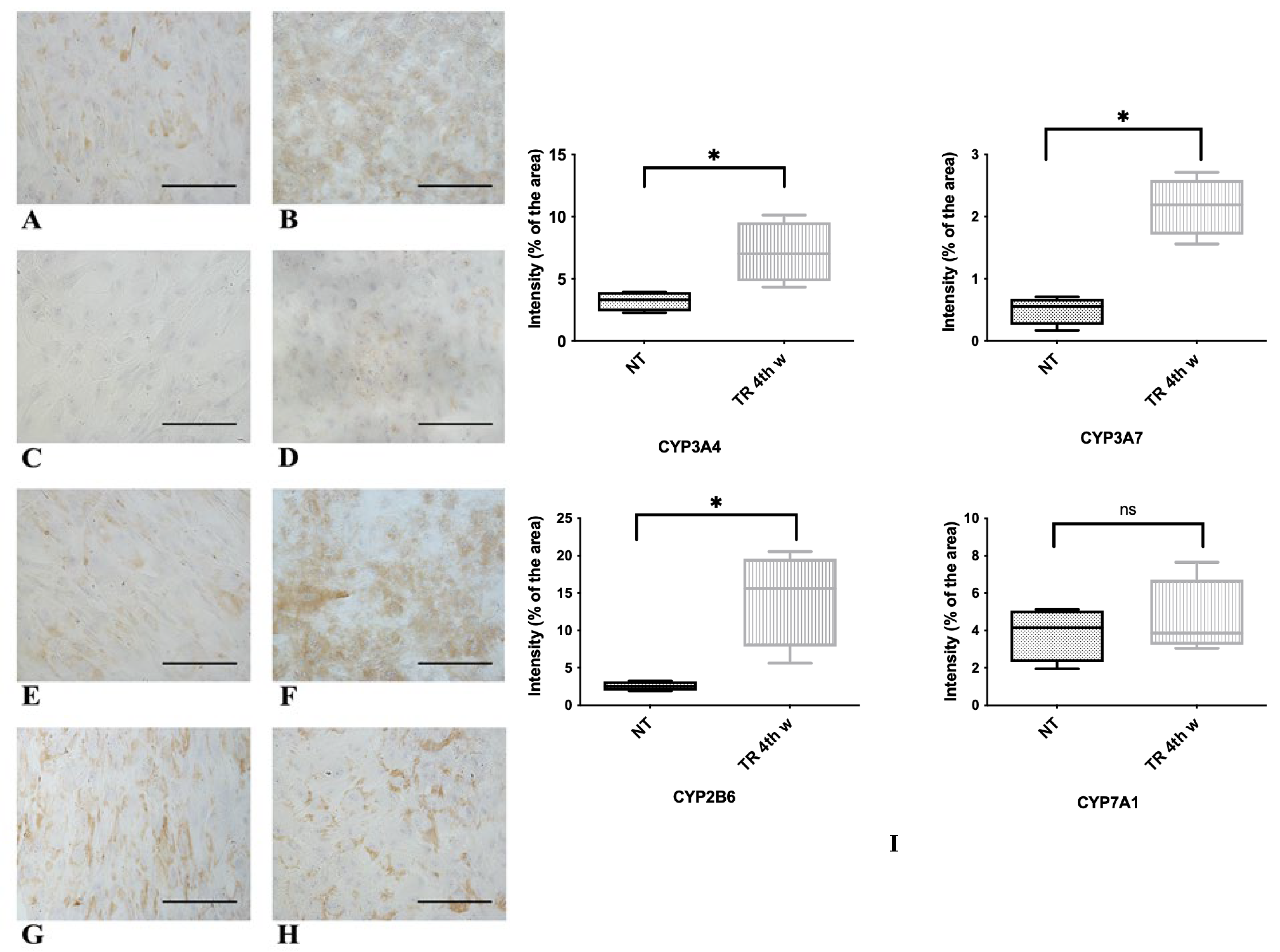
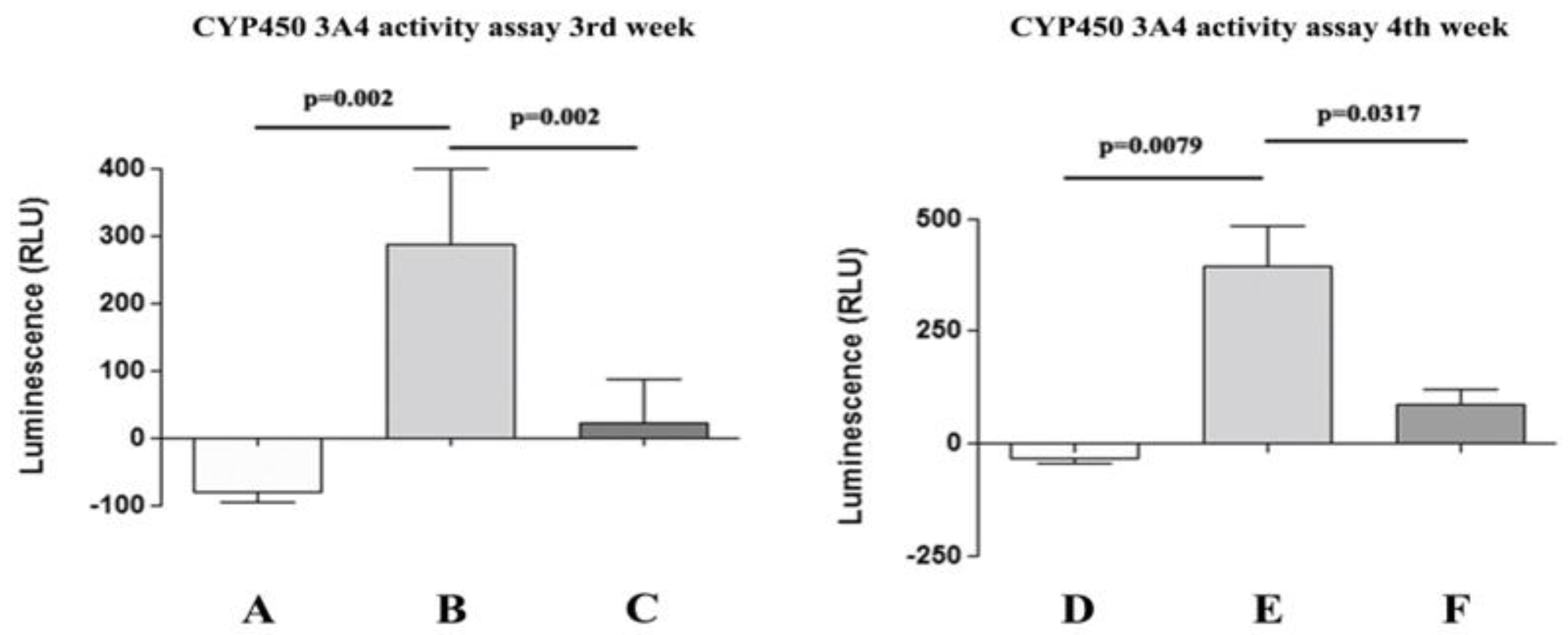
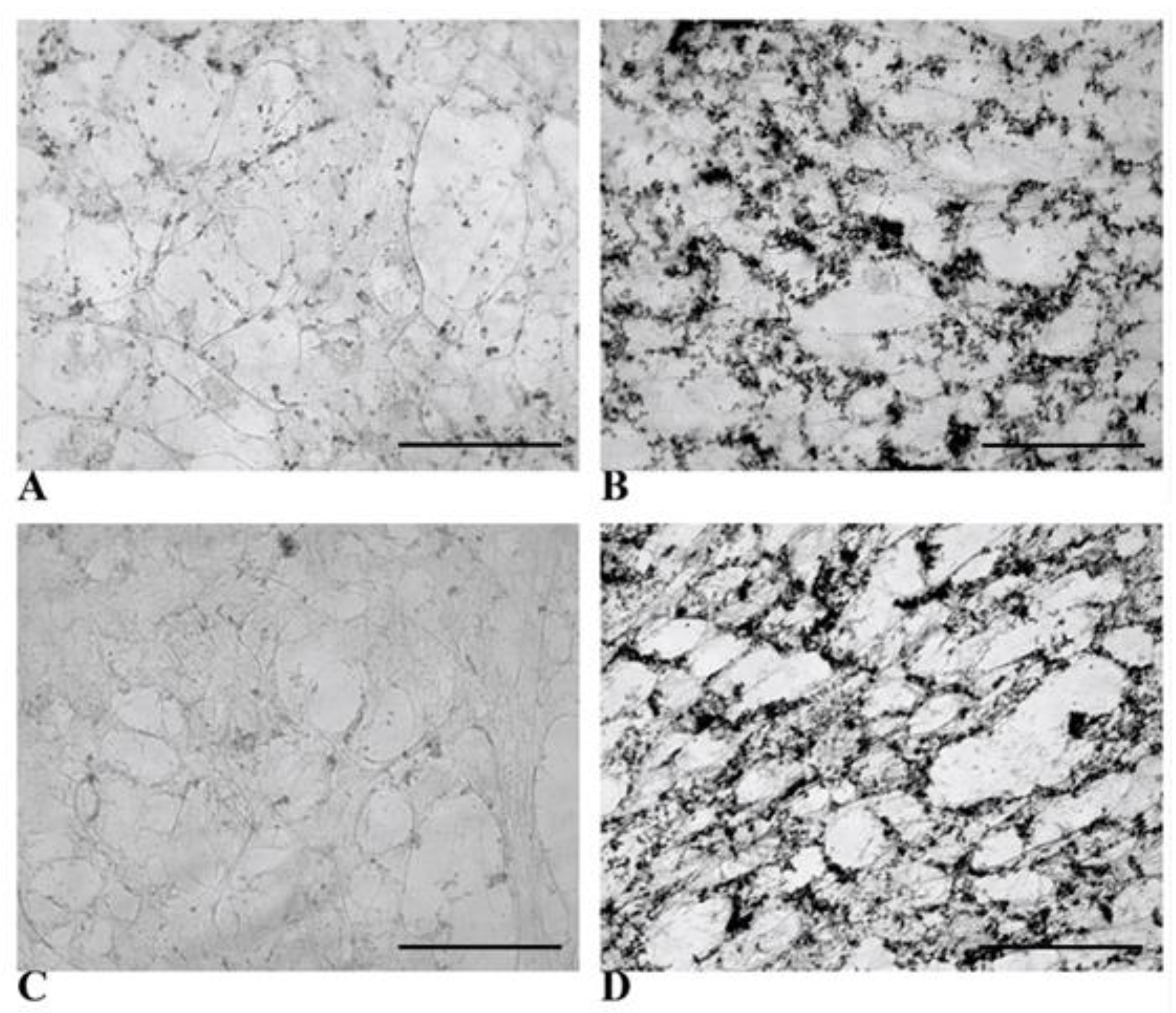
3.3. Functional Features Acquired by WJ-MSCs After Hepatic Differentiation
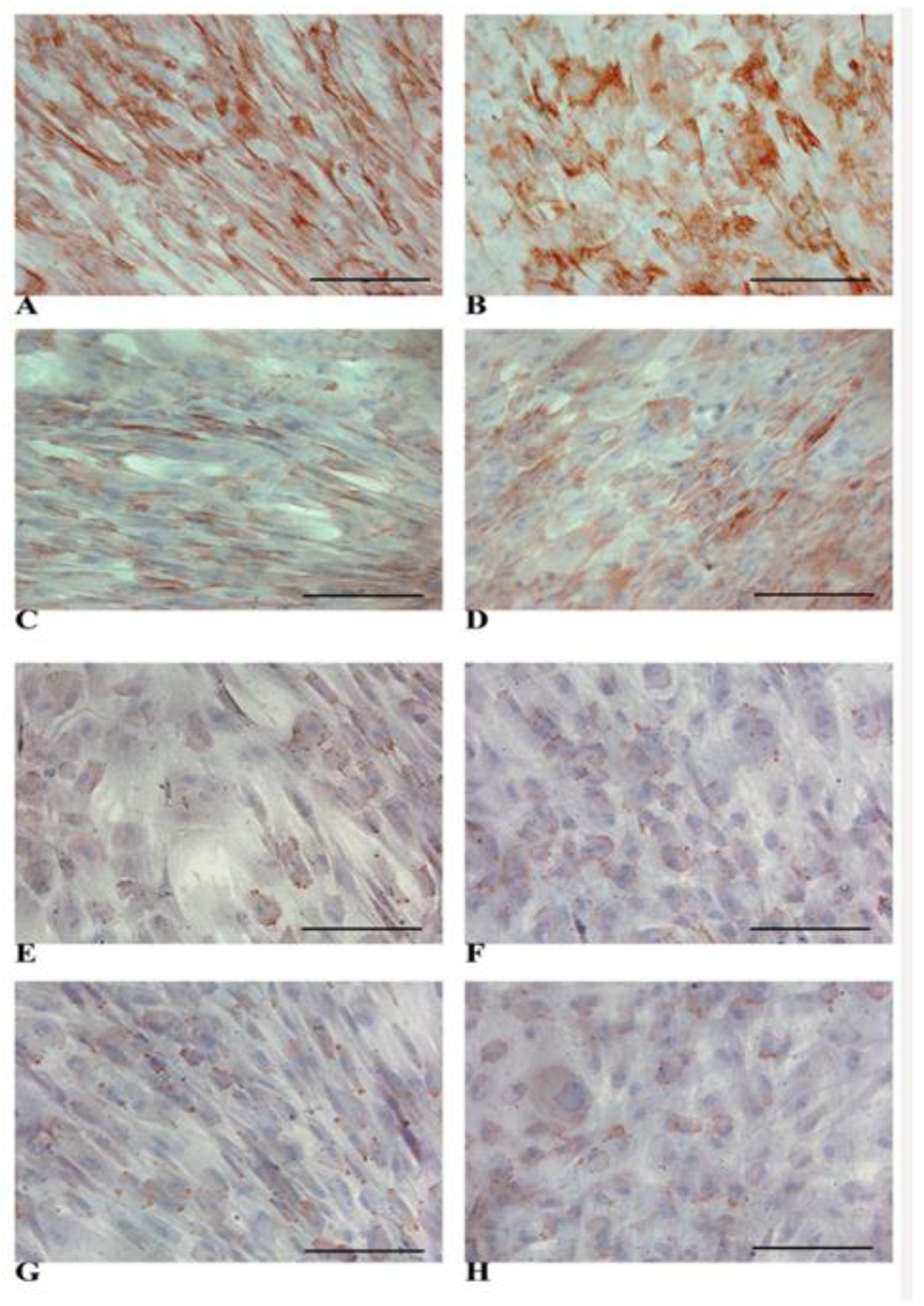
3.4. HLCs Maintain the Immunomodulatory Molecules Responsible for Immune Tolerance
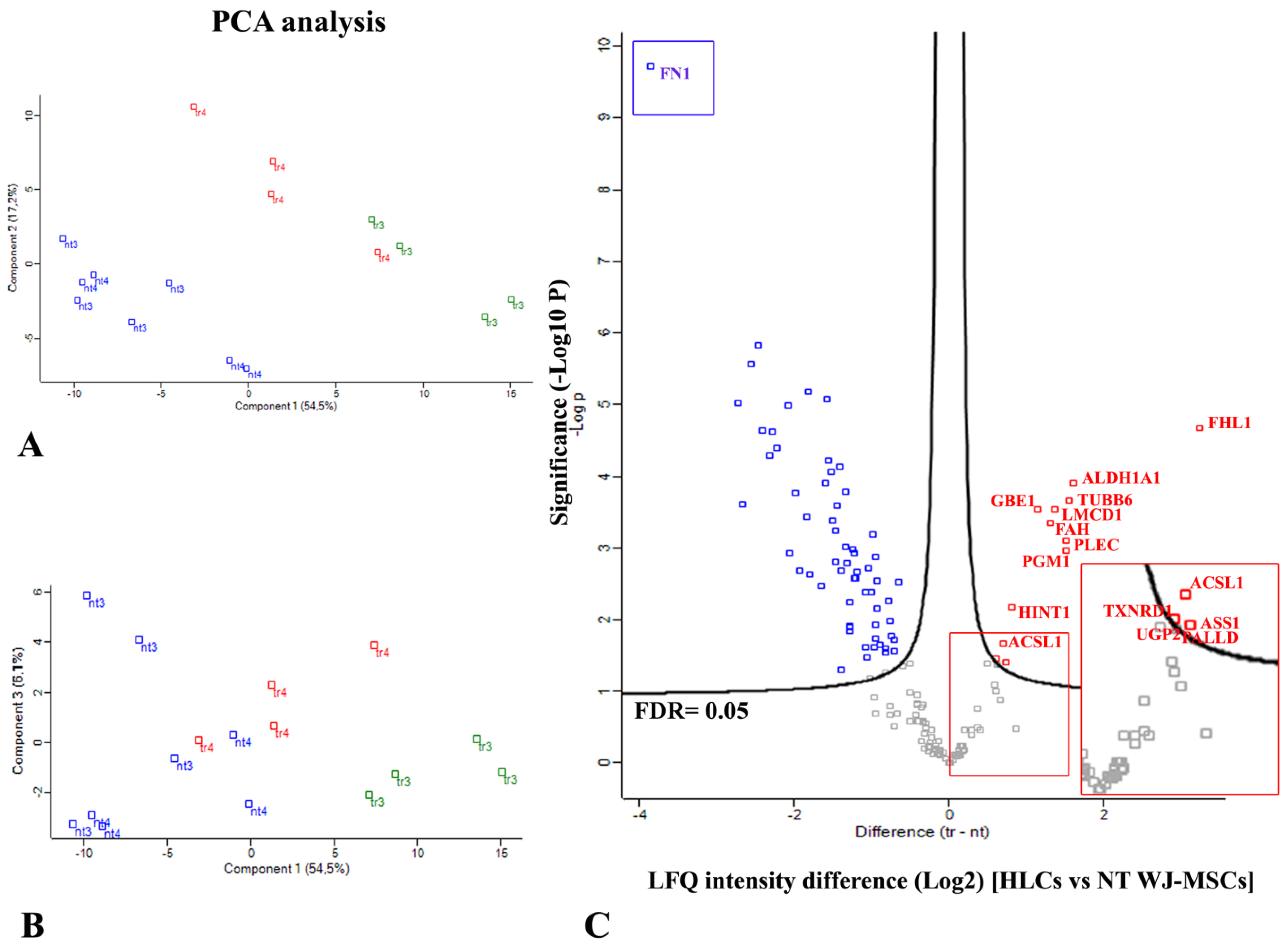
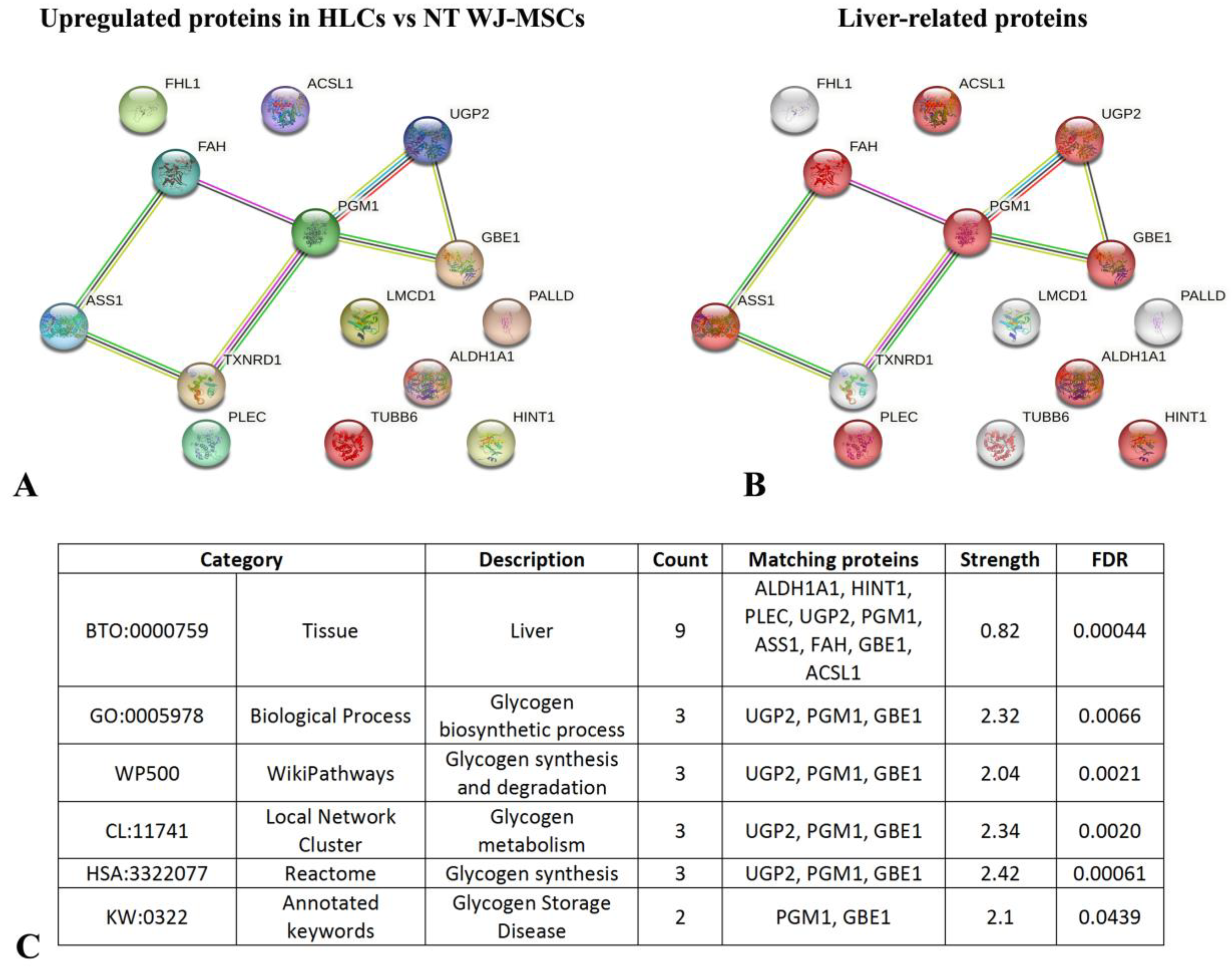
3.5. HPLC-MS/MS Proteome Analyses Identified Novel Intra- and Extracellular Hepatic Proteins in HLCs Derived from WJ-MSCs
Quantitative Analysis of Proteins Differentially Expressed in HLCs Revealed an Increase in Liver-Enriched Enzymes with Respect to NT WJ-MSCs
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- European Association for the Study of the Liver. EASL Clinical Practice Guidelines: Liver transplantation. J. Hepatol. 2016, 64, 433–485. [Google Scholar] [CrossRef] [PubMed]
- Baganate, F.; Beal, E.W.; Tumin, D.; Azoulay, D.; Mumtaz, K.; Black, S.M.; Washburn, K.; Pawlik, T.M. Early mortality after liver transplantation: Defining the course and the cause. Surgery 2018, 164, 694–704. [Google Scholar] [CrossRef] [PubMed]
- Nicolas, C.T.; Wang, Y.; Nyberg, S.L. Cell therapy in chronic liver disease. Curr. Opin. Gastroenterol. 2016, 32, 189–194. [Google Scholar] [CrossRef] [PubMed]
- La Rocca, G.; Anzalone, R. Perinatal stem cells revisited: Directions and indications at the crossroads between tissue regeneration and repair. Curr. Stem Cell Res. Ther. 2013, 8, 2–5. [Google Scholar] [CrossRef] [PubMed]
- Anzalone, R.; Lo Iacono, M.; Corrao, S.; Corsello, T.; Farina, F.; La Rocca, G. Recent patents and advances in hepatocyte-like cells differentiation by perinatal stem cells. Rec. Pat. Regen. Med. 2013, 3, 227–236. [Google Scholar] [CrossRef]
- Afshari, A.; Shamdani, S.; Uzan, G.; Naserian, S.; Azarpira, N. Different approaches for transformation of mesenchymal stem cells into hepatocyte-like cells. Stem Cell Res. Ther. 2020, 11, 54. [Google Scholar] [CrossRef]
- Shahrbaf, M.A.; Nouri, M.; Zarrabi, M.; Gramignoli, R.; Vosough, M. Extraembryonic Mesenchymal Stromal/Stem Cells in Liver Diseases: A Critical Revision of Promising Advanced Therapy Medicinal Products. Cells 2022, 11, 1074. [Google Scholar] [CrossRef]
- Mallis, P.; Michalopoulos, E.; Chatzistamatiou, T.; Giokas, C.S. Interplay between mesenchymal stromal cells and immune system: Clinical applications in immune-related diseases. Explor. Immunol. 2021, 1, 112–139. [Google Scholar] [CrossRef]
- Zhao, L.; Chen, S.; Shi, X.; Cao, H.; Li, L. A pooled analysis of mesenchymal stem cell-based therapy for liver disease. Stem Cell Res. Ther. 2018, 9, 72. [Google Scholar] [CrossRef]
- Jing, Y.T.; Vijesh, V.; Yu, H.L.; Padma, M.; James, C.; Ursula, M. Immunogenicity and Immunomodulatory Properties of Hepatocyte-like Cells Derived from Human Amniotic Epithelial Cells. Curr. Stem Cell Res. Ther. 2013, 8, 91–99. [Google Scholar]
- Li, Y.; Wu, Q.; Wang, Y.; Li, L.; Chen, F.; Shi, Y.; Bu, H.; Bao, J. Immunogenicity of hepatic differentiated human umbilical cord mesenchymal stem cells promoted by porcine decellularized liver scaffolds. Xenotransplantation 2017, 24, e12287. [Google Scholar] [CrossRef] [PubMed]
- Yamana, H.; Inagaki, A.; Imura, T.; Nakamura, Y.; Nishimaki, H.; Katano, T.; Ohashi, K.; Miyagi, S.; Kamei, T.; Unno, M.; et al. Cotransplantation With Adipose Tissue-derived Stem Cells Improves Engraftment of Transplanted Hepatocytes. Transplantation 2022, 106, 1963–1973. [Google Scholar] [CrossRef] [PubMed]
- Kong, D.; Xu, H.; Chen, M.; Yu, Y.; Qian, Y.; Qin, T.; Tong, Y.; Xia, Q.; Hang, H. Co-encapsulation of HNF4α overexpressing UMSCs and human primary hepatocytes ameliorates mouse acute liver failure. Stem Cell Res. Ther. 2020, 11, 449. [Google Scholar] [CrossRef]
- Soncini, M.; Vertua, E.; Gibelli, L.; Zorzi, F.; Denegri, M.; Albertini, A.; Wengler, G.S.; Parolini, O. Isolation and characterization of mesenchymal cells from human fetal membranes. J. Tissue Eng. Regen. Med. 2007, 1, 296–305. [Google Scholar] [CrossRef]
- Zagoura, D.S.; Roubelakis, M.G.; Bitsika, V.; Trohatou, O.; Pappa, K.I.; Kapelouzou, A.; Antsaklis, A.; Anagnou, N.P. Therapeutic potential of a distinct population of human amniotic fluid mesenchymal stem cells and their secreted molecules in mice with acute hepatic failure. Gut 2012, 61, 894–906. [Google Scholar] [CrossRef]
- Jin, H.J.; Bae, Y.K.; Kim, M.; Kwon, S.J.; Jeon, H.B.; Choi, S.J.; Kim, S.W.; Yang, Y.S.; Oh, W.; Chang, J.W. Comparative analysis of human mesenchymal stem cells from bone marrow, adipose tissue, and umbilical cord blood as sources of cell therapy. Int. J. Mol. Sci. 2013, 14, 17986–18001. [Google Scholar] [CrossRef]
