Recent Applications of Artificial Intelligence from Histopathologic Image-Based Prediction of Microsatellite Instability in Solid Cancers: A Systematic Review
Abstract
:Simple Summary
Abstract
1. Introduction
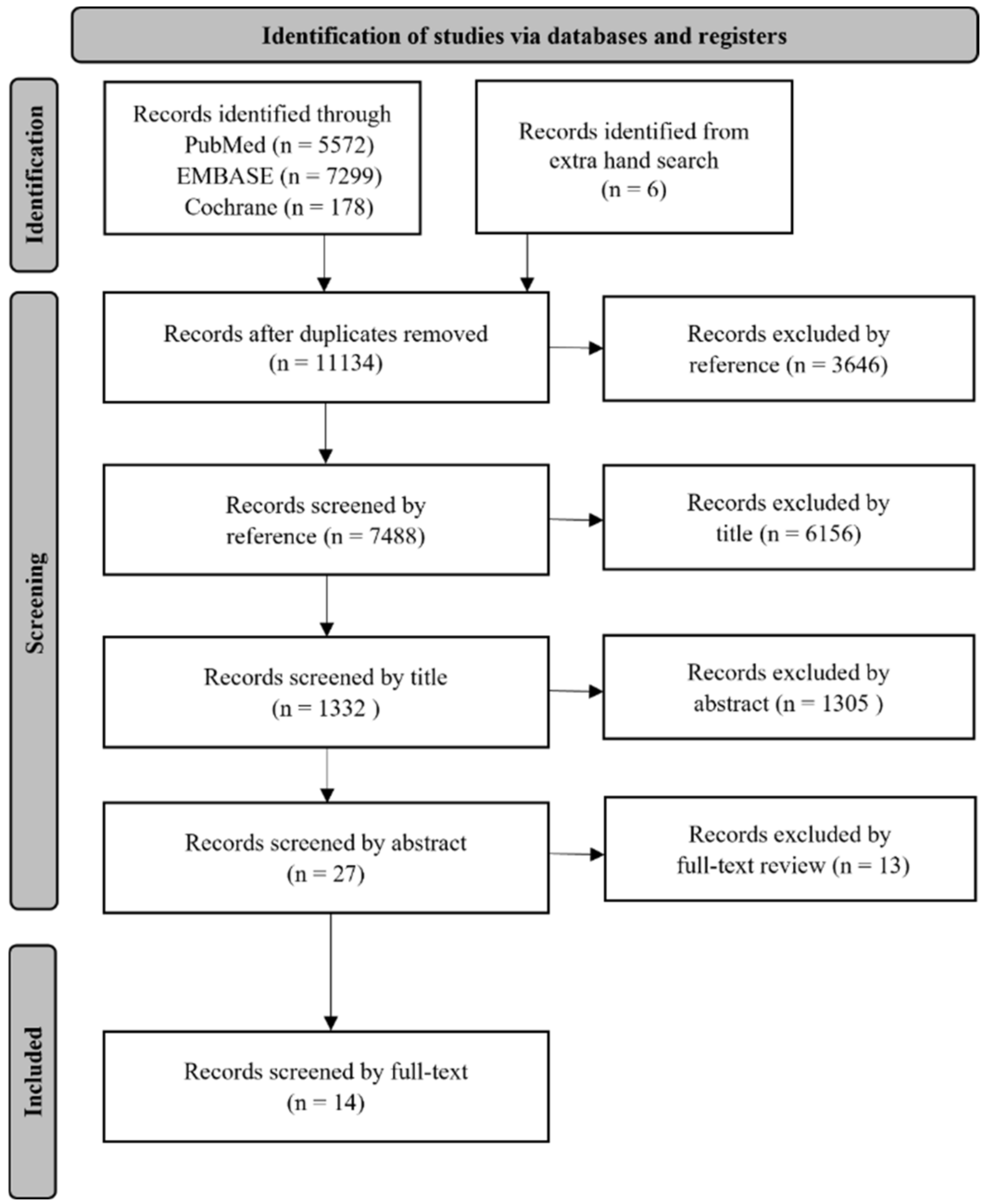
2. Materials and Methods
2.1. Search Strategy
2.2. Article Selection and Data Extraction and Analysis
3. Results
3.1. Characteristics of Eligible Study
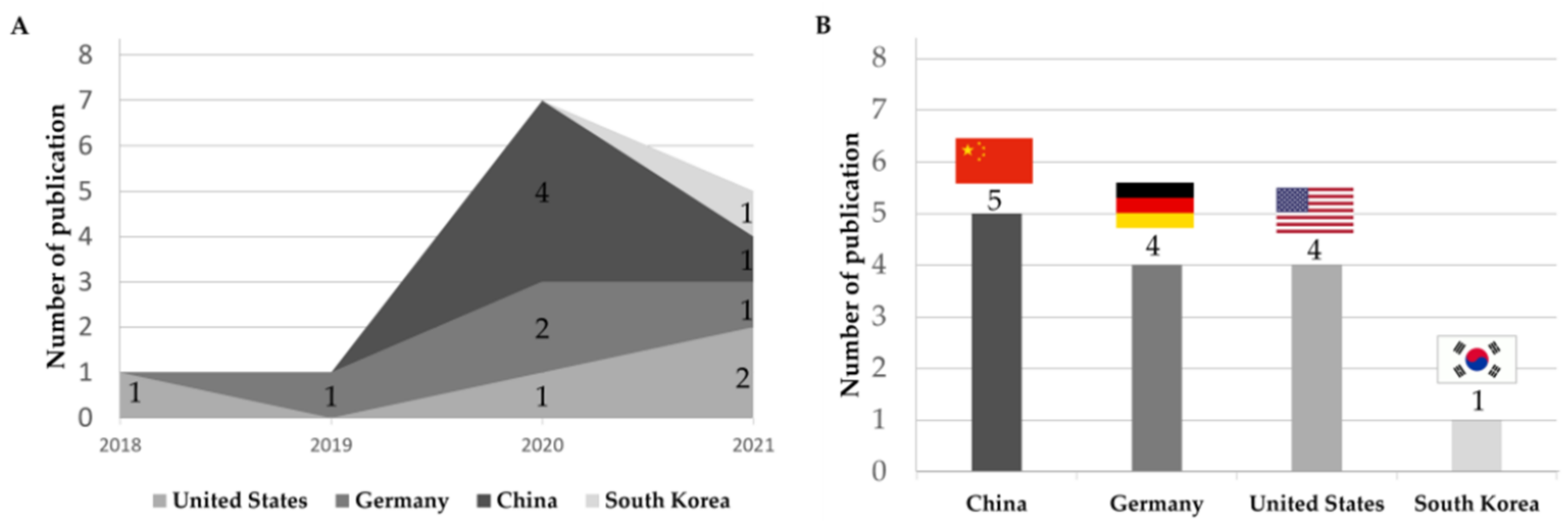
3.2. Yearly and Country-Wise Trend of Publication
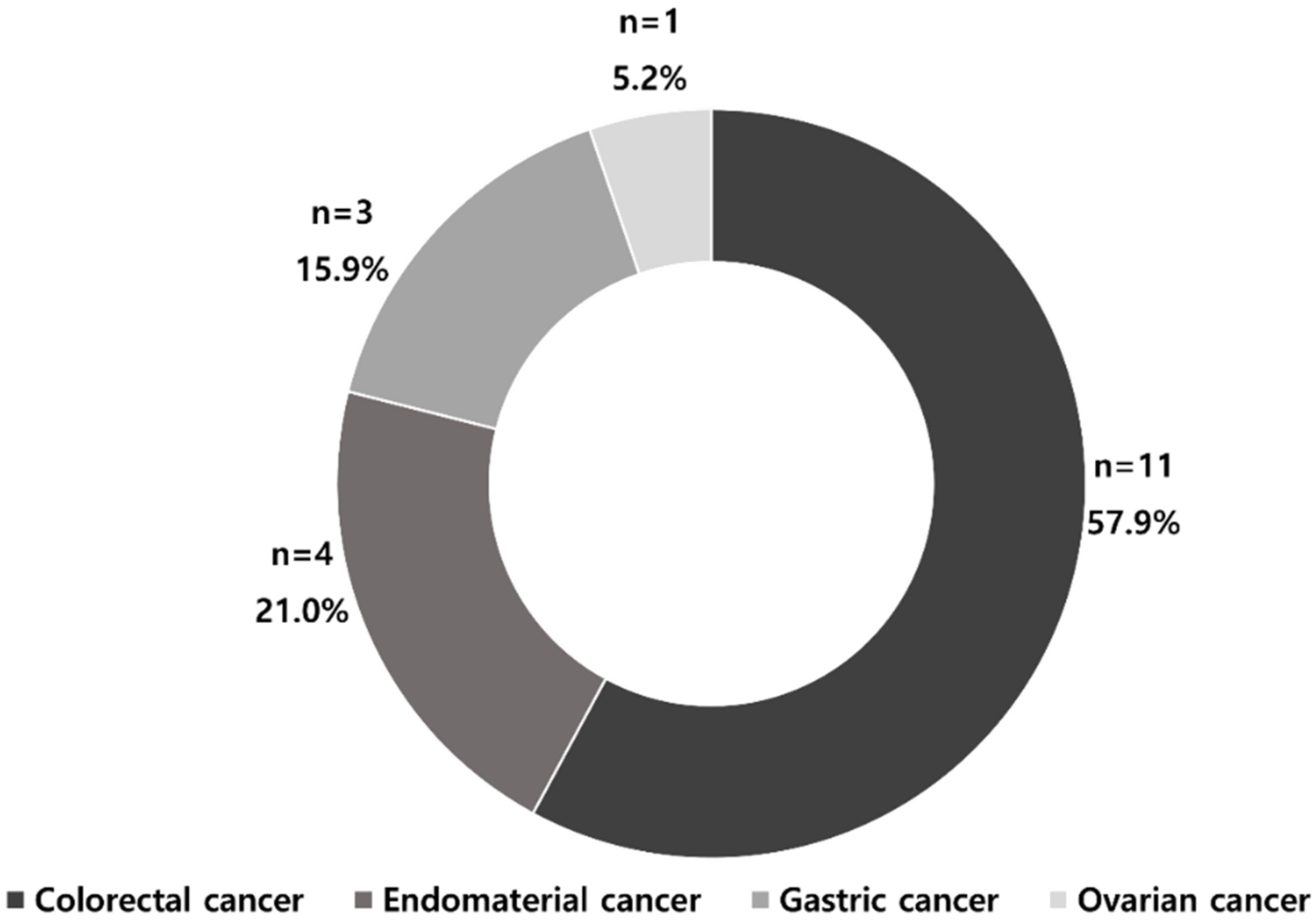
3.3. MSI Prediction Models by Cancer Types
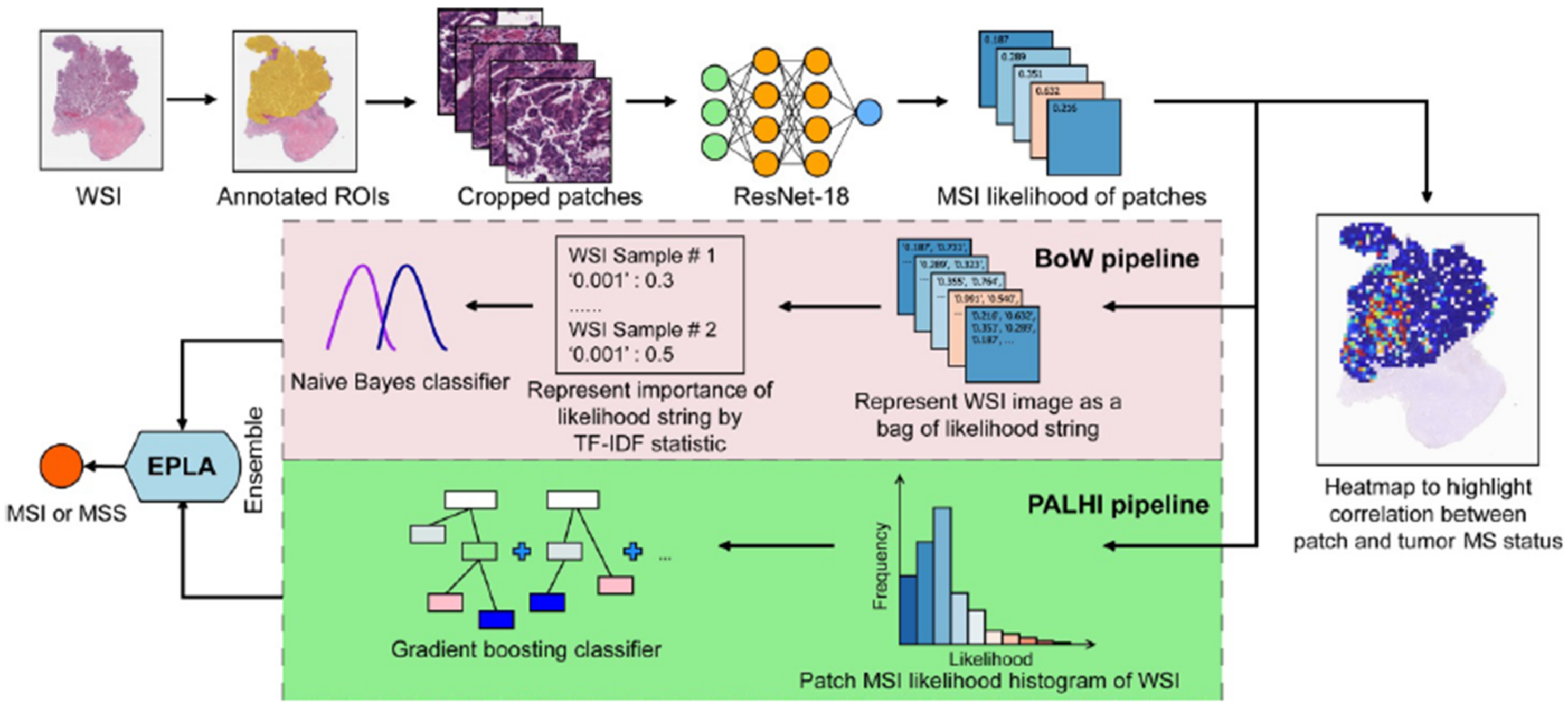
3.4. Prediction of MSI Status in CRC
