Predictive Toxicology
A topical collection in Toxics (ISSN 2305-6304). This collection belongs to the section "Novel Methods in Toxicology Research".
Viewed by 9767Editor
Interests: bioinformatics; drug-induced liver injury; drug safety; biomarker discovery; toxicogenomics
Special Issues, Collections and Topics in MDPI journals
Topical Collection Information
Dear Colleagues,
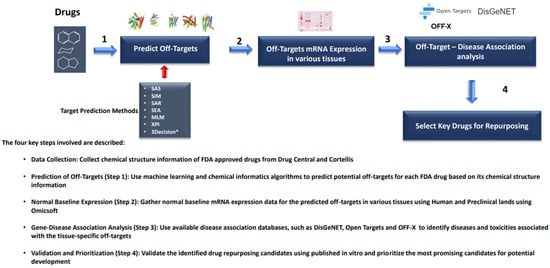
The recent advances of toxicogenomics, high-throughput screening, stem cells, and image analysis are creating unique opportunities to improve our ability to predict risk in humans and the development of predictive toxicology. These modern biotechnologies are producing big toxicological data and require advanced artificial intelligence technologies to evaluate the potential for predicting toxicity. The application of conventional machine learning algorithms, such as logical regression, decision tree, and support vector machines, have largely enhanced our capability to recover useful knowledge from the increasing volume of toxicity data. A recent study reported by researchers from John Hopkins University, demonstrated that using artificial intelligent algorithms trained on chemical-safety, big data could be more predictive and outperform expensive animals studies on some toxicities. Notably, the development of deep learning techniques, with the help of advanced computer technologies (e.g., the use of graphical processing units (GPU)) and complicated neural network algorithms, have brought about breakthroughs in computer vision and pattern recognition, image and speech recognition, drug discovery, and toxicology. In several public scientific challenges, including the Merck-sponsored Kaggle competition in 2012 and the Tox21 Data Challenge in 2015, deep learning algorithms demonstrated a superior predictive performance to convenient machine learning algorithms.
In this Topical Collection, we focus on exploring the relationship between the toxicity of xenobiotics and their chemical structures, disturbed cellular, and molecular pathways by the application of artificial intelligent methods to improve the prediction of toxicity risk. In addition, we especially encourage submissions on applying deep learning techniques to process datasets from high-dimensional gene expression, image and high-throughput screening, and chemical structures.
You may choose our Joint Topical Collection in IJERPH.
Dr. Minjun Chen
Collection Editor
Manuscript Submission Information
Manuscripts should be submitted online at www.mdpi.com by registering and logging in to this website. Once you are registered, click here to go to the submission form. Manuscripts can be submitted until the deadline. All submissions that pass pre-check are peer-reviewed. Accepted papers will be published continuously in the journal (as soon as accepted) and will be listed together on the collection website. Research articles, review articles as well as short communications are invited. For planned papers, a title and short abstract (about 100 words) can be sent to the Editorial Office for announcement on this website.
Submitted manuscripts should not have been published previously, nor be under consideration for publication elsewhere (except conference proceedings papers). All manuscripts are thoroughly refereed through a single-blind peer-review process. A guide for authors and other relevant information for submission of manuscripts is available on the Instructions for Authors page. Toxics is an international peer-reviewed open access monthly journal published by MDPI.
Please visit the Instructions for Authors page before submitting a manuscript. The Article Processing Charge (APC) for publication in this open access journal is 2600 CHF (Swiss Francs). Submitted papers should be well formatted and use good English. Authors may use MDPI's English editing service prior to publication or during author revisions.
Keywords
- predictive toxicology
- artificial intelligence
- big data
- machine learning
- deep learning
- toxicogenomics
- high throughput screening
- image analysis
- chemical structure