- Salehinejad, P.; Moshrefi, M.; Eslaminejad, T. An Overview on Mesenchymal Stem Cells Derived from Extraembryonic Tissues: Supplement Sources and Isolation Methods. Stem Cells Cloning 2020, 13, 57–65. [Google Scholar] [CrossRef]
- Corsello, T.; Amico, G.; Corrao, S.; Anzalone, R.; Timoneri, F.; Lo Iacono, M.; Russo, E.; Spatola, G.F.; Uzzo, M.L.; Giuffrè, M.; et al. Whaton’s jelly mesenchymal stromal cells from human umbilical cord: A close-up on immunomodulatory molecules featured in situ and in vitro. Stem Cell Rev. Rep. 2019, 15, 900–918. [Google Scholar] [CrossRef]
- Mahmoud, R.; Bassiouny, M.; Badawy, A.; Darwish, A.; Yahia, S.; El-Tantawy, N. Maternal and neonatal factors’ effects on wharton’s jelly mesenchymal stem cell yield. Sci. Rep. 2024, 14, 24376. [Google Scholar] [CrossRef]
- Can, A.; Karahuseyinoglu, S. Concise review: Human umbilical cord stroma with regard to the source of fetus-derived stem cells. Stem Cells 2007, 25, 2886–2895. [Google Scholar] [CrossRef]
- Anzalone, R.; Lo Iacono, M.; Loria, T.; Di Stefano, A.; Giannuzzi, P.; Farina, F.; La Rocca, G. Wharton’s jelly mesenchymal stem cells as candidates for beta cells regeneration: Extending the differentiative and immunomodulatory benefits of adult mesenchymal stem cells for the treatment of type 1 diabetes. Stem Cell Rev. 2011, 7, 342–363. [Google Scholar] [CrossRef] [PubMed]
- La Rocca, G. Editorial—Connecting the Dots: The Promises of Wharton’s Jelly Mesenchymal Stem Cells for Tissue Repair and Regeneration. Open Tissue Eng. Regen. Med. J. 2011, 4, 3–5. [Google Scholar] [CrossRef]
- Ertl, J.; Pichlsberger, M.; Tuca, A.C.; Wurzer, P.; Fuchs, J.; Geyer, S.H.; Maurer-Gesek, B.; Weninger, W.J.; Pfeiffer, D.; Bubalo, V.; et al. Comparative study of regenerative effects of mesenchymal stem cells derived from placental amnion, chorion and umbilical cord on dermal wounds. Placenta 2018, 65, 37–46. [Google Scholar] [CrossRef]
- Lo Iacono, M.; Anzalone, R.; La Rocca, G.; Baiamonte, E.; Maggio, A.; Acuto, S. Wharton’s Jelly Mesenchymal Stromal Cells as a Feeder Layer for the Ex Vivo Expansion of Hematopoietic Stem and Progenitor Cells: A Review. Stem Cell Rev. 2017, 13, 35–49. [Google Scholar] [CrossRef]
- Lo Iacono, M.; Russo, E.; Anzalone, R.; Baiamonte, E.; Alberti, G.; Gerbino, A.; Maggio, A.; La Rocca, G.; Acuto, S. Wharton’s Jelly Mesenchymal Stromal Cells Support the Expansion of Cord Blood-derived CD34(+) Cells Mimicking a Hematopoietic Niche in a Direct Cell-cell Contact Culture System. Cell Transplant. 2018, 27, 117–129. [Google Scholar] [CrossRef]
- La Rocca, G.; Lo Iacono, M.; Corsello, T.; Corrao, S.; Farina, F.; Anzalone, R. Human Wharton’s jelly mesenchymal stem cells maintain the expression of key immunomodulatory molecules when subjected to osteogenic, adipogenic and chondrogenic differentiation in vitro: New perspectives for cellular therapy. Curr. Stem Cell Res. Ther. 2013, 8, 100–113. [Google Scholar] [CrossRef]
- La Rocca, G.; Anzalone, R.; Corrao, S.; Magno, F.; Loria, T.; Lo Iacono, M.; Di Stefano, A.; Giannuzzi, P.; Marasa, L.; Cappello, F.; et al. Isolation and characterization of Oct-4+/HLA-G+ mesenchymal stem cells from human umbilical cord matrix: Differentiation potential and detection of new markers. Histochem. Cell Biol. 2009, 131, 267–282. [Google Scholar] [CrossRef]
- Weiss, M.L.; Anderson, C.; Medicetty, S.; Seshareddy, K.B.; Weiss, R.J.; VanderWerff, I.; Troyer, D.; McIntosh, K.R. Immune properties of human umbilical cord Wharton’s jelly-derived cells. Stem Cells 2008, 26, 2865–2874. [Google Scholar] [CrossRef]
- Silini, A.R.; Di Pietro, R.; Lang-Olip, I.; Alviano, F.; Banerjee, A.; Basile, M.; Borutinskaite, V.; Eissner, G.; Gellhaus, A.; Giebel, B.; et al. Where Do We Stand? A Roadmap of the Human Placenta and Consensus for Tissue and Cell Nomenclature. Front. Bioeng. Biotechnol. 2020, 8, 610544. [Google Scholar] [CrossRef]
- van Megen, K.M.; van’t Wout, E.T.; Lages Motta, J.; Dekker, B.; Nikolic, T.; Roep, B.O. Activated Mesenchymal Stromal Cells Process and Present Antigens Regulating Adaptive Immunity. Front. Immunol. 2019, 10, 694. [Google Scholar] [CrossRef]
- Le Blanc, K.; Tammik, C.; Rosendahl, K.; Zetterberg, E.; Ringdén, O. HLA expression and immunologic properties of differentiated and undifferentiated mesenchymal stem cells. Exp. Hematol. 2003, 31, 890–896. [Google Scholar] [CrossRef] [PubMed]
- Kwak, J.; Choi, W.; Bae, Y.; Kim, M.; Choi, S.; Oh, W.; Jin, H. HLA-A2 Promotes the Therapeutic Effect of Umbilical Cord Blood-Derived Mesenchymal Stem Cells in Hyperoxic Lung Injury. Bioengineering 2022, 9, 177. [Google Scholar] [CrossRef] [PubMed]
- Anzalone, R.; Corrao, S.; Lo Iacono, M.; Loria, T.; Corsello, T.; Cappello, F.; Di Stefano, A.; Giannuzzi, P.; Zummo, G.; Farina, F.; et al. Isolation and characterization of CD276+/HLA-E+ human subendocardial mesenchymal stem cells from chronic heart failure patients: Analysis of differentiative potential and immunomodulatory markers expression. Stem Cells Dev. 2013, 22, 1–17. [Google Scholar] [CrossRef]