| Author | Year | Country | AI Model | Training and Validation Data Set/WSIs/No. of Patients (n) | Pixel Levels | Additional Methodology for Validating MSI | Performance Metrics | External Validation Dataset/WSIs/No. of Patients (n) | External Validation Result | Ref. |
|---|---|---|---|---|---|---|---|---|---|---|
| Zhang | 2018 | USA | Inception-V3- | TCGA/NC/585 | 1000 × 1000 | NC | ACC: 98.3% | NS | NS | [51] |
| Kather | 2019 | Germany | ResNet18 | TCGA-FFPE/360/NC | NC | PCR | AUC: 0.77 | DACHS-FFPE, n = 378 | AUC: 0.84 | [29] |
| TCGA-FSS/387/NC | NC | PCR | AUC: 0.84 | DACHS-FFPE, n = 378 | AUC: 0.61 | |||||
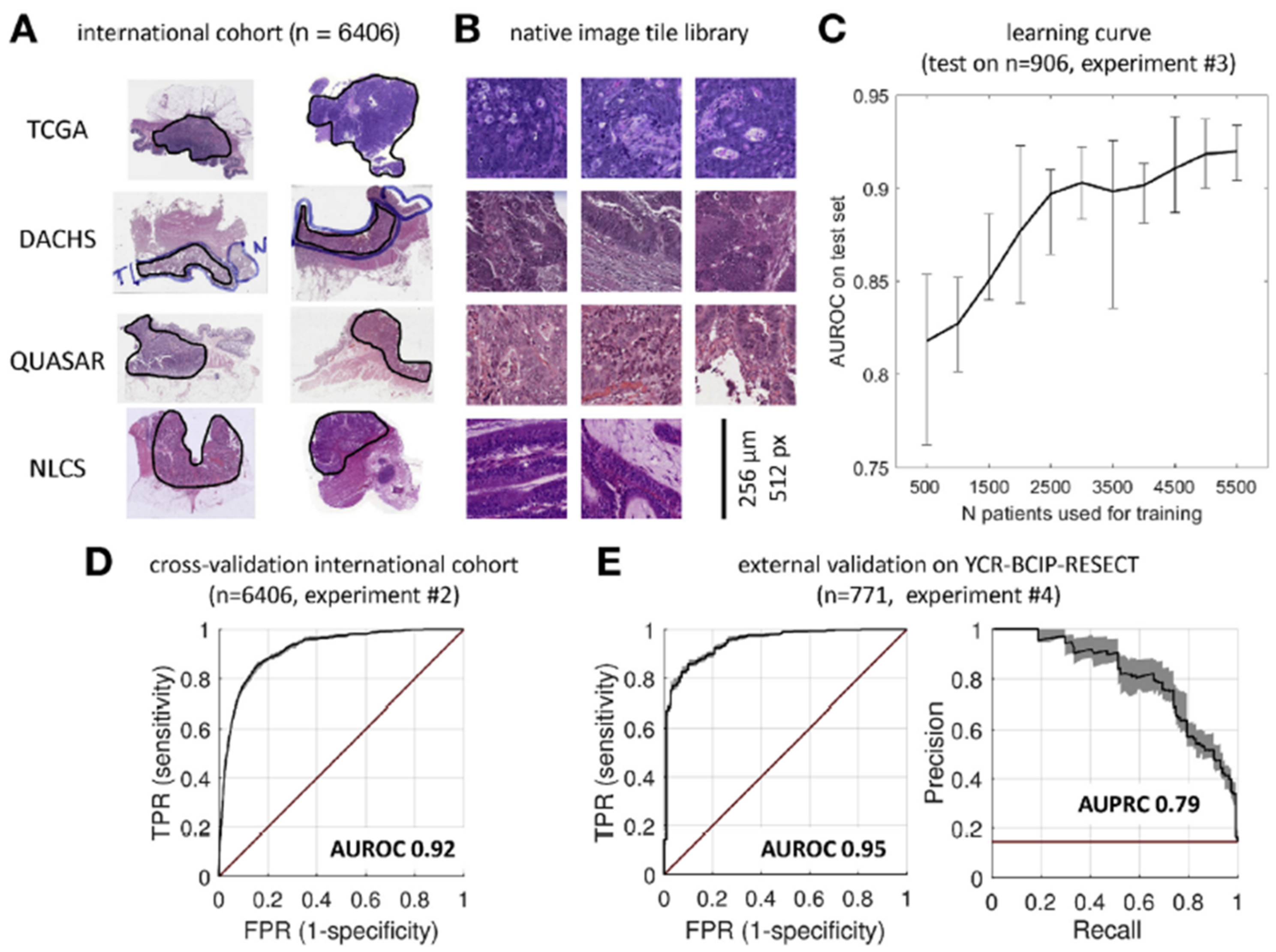
| Echle | 2020 | Germany | ShuffleNet | TCGA, DACHS, QUASAR, NLCS/6406/6406 | 512 × 512 | PCR/IHC | AUC: 0.92 Specificity: 67.0% Sensitivity: 95.0% | YCR-BCIP-RESECT, n = 771 | AUC: 0.95 | [30] |
