Skeletal Muscle Differentiation and Epigenetics
A topical collection in Cells (ISSN 2073-4409). This collection belongs to the section "Cell and Gene Therapy".
Submission Status: Closed | Viewed by 45058Editor
Interests: skeletal muscle differentiation; cell cycle inhibitors; MyoD; gene expression; transcriptional control; chromatin architecture; chromatin dynamics; epigenetics; long noncoding RNAs
Topical Collection Information
Dear Colleagues,
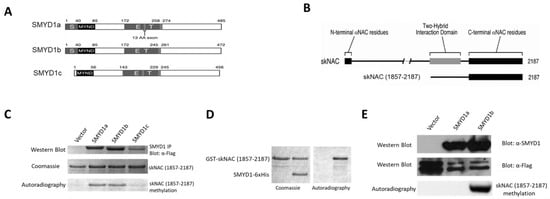
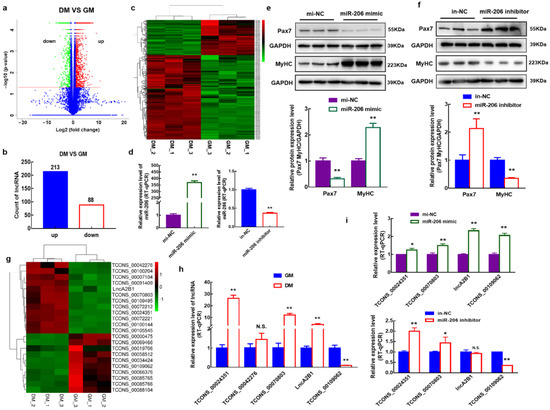
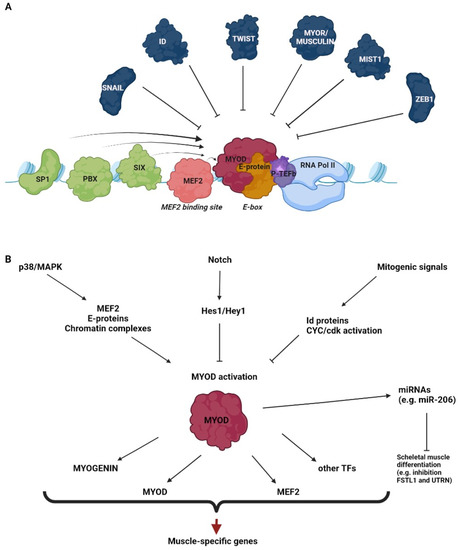
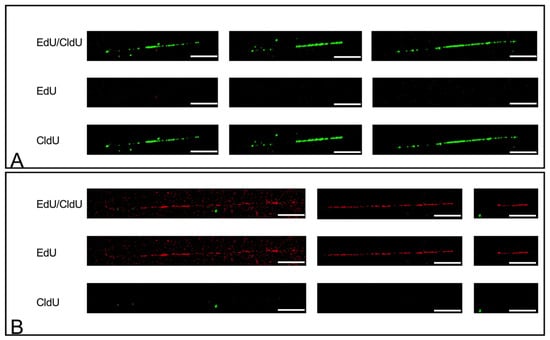
Skeletal myogenesis is a well-characterized process, both as regards the developmental phases of muscle formation and the adult phase of muscle regeneration. The commitment of mesodermal precursors to the myogenic lineage and the terminal differentiation of myoblasts into myofibers are regulated at multiple levels, ranging from pre-transcriptional to post-translational mechanisms. Special attention is being paid to the diverse epigenetic strategies by which muscle-specific patterns of gene expression are generated and maintained. Considerable evidence has been accumulated, showing that myogenic transcription factors, such as the prototypical pioneer factor MyoD, work in concert with chromatin modifiers in order to establish an open chromatin environment permissive for the transcriptional activation of muscle-specific genes. More recently, genome-wide studies correlating transcription factor binding, 3D chromatin dynamics, and gene expression have provided further insight into the molecular events underlying the coordinate activation or repression of entire sets of genes during myogenesis. Despite the advances in our understanding of these complex processes, many aspects of the epigenetics of skeletal muscle differentiation remain to be elucidated.
This Topical Collection will present a collection of recent original research papers and review articles in all areas of this field. Potential subjects include, but are not limited to, the identification and characterization of novel epigenetic players as well as of novel functional interactions of myogenic factors with chromatin-modifying enzymes, chromatin remodelers, regulatory noncoding RNAs, and chromatin architectural proteins. Additional topics of interest are the roles of extracellular and intracellular signaling in the modulation of chromatin function and the dysregulation of epigenetic networks in skeletal muscle pathologies, with a view to developing new therapeutic approaches based on the manipulation of specific regulatory pathways.
Prof. Dr. Rossella Maione
Collection Editor
Manuscript Submission Information
Manuscripts should be submitted online at www.mdpi.com by registering and logging in to this website. Once you are registered, click here to go to the submission form. Manuscripts can be submitted until the deadline. All submissions that pass pre-check are peer-reviewed. Accepted papers will be published continuously in the journal (as soon as accepted) and will be listed together on the collection website. Research articles, review articles as well as short communications are invited. For planned papers, a title and short abstract (about 100 words) can be sent to the Editorial Office for announcement on this website.
Submitted manuscripts should not have been published previously, nor be under consideration for publication elsewhere (except conference proceedings papers). All manuscripts are thoroughly refereed through a single-blind peer-review process. A guide for authors and other relevant information for submission of manuscripts is available on the Instructions for Authors page. Cells is an international peer-reviewed open access semimonthly journal published by MDPI.
Please visit the Instructions for Authors page before submitting a manuscript. The Article Processing Charge (APC) for publication in this open access journal is 2700 CHF (Swiss Francs). Submitted papers should be well formatted and use good English. Authors may use MDPI's English editing service prior to publication or during author revisions.
Keywords
- skeletal muscle differentiation and regeneration
- epigenetic control of gene expression
- chromatin stucture and architecture
- DNA methylation
- histone modifications
- nucleosome remodeling
- noncoding RNAs
- muscle regulatory factors
- signal transduction
Related Special Issue
- Skeletal Muscle Differentiation and Epigenetics - Volume II in Cells (2 articles)