- Vigdorovich, V.; Ramagopal, U.A.; Lazar-Molnar, E.; Sylvestre, E.; Lee, J.S.; Hofmeyer, K.A.; Zang, X.; Nathenson, S.G.; Almo, S.C. Structure and T cell inhibition properties of B7 family member, B7-H3. Structure 2013, 21, 707–717. [Google Scholar] [CrossRef]
- Tipnis, S.; Viswanathan, C.; Majumdar, A.S. Immunosuppressive properties of human umbilical cord-derived mesenchymal stem cells: Role of B7-H1 and IDO. Immunol. Cell Biol. 2010, 88, 795–806. [Google Scholar] [CrossRef]
- Anzalone, R.; Lo Iacono, M.; Corrao, S.; Magno, F.; Loria, T.; Cappello, F.; Zummo, G.; Farina, F.; La Rocca, G. New emerging potentials for human Wharton’s jelly mesenchymal stem cells: Immunological features and hepatocyte-like differentiative capacity. Stem Cells Dev. 2010, 19, 423–438. [Google Scholar] [CrossRef]
- Bharti, D.; Shivakumar, S.B.; Park, J.K.; Ullah, I.; Subbarao, R.B.; Park, J.S.; Lee, S.L.; Park, B.W.; Rho, G.J. Comparative analysis of human Wharton’s jelly mesenchymal stem cells derived from different parts of the same umbilical cord. Cell Tissue Res. 2018, 372, 51–65. [Google Scholar] [CrossRef]
- Sokal, E.M.; Trivedi, P.; Cheeseman, P.; Portmann, B.; Mowat, A.P. The application of quantitative cytochemistry to study the acinar distribution of enzymatic activities in human liver biopsy sections. J. Hepatol. 1989, 9, 42–48. [Google Scholar] [CrossRef]
- Khuu, D.N.; Scheers, I.; Ehnert, S.; Jazouli, N.; Nyabi, O.; Buc-Calderon, P.; Meulemans, A.; Nussler, A.; Sokal, E.; Najimi, M. In vitro differentiated adult human liver progenitor cells display mature hepatic metabolic functions: A potential tool for in vitro pharmacotoxicological testing. Cell Transplant. 2011, 20, 287–302. [Google Scholar] [CrossRef]
- Dominici, M.; Le Blanc, K.; Mueller, I.; Slaper-Cortenbach, I.; Marini, F.; Krause, D.; Deans, R.; Keating, A.; Prockop, D.; Horwitz, E. Minimal criteria for defining multipotent mesenchymal stromal cells. The International Society for Cellular Therapy position statement. Cytotherapy 2006, 8, 315–317. [Google Scholar] [CrossRef]
- Buyl, K.; De Kock, J.; Najar, M.; Lagneaux, L.; Branson, S.; Rogiers, V.; Vanhaecke, T. Characterization of hepatic markers in human Wharton’s Jelly-derived mesenchymal stem cells. Toxicol. In Vitro 2014, 28, 113–119. [Google Scholar] [CrossRef] [PubMed]
- Willebrords, J.; Crespo Yanguas, S.; Maes, M.; Decrock, E.; Wang, N.; Leybaert, L.; da Silva, T.C.; Veloso Alves Pereira, I.; Jaeschke, H.; Cogliati, B.; et al. Structure, Regulation and Function of Gap Junctions in Liver. Cell Commun. Adhes. 2015, 22, 29–37. [Google Scholar] [CrossRef] [PubMed]
- Michalik, M.; Gladys, A.; Czekaj, P. Differentiation of Cells Isolated from Afterbirth Tissues into Hepatocyte-Like Cells and Their Potential Clinical Application in Liver Regeneration. Stem Cell Rev. Rep. 2021, 17, 581–603. [Google Scholar] [CrossRef] [PubMed]
- Campard, D.; Lysy, P.A.; Najimi, M.; Sokal, E.M. Native umbilical cord matrix stem cells express hepatic markers and differentiate into hepatocyte-like cells. Gastroenterology 2008, 134, 833–848. [Google Scholar] [CrossRef]
- Guengerich, F.P. Intersection of the Roles of Cytochrome P450 Enzymes with Xenobiotic and Endogenous Substrates: Relevance to Toxicity and Drug Interactions. Chem. Res. Toxicol. 2017, 30, 2–12. [Google Scholar] [CrossRef]
- Hutzler, J.M.; Yang, Y.S.; Albaugh, D.; Fullenwider, C.L.; Schmenk, J.; Fisher, M.B. Characterization of aldehyde oxidase enzyme activity in cryopreserved human hepatocytes. Drug Metab. Dispos. 2012, 40, 267–275. [Google Scholar] [CrossRef]
- Lewinska, M.; Zmrzljak, U.P.; Rozman, D. Low nucleotide variability of CYP51A1 in humans: Meta-analysis of cholesterol and bile acid synthesis and xenobiotic metabolism pathways. Acta Chim. Slov. 2013, 60, 875–883. [Google Scholar]
- Decker, M.; Arand, M.; Cronin, A. Mammalian epoxide hydrolases in xenobiotic metabolism and signalling. Arch. Toxicol. 2009, 83, 297–318. [Google Scholar] [CrossRef]
- Koukouritaki, S.B.; Simpson, P.; Yeung, C.K.; Rettie, A.E.; Hines, R.N. Human hepatic flavin-containing monooxygenases 1 (FMO1) and 3 (FMO3) developmental expression. Pediatr. Res. 2002, 51, 236–243. [Google Scholar] [CrossRef]
- Soderdahl, T.; Kuppers-Munther, B.; Heins, N.; Edsbagge, J.; Bjorquist, P.; Cotgreave, I.; Jernstrom, B. Glutathione transferases in hepatocyte-like cells derived from human embryonic stem cells. Toxicol. In Vitro 2007, 21, 929–937. [Google Scholar] [CrossRef]
- Yin, Z.L.; Dahlstrom, J.E.; Le Couteur, D.G.; Board, P.G. Immunohistochemistry of omega class glutathione S-transferase in human tissues. J. Histochem. Cytochem. 2001, 49, 983–987. [Google Scholar] [CrossRef] [PubMed]