| YCR-BCIP-BIOPSY, n = 1531 | AUC: 0.78 | |||||||||
| Cao | 2020 | China | ResNet18 | TCGA-FSS/429/429 | 224 × 224 | NGS/PCR | AUC: 0.88 Specificity: 77.0% Sensitivity: 91.0% | Asian-CRC-FFPE, n = 785 | AUC: 0.64 | [50] |
| Ke | 2020 | China | AlexNet | TCGA/747/NC | 224 × 224 | NC | MSI score: 0.90 | NS | NS | [52] |
| Kather | 2020 | Germany | ShuffleNet | TCGA/NC/426, | 512 × 512 | PCR | NC | DACHS, n = 379 | AUC: 0.89 | [53] |
| Schmauch | 2020 | USA | ResNet50 | TCGA/NC/465 | 224 × 224 | PCR | AUC: 0.82 | NS | NS | [54] |
| Zhu | 2020 | China | ResNet18 | TCGA-FFPE: 360 | NC | NC | AUC: 0.81 | NS | NS | [55] |
| TCGA-FSS: 385 | NC | NC | AUC: 0.84 | |||||||
| Yamashita | 2021 | USA | MSINet | In-house sample/100/100 | 224 × 224 | PCR | AUC: 0.93 | TCGA/484/479 | AUC: 0.77 | [49] |
| Krause | 2021 | Germany | ShuffleNet | TCGA-FFPE, n = 398 | 512 × 512 | PCR | AUC: 0.74 | NS | NS | [56] |
| Lee | 2021 | South Korea | Inception-V3- | TCGA and SMH/1920/500 | 360 × 360 | PCR/IHC | AUC: 0.89 | NC | AUC: 0.97 | [48] |
3.5. Prediction of MSI Status in Endometrial, Gastric, and Ovarian Cancers
| Organ /Cancers | Author | Year | Country | AI-Based Model | Data Set/WSIs/No. of Patients (n) | Pixel Level | Additional Methodology for Validating MSI | Performance Metrics | External Validation Dataset/WSIs/No. of Patients (n) | External Validation Result | Ref. |
|---|---|---|---|---|---|---|---|---|---|---|---|
| Endometrial cancer | Zhang | 2018 | USA | Inception-V3 | TCGA-UCEC and CRC/1141/NC | 1000 × 1000 | NC | ACC: 84.2% | NS | NS | [51] |
| Kather | 2019 | Germany | ResNet18 | TCGA-FFPE/NC/492 | NC | PCR | AUC: 0.75 | NS | NS | [29] | |
| Wang | 2020 | China | ResNet18 | TCGA/NC/516 | 512 × 512 | NC | AUC: 0.73 | NS | NS | [59] | |
| Hong | 2021 | USA | InceptionResNetVI | TCGA, CPTAC/496/456 | 299 × 299 | PCR/NGS | AUC: 0.82 | NYU-H/137/41 | AUC: 0.66 | [57] | |
| Gastric cancer | Kather | 2019 | Germany | ResNet18 | TCGA-FFPE/NC/315 | NC | PCR | AUC: 0.81 | KCCH-FFPE-Japan/NC/185 | AUC: 0.69 | [29] |
| Zhu | 2020 | China | ResNet18 | TCGA-FFPE/285/NC | NC | NC | AUC