- Zelko, I.; Negishi, M. Phenobarbital-elicited activation of nuclear receptor CAR in induction of cytochrome P450 genes. Biochem. Biophys. Res. Commun. 2000, 277, 1–6. [Google Scholar] [CrossRef] [PubMed]
- Lu, D.; Xie, Q.; Wu, B. N-glucuronidation catalyzed by UGT1A4 and UGT2B10 in human liver microsomes: Assay optimization and substrate identification. J. Pharm. Biomed. Anal. 2017, 145, 692–703. [Google Scholar] [CrossRef]
- O’Shaughnessy, P.J.; Monteiro, A.; Bhattacharya, S.; Fraser, M.J.; Fowler, P.A. Steroidogenic enzyme expression in the human fetal liver and potential role in the endocrinology of pregnancy. Mol. Hum. Reprod. 2013, 19, 177–187. [Google Scholar] [CrossRef]
- Langmann, T.; Klucken, J.; Reil, M.; Liebisch, G.; Luciani, M.F.; Chimini, G.; Kaminski, W.E.; Schmitz, G. Molecular cloning of the human ATP-binding cassette transporter 1 (hABC1): Evidence for sterol-dependent regulation in macrophages. Biochem. Biophys. Res. Commun. 1999, 257, 29–33. [Google Scholar] [CrossRef]
- Bhattacharjee, J.; Ietta, F.; Giacomello, E.; Bechi, N.; Romagnoli, R.; Fava, A.; Paulesu, L. Expression and localization of ATP binding cassette transporter A1 (ABCA1) in first trimester and term human placenta. Placenta 2010, 31, 423–430. [Google Scholar] [CrossRef]
- Kaminski, W.E.; Wenzel, J.J.; Piehler, A.; Langmann, T.; Schmitz, G. ABCA6, a novel a subclass ABC transporter. Biochem. Biophys. Res. Commun. 2001, 285, 1295–1301. [Google Scholar] [CrossRef]
- Zhao, Y.; Ishigami, M.; Nagao, K.; Hanada, K.; Kono, N.; Arai, H.; Matsuo, M.; Kioka, N.; Ueda, K. ABCB4 exports phosphatidylcholine in a sphingomyelin-dependent manner. J. Lipid Res. 2015, 56, 644–652. [Google Scholar] [CrossRef]
- Aoyama, T.; Souri, M.; Ushikubo, S.; Kamijo, T.; Yamaguchi, S.; Kelley, R.I.; Rhead, W.J.; Uetake, K.; Tanaka, K.; Hashimoto, T. Purification of human very-long-chain acyl-coenzyme A dehydrogenase and characterization of its deficiency in seven patients. J. Clin. Investig. 1995, 95, 2465–2473. [Google Scholar] [CrossRef]
- Parini, P.; Davis, M.; Lada, A.T.; Erickson, S.K.; Wright, T.L.; Gustafsson, U.; Sahlin, S.; Einarsson, C.; Eriksson, M.; Angelin, B.; et al. ACAT2 is localized to hepatocytes and is the major cholesterol-esterifying enzyme in human liver. Circulation 2004, 110, 2017–2023. [Google Scholar] [CrossRef]
- Singh, A.B.; Kan, C.F.; Dong, B.; Liu, J. SREBP2 Activation Induces Hepatic Long-chain Acyl-CoA Synthetase 1 (ACSL1) Expression in Vivo and in Vitro through a Sterol Regulatory Element (SRE) Motif of the ACSL1 C-promoter. J. Biol. Chem. 2016, 291, 5373–5384. [Google Scholar] [CrossRef] [PubMed]
- Hess, A.L.; Carayol, J.; Blaedel, T.; Hager, J.; Di Cara, A.; Astrup, A.; Saris, W.H.M.; Larsen, L.H.; Valsesia, A. Analysis of circulating angiopoietin-like protein 3 and genetic variants in lipid metabolism and liver health: The DiOGenes study. Genes. Nutr. 2018, 13, 7. [Google Scholar] [CrossRef] [PubMed]
- Houten, S.M.; Denis, S.; Argmann, C.A.; Jia, Y.; Ferdinandusse, S.; Reddy, J.K.; Wanders, R.J. Peroxisomal L-bifunctional enzyme (Ehhadh) is essential for the production of medium-chain dicarboxylic acids. J. Lipid Res. 2012, 53, 1296–1303. [Google Scholar] [CrossRef]
- Qin, X.; Gao, B. The complement system in liver diseases. Cell Mol. Immunol. 2006, 3, 333–340. [Google Scholar]
- Luo, W.; Vik, D.P. Regulation of complement factor H in a human liver cell line by interferon-gamma. Scand. J. Immunol. 1999, 49, 487–494. [Google Scholar] [CrossRef]
- Lung, T.; Sakem, B.; Risch, L.; Wurzner, R.; Colucci, G.; Cerny, A.; Nydegger, U. The complement system in liver diseases: Evidence-based approach and therapeutic options. J. Transl. Autoimmun. 2019, 2, 100017. [Google Scholar] [CrossRef]
- Agarwal, B.; Wright, G.; Gatt, A.; Riddell, A.; Vemala, V.; Mallett, S.; Chowdary, P.; Davenport, A.; Jalan, R.; Burroughs, A. Evaluation of coagulation abnormalities in acute liver failure. J. Hepatol. 2012, 57, 780–786. [Google Scholar] [CrossRef]
- Prendergast, G.C.; Metz, R.; Muller, A.J.; Merlo, L.M.; Mandik-Nayak, L. IDO2 in Immunomodulation and Autoimmune Disease. Front. Immunol. 2014, 5, 585. [Google Scholar] [CrossRef]
- Potikha, T.; Ella, E.; Cerliani, J.P.; Mizrahi, L.; Pappo, O.; Rabinovich, G.A.; Galun, E.; Goldenberg, D.S. Galectin-1 is essential for efficient liver regeneration following hepatectomy. Oncotarget 2016, 7, 31738–31754. [Google Scholar] [CrossRef]
- Sioud, M.; Mobergslien, A.; Boudabous, A.; Floisand, Y. Evidence for the involvement of galectin-3 in mesenchymal stem cell suppression of allogeneic T-cell proliferation. Scand. J. Immunol. 2010, 71, 267–274. [Google Scholar] [CrossRef]
- Zhou, Y.J.; Gao, J.; Yang, H.M.; Yuan, X.L.; Chen, T.X.; He, Z.J. The role of the lactadherin in promoting intestinal DCs development in vivo and vitro. Clin. Dev. Immunol. 2010, 2010, 357541. [Google Scholar] [CrossRef] [PubMed]
- Singh, Y.; Chen, H.; Zhou, Y.; Foller, M.; Mak, T.W.; Salker, M.S.; Lang, F. Differential effect of DJ-1/PARK7 on development of natural and induced regulatory T cells. Sci. Rep. 2015, 5, 17723. [Google Scholar] [CrossRef] [PubMed]
- Skornicka, E.L.; Kiyatkina, N.; Weber, M.C.; Tykocinski, M.L.; Koo, P.H. Pregnancy zone protein is a carrier and modulator of placental protein-14 in T-cell growth and cytokine production. Cell Immunol. 2004, 232, 144–156. [Google Scholar] [CrossRef] [PubMed]
- Daveau, M.; Jean, L.; Soury, E.; Olivier, E.; Masson, S.; Lyoumi, S.; Chan, P.; Hiron, M.; Lebreton, J.P.; Husson, A.; et al. Hepatic and extra-hepatic transcription of inter-alpha-inhibitor family genes under normal or acute inflammatory conditions in rat. Arch. Biochem. Biophys. 1998, 350, 315–323. [Google Scholar] [CrossRef]
- Ventura, G.; De Bandt, J.P.; Segaud, F.; Perret, C.; Robic, D.; Levillain, O.; Le Plenier, S.; Godard, C.; Cynober, L.; Moinard, C. Overexpression of ornithine aminotransferase: Consequences on amino acid homeostasis. Br. J. Nutr. 2009, 101, 843–851. [Google Scholar] [CrossRef]
- Madiraju, A.K.; Alves, T.; Zhao, X.; Cline, G.W.; Zhang, D.; Bhanot, S.; Samuel, V.T.; Kibbey, R.G.; Shulman, G.I. Argininosuccinate synthetase regulates hepatic AMPK linking protein catabolism and ureagenesis to hepatic lipid metabolism. Proc. Natl. Acad. Sci. USA 2016, 113, E3423–E3430. [Google Scholar] [CrossRef]
- Crabb, D.W.; Matsumoto, M.; Chang, D.; You, M. Overview of the role of alcohol dehydrogenase and aldehyde dehydrogenase and their variants in the genesis of alcohol-related pathology. Proc. Nutr. Soc. 2004, 63, 49–63. [Google Scholar] [CrossRef]
- Liu, M.; Sun, L.Y. Liver Transplantation for Glycogen Storage Disease Type IV. Front. Pediatr. 2021, 19, 633822. [Google Scholar] [CrossRef]
- Ichimoto, K.; Fujisawa, T.; Shimura, M.; Fushimi, T.; Tajika, M.; Matsunaga, A.; Ogawa-Tominaga, M.; Akiyama, N.; Naruke, Y.; Horie, H.; et al. Two cases of a non-progressive hepatic form of glycogen storage disease type IV with atypical liver pathology. Mol. Genet. Metab. Rep. 2020, 24, 100601. [Google Scholar] [CrossRef]
- Chou, J.Y.; Jun, H.S.; Mansfield, B.C. Type I glycogen storage diseases: Disorders of the glucose-6-phosphatase/glucose-6-phosphate transporter complexes. J. Inherit. Metab. Dis. 2015, 38, 511–519. [Google Scholar] [CrossRef]
- Rake, J.P.; Visser, G.; Labrune, P.; Leonard, J.V.; Ullrich, K.; Smit, G.P.A. Glycogen storage disease type I: Diagnosis, management, clinical course and outcome. Results of the European Study on Glycogen Storage Disease Type I (ESGSD I). Euro J. Pediatr. 2002, 161 (Suppl. 1), S20–S34. [Google Scholar] [CrossRef]
- Koch, R.L.; Soler-Alfonso, C.; Kiely, B.T.; Asai, A.; Smith, A.L.; Bali, D.S.; Kang, P.B.; Landstrom, A.P.; Akman, H.O.; Burrow, T.A.; et al. Burrow Diagnosis and management of glycogen storage disease type IV, including adult polyglucosan body disease: A clinical practice resource. Mol. Genet. Metab. 2023, 38, 107525. [Google Scholar] [CrossRef]
- Arutyunyan, I.; Elchaninov, A.; Fatkhudinov, T.; Makarov, A.; Kananykhina, E.; Usman, N.; Bolshakova, G.; Glinkina, V.; Goldshtein, D.; Sukhikh, G. Elimination of allogeneic multipotent stromal cells by host macrophages in different models of regeneration. Int. J. Clin. Exp. Pathol. 2015, 8, 4469–4480. [Google Scholar]
- Shi, Y.; Hu, G.; Su, J.; Li, W.; Chen, Q.; Shou, P.; Xu, C.; Chen, X.; Huang, Y.; Zhu, Z.; et al. Mesenchymal stem cells: A new strategy for immunosuppression and tissue repair. Cell Res. 2010, 20, 510–518. [Google Scholar] [CrossRef]
- Wen, F.; Yang, G.; Yu, S.; Liu, H.; Liao, N.; Liu, Z. Mesenchymal stem cell therapy for liver transplantation: Clinical progress and immunomodulatory properties. Stem Cell Res. Ther. 2024, 15, 320. [Google Scholar] [CrossRef]
- Zhao, Q.; Ren, H.; Li, X.; Chen, Z.; Zhang, X.; Gong, W.; Liu, Y.; Pang, T.; Han, Z.C. Differentiation of human umbilical cord mesenchymal stromal cells into low immunogenic hepatocyte-like cells. Cytotherapy 2009, 414–426. [Google Scholar] [CrossRef]
- Zhang, Y.N.; Lie, P.C.; Wei, X. Differentiation of mesenchymal stromal cells derived from umbilical cord Wharton’s jelly into hepatocyte-like cells. Cytotherapy 2009, 11, 548–558. [Google Scholar] [CrossRef]
- Zhang, Z.; Lin, H.; Shi, M.; Xu, R.; Fu, J.; Lv, J.; Chen, L.; Lv, S.; Li, Y.; Yu, S.; et al. Human umbilical cord mesenchymal stem cells improve liver function and ascites in decompensated liver cirrhosis patients. J. Gastroenterol. Hepatol. 2012, 27 (Suppl. 2), 112–120. [Google Scholar] [CrossRef]
- Shi, M.; Zhang, Z.; Xu, R.; Lin, H.; Fu, J.; Zou, Z.; Zhang, A.; Shi, J.; Chen, L.; Lv, S.; et al. Human mesenchymal stem cell transfusion is safe and improves liver function in acute-on-chronic liver failure patients. Stem Cells Transl. Med. 2012, 1, 725–731. [Google Scholar] [CrossRef]
- Yan, Y.; Xu, W.; Qian, H.; Si, Y.; Zhu, W.; Cao, H.; Zhou, H.; Mao, F. Mesenchymal stem cells from human umbilical cords ameliorate mouse hepatic injury in vivo. Liver Int. 2009, 29, 356–365. [Google Scholar] [CrossRef]






| Biological Process | Name | Accession Number | Gene | 3rd Week NT WJ-MSCs | 3rd Week HLCs | 4th Week NT WJ-MSCs | 4th Week HLCs |
|---|---|---|---|---|---|---|---|
| Xenobiotic and drug metabolism process | Aflatoxin B1 aldehyde reductase member 2 (Fragment) | H3BLU7 | AKR7A2 | − | + | − | + |
| Aldehyde oxidase (**) | Q06278 | AOX1 | − | + | − | − | |
| Cytochrome P450 1B1 | Q16678 | CYP1B1 | − | − | − | + | |
| Cytochrome P450 2A13 | Q16696 | CYP2A13 | − | − | − | + | |
| Cytochrome P450 2S1 | Q96SQ9 | CYP2S1 | + | + | − | − | |
| Dimethylaniline monooxygenase [N-oxide-forming] 3 (**) | P31513 | FMO3 | − | − | − | + | |
| Epoxide hydrolase 1 (*) | P07099 | EPHX1 | + | + | + | + | |
| Glutathione S-transferase Mu 1 (**) | P09488 | GSTM1 | − | − | − | + | |
| Glutathione S-transferase omega-1 (**) | P78417 | GSTO1 | + | + | + | + | |
| Glutathione S-transferase P (**) | P09211 | GSTP1 | + | + | + | + | |
| Lanosterol 14-alpha demethylase (**) | Q16850 | CYP51A1 | − | + | − | + | |
| Nuclear receptor subfamily 1 group I member 3 (**) | E9PCF2 | NR1I3 | − | − | − | + | |
| UDP-glucuronosyltransferase 1–4 (**) | P22310 | UGT1A4 | - | + | − | − | |
| Lipid metabolic process | Acetyl-CoA acetyltransferase, cytosolic (**) | Q9BWD1 | ACAT2 | − | + | − | + |
| Acetyl-CoA acetyltransferase, mitochondrial | P24752 | ACAT1 | − | + | − | + | |
| Acyl-coenzyme A thioesterase 2, mitochondrial | P49753 | ACOT2 | + | + | − | − | |
| Angiopoietin-related protein 3 (**) | Q9Y5C1 | ANGPTL3 | − | + | − | − | |
| ATP-binding cassette sub-family A member 1 (**) (§) | O95477 | ABCA1 | − | − | − | + | |
| Carboxylesterase 3 | Q6UWW8 | CES3 | − | + | − | − | |
| Enoyl-CoA hydratase domain-containing protein 2, mitochondrial (Fragment) (**) | F5GWU3 | ECHDC2 | − | − | − | + | |
| Erlin-1 (**) | O75477 | ERLIN1 | − | − | − | + | |
| Fatty acid synthase | P49327 | FASN | − | − | − | + | |
| Heme oxygenase 2 (**) | P30519 | HMOX2 | − | + | − | − | |
| Hydroxyacyl-coenzyme A dehydrogenase, mitochondrial | Q16836 | HADH | − | + | − | − | |
| Long-chain-fatty-acid--CoA ligase 1 (**) | E7EPM6 | ACSL1 | − | + | − | + | |
| Long-chain-fatty-acid--CoA ligase 3 (**) | O95573 | ACSL3 | − | + | − | + | |
| Mevalonate kinase (**) | F5H8H2 | MVK | − | − | − | + | |
| Peroxisomal bifunctional enzyme (**) | Q08426 | EHHADH | − | − | − | + | |
| Phosphatidylcholine translocator ABCB4 | P21439 | ABCB4 | − | + | − | − | |
| Very long-chain specific acyl-CoA dehydrogenase, mitochondrial (**) | P49748 | ACADVL | − | + | + | + | |
| Very-long-chain (3R)-3-hydroxyacyl-CoA dehydratase 3 (**) | Q9P035 | HACD3 | + | − | − | + | |
| Very long-chain acyl-CoA synthetase | O14975 | SLC27A2 | − | − | − | + | |
| Cholesterol, bile and carnitine metabolic process | 4-trimethylaminobutyraldehyde dehydrogenase (**) | P49189 | ALDH9A1 | − | + | − | + |
| 7-dehydrocholesterol reductase (**) | Q9UBM7 | DHCR7 | + | + | + | + | |
| Delta (24)-sterol reductase (**) | Q15392 | DHCR24 | − | + | − | + | |
| 24-hydroxycholesterol 7-alpha-hydroxylase (**) | Q9NYL5 | CYP39A1 | − | + | − | − | |
| Cholesterol side-chain cleavage enzyme, mitochondrial | P05108 | CYP11A1 | − | + | − | − | |
| Bile acid receptor (**) | Q96RI1 | NR1H4 | − | + | − | − | |
| Epididymal secretory protein E1 (**) | G3V3E8 | NPC2 | + | + | + | + | |
| Leptin receptor | P48357 | LEPR | − | − | − | + | |
| Glucose metabolic process | 1,4-alpha-glucan-branching enzyme (**) | Q04446 | GBE1 | + | + | + | + |
| Alpha-enolase | P06733 | ENO1 | + | + | + | + | |
| Alpha-N-acetylglucosaminidase | P54802 | NAGLU | + | + | − | + | |
| ATP-dependent 6-phosphofructokinase, liver type (**) | P17858 | PFKL | + | + | + | + | |
| Glycogen debranching enzyme (**) | P35573 | AGL | − | + | + | + | |
| Glycogen phosphorylase, liver form (**) | P06737 | PYGL | − | − | − | + | |
| Glycogen [starch] synthase, muscle | P13807 | GYS1 | − | + | − | + | |
| Insulin receptor substrate 2 (**) | Q9Y4H2 | IRS2 | − | − | − | + | |
| L-lactate dehydrogenase A chain | P00338 | LDHA | + | + | + | + | |
| Phosphoenolpyruvate carboxykinase [GTP], mitochondrial (**) | Q16822 | PCK2 | − | + | − | + | |
| Phosphoglucomutase-1 (**) | P36871 | PGM1 | + | + | + | + | |
| Thioredoxin, mitochondrial | Q99757 | TXN2 | − | − | − | + | |
| Insulin-like growth factor-binding protein 4 | P22692 | IGFBP4 | − | − | − | + | |
| Non-POU domain-containing octamer−binding protein | Q15233 | NONO | − | − | − | + | |
| Protein deglycase DJ-1 | Q99497 | PARK7 | + | + | + | + | |
| Prothrombin | P00734 | F2 | + | − | + | + | |
| Amino acid metabolic process | Amine oxidase [flavin-containing] A | P21397 | MAOA | − | + | − | + |
| Aminopeptidase N (CD13) | P15144 | ANPEP | + | + | + | + | |
| Alanine aminotransferase 1 (**) | P24298 | GPT | − | + | − | − | |
| Aspartate aminotransferase, cytoplasmic (**) | P17174 | GOT1 | − | + | − | + | |
| Aspartate aminotransferase, mitochondrial (**) | P00505 | GOT2 | + | + | + | + | |
| Carnosine N-methyltransferase | Q8N4J0 | CARNMT1 | − | − | − | + | |
| Cathepsin D | P07339 | CTSD | − | − | − | + | |
| Neprilysin (CD10) | P08473 | MME | + | + | + | + | |
| Phosphoserine aminotransferase | Q9Y617 | PSAT1 | + | + | + | + | |
| Putative aspartate aminotransferase, cytoplasmic 2 | Q8NHS2 | GOT1L1 | − | − | − | + | |
| Urea cycle | Argininosuccinate synthase (**) (§) | P00966 | ASS1 | + | + | + | + |
| Mitochondrial ornithine transporter 1 (**) | Q9Y619 | SLC25A15 | − | − | − | + | |
| Ornithine aminotransferase, mitochondrial (**) | P04181 | OAT | + | + | + | + | |
| Alcohol metabolic process | Alcohol dehydrogenase class-3 (**) | P11766 | ADH5 | + | + | + | + |
| Aldehyde dehydrogenase, mitochondrial (**) | P05091 | ALDH2 | − | + | − | − | |
| Binding | Albumin | P02768 | ALB | + | + | + | + |
| Apolipoprotein L5 | Q9BWW9 | APOL5 | − | − | − | + | |
| ATP-binding cassette sub-family A member 6 (**) | Q8N139 | ABCA6 | − | − | − | + | |
| CREB-binding protein | Q92793 | CREBBP | − | − | − | + | |
| Cytochrome b-c1 complex subunit 1, mitochondrial | P31930 | UQCRC1 | + | + | − | + | |
| Cytochrome c1, heme protein, mitochondrial (**) | P08574 | CYC1 | − | + | − | − | |
| Cytochrome c oxidase subunit 2 | P00403 | MT-CO2 | + | + | − | − | |
| Cytochrome c oxidase subunit 4 isoform 1, mitochondrial | P13073 | COX4I1 | + | + | − | + | |
| Cytochrome c oxidase subunit 5A, mitochondrial | P20674 | COX5A | + | + | + | + | |
| Exportin-T | O43592 | XPOT | − | − | − | + | |
| Filamin-binding LIM protein 1 | Q8WUP2 | FBLIM1 | + | + | − | + | |
| Hemoglobin subunit gamma-2 (**) | E9PBW4 | HBG2 | − | + | − | − | |
| Immediate early response 3-interacting protein 1 | Q9Y5U9 | IER3IP1 | − | − | − | + | |
| Insulin-like growth factor-binding protein 7 (**) | Q16270 | IGFBP7 | + | + | + | + | |
| Metallothionein-2 (**) | P02795 | MT2A | − | − | − | + | |
| Multidrug resistance-associated protein 5 | O15440 | ABCC5 | − | − | − | + | |
| Poly(rC)-binding protein 1 | Q15365 | PCBP1 | + | + | + | + | |
| Putative cytochrome b-c1 complex subunit Rieske-like protein 1 | P0C7P4 | UQCRFS1P1 | − | + | − | − | |
| Transferrin | P02786 | TFRC | + | − | − | + | |
| Cellular and apoptotic process | 5′-nucleotidase (CD73) | P21589 | NT5E | + | + | + | + |
| Adipocyte plasma membrane-associated protein | Q9HDC9 | APMAP | − | + | − | − | |
| Carnosine N-methyltransferase | Q8N4J0 | CARNMT1 | − | − | − | + | |
| Cytochrome P450 1B1 | Q16678 | CYP1B1 | − | − | − | + | |
| Death-inducer obliterator 1 (**) | Q9BTC0 | DIDO1 | − | + | − | − | |
| Eukaryotic translation initiation factor 3 subunit F | O00303 | EIF3F | − | + | − | + | |
| Gelsolin (**) | P06396 | GSN | + | + | − | + | |
| Inter-alpha-trypsin inhibitor heavy chain H4 (**) | Q14624 | ITIH4 | − | + | − | − | |
| mRNA-capping enzyme | O60942 | RNGTT | − | − | − | + | |
| multiple epidermal growth factor-like domains protein 8 | Q7Z7M0 | MEGF8 | + | + | + | + | |
| PH domain leucine-rich repeat-containing protein phosphatase 1 | O60346 | PHLPP1 | − | − | − | + | |
| Protein-glutamine gamma-glutamyltransferase 2 | P21980 | TGM2 | + | + | + | + | |
| Retinoid-inducible serine carboxypeptidase | Q9HB40 | SCPEP1 | + | − | + | + | |
| Retinol dehydrogenase 11 | Q8TC12 | RDH11 | − | + | − | + | |
| Proliferation/differentiation and liver development | Dapper homolog 2 | Q5SW24 | DACT2 | − | − | − | + |
| Fascin (**) | Q16658 | FSCN1 | + | + | + | + | |
| Fibroblast growth factor receptor 2 | D2CGD1 | FGFR2 | − | − | − | + | |
| Growth/differentiation factor 8 | O14793 | MSTN | − | − | − | + | |
| H2.0-like homeobox protein (**) | Q14774 | HLX | − | − | − | + | |
| Hepatocyte growth factor receptor (**) (§) | P08581 | MET | − | + | − | − | |
| Hepatocyte nuclear factor 1-alpha | U3KQS6 | HNF1A | − | + | − | − | |
| Hepatocyte nuclear factor 6 | Q9UBC0 | ONECUT1 | − | + | + | − | |
| Latent-transforming growth factor beta-binding protein 3 (**) (§) | Q9NS15 | LTBP3 | + | + | − | − | |
| Prospero homeobox protein 1 (**) (§) | Q92786 | PROX1 | − | − | − | + | |
| Transcription factor SOX-4 (**) | Q06945 | SOX4 | − | − | − | + | |
| Transgelin | Q01995 | TAGLN | + | + | + | + | |
| Immune system process | Beta-2-microglobulin | P61769 | B2M | + | − | + | + |
| Complement C1r subcomponent | B4DPQ0 | C1R | − | + | − | − | |
| Complement C2 (Fragment) | H0Y868 | C2 | − | − | − | + | |
| Complement C3 (Fragment) | M0R0Q9 | C3 | − | + | − | − | |
| Complement C5 | P01031 | C5 | − | − | − | + | |
| Complement factor H | P08603 | CFH | − | + | − | − | |
| Fibronectin | P02751 | FN1 | + | + | + | + | |
| Galectin-1 | P09382 | LGALS1 | + | + | + | + | |
| Galectin-3 | P17931 | LGALS3 | + | + | − | + | |
| Guanylate-binding protein 1 | P32455 | GBP1 | − | + | − | + | |
| HLA class I histocompatibility antigen, A-29 alpha chain | P30512 | HLA-A | − | + | − | − | |
| HLA class I histocompatibility antigen, A-68 alpha chain | P01891 | HLA-A | − | + | − | + | |
| Indoleamine 2,3-dioxygenase 2 | Q6ZQW0 | IDO2 | − | − | − | + | |
| Indoleamine-pyrrole 2,3-dioxygenase | P14902 | IDO1 | − | + | − | − | |
| Kallikrein-2 (Fragment) | A0A075B7A6 | KLK2 | − | − | − | + | |
| Non-POU domain-containing octamer-binding protein | Q15233 | NONO | − | − | − | + | |
| Protein deglycase DJ-1 | Q99497 | PARK7 | + | + | + | + | |
| Prothrombin | P00734 | F2 | + | − | + | + | |
| Other functions | Actin, cytoplasmic 1 | P60709 | ACTB | + | + | + | − |
| Adenylate cyclase type 4 | Q8NFM4 | ADCY4 | − | + | − | + | |
| Adenylyl cyclase-associated protein 1 | Q01518 | CAP1 | + | + | + | + | |
| Amyloid-like protein 2 | Q06481 | APLP2 | − | − | − | + | |
| Calretinin | P22676 | CALB2 | − | − | − | + | |
| Calumenin | O43852 | CALU | + | + | + | + | |
| CD44 antigen | H0YDX6 | CD44 | + | + | + | + | |
| Cytokeratin 18 | P05783 | KRT18 | + | + | + | + | |
| Cytokeratin 19 | P08727 | KRT19 | + | − | + | + | |
| Cytokeratin 8 | P05787 | KRT8 | + | + | + | + | |
| Desmin | P17661 | DES | − | + | − | + | |
| Fibrillin-1 | P35555 | FBN1 | − | + | + | − | |
| Filamin-binding LIM protein 1 | Q8WUP2 | FBLIM1 | + | + | − | + | |
| Growth arrest-specific protein 2 | O43903 | GAS2 | − | − | − | + | |
| Growth factor receptor-bound protein 7 | Q14451 | GRB7 | − | − | − | + | |
| Heat shock-related 70 kDa protein 2 | P54652 | HSPA2 | − | + | − | + | |
| Integrin beta-1 (CD29) | P05556 | ITGB1 | + | + | + | + | |
| Kalirin | O60229 | KALRN | − | − | − | + | |
| Lactadherin | Q08431 | MFGE8 | + | + | + | + | |
| Laminin subunit alpha-1 | P25391 | LAMA1 | − | + | − | − | |
| Lipoma-preferred partner | Q93052 | LPP | − | + | − | + | |
| Plastin-3 | P13797 | PLS3 | + | + | + | + | |
| Platelet-derived growth factor receptor beta | P09619 | PDGFRB | − | + | − | + | |
| Plectin | Q15149 | PLEC | + | + | + | + | |
| PMS1 protein homolog 1 | P54277 | PMS1 | − | − | − | + | |
| Pregnancy zone protein | P20742 | PZP | − | + | − | − | |
| Procollagen-lysine,2-oxoglutarate 5-dioxygenase 1 | Q02809 | PLOD1 | + | + | + | + | |
| Septin-9 | Q9UHD8 | SEPT9 | − | + | − | + | |
| Sparc | P09486 | SPARC | + | + | + | + | |
| T-complex protein 1 subunit gamma | P49368 | CCT3 | + | + | − | + | |
| Tenascin-R | Q92752 | TNR | − | − | − | + | |
| Thy-1 membrane glycoprotein (CD90) | P04216 | THY1 | + | + | + | + | |
| UDP-glucose:glycoprotein glucosyltransferase 1 | Q9NYU2 | UGGT1 | − | + | + | + |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lo Iacono, M.; Corrao, S.; Alberti, G.; Amico, G.; Timoneri, F.; Russo, E.; Cucina, A.; Indelicato, S.; Rappa, F.; Corsello, T.; et al. Characterization and Proteomic Profiling of Hepatocyte-like Cells Derived from Human Wharton’s Jelly Mesenchymal Stromal Cells: De Novo Expression of Liver-Specific Enzymes. Biology 2025, 14, 124. https://doi.org/10.3390/biology14020124
Lo Iacono M, Corrao S, Alberti G, Amico G, Timoneri F, Russo E, Cucina A, Indelicato S, Rappa F, Corsello T, et al. Characterization and Proteomic Profiling of Hepatocyte-like Cells Derived from Human Wharton’s Jelly Mesenchymal Stromal Cells: De Novo Expression of Liver-Specific Enzymes. Biology. 2025; 14(2):124. https://doi.org/10.3390/biology14020124
Chicago/Turabian StyleLo Iacono, Melania, Simona Corrao, Giusi Alberti, Giandomenico Amico, Francesca Timoneri, Eleonora Russo, Annamaria Cucina, Sergio Indelicato, Francesca Rappa, Tiziana Corsello, and et al. 2025. "Characterization and Proteomic Profiling of Hepatocyte-like Cells Derived from Human Wharton’s Jelly Mesenchymal Stromal Cells: De Novo Expression of Liver-Specific Enzymes" Biology 14, no. 2: 124. https://doi.org/10.3390/biology14020124
APA StyleLo Iacono, M., Corrao, S., Alberti, G., Amico, G., Timoneri, F., Russo, E., Cucina, A., Indelicato, S., Rappa, F., Corsello, T., Saieva, S., Di Stefano, A., Di Gaudio, F., Conaldi, P. G., & La Rocca, G. (2025). Characterization and Proteomic Profiling of Hepatocyte-like Cells Derived from Human Wharton’s Jelly Mesenchymal Stromal Cells: De Novo Expression of Liver-Specific Enzymes. Biology, 14(2), 124. https://doi.org/10.3390/biology14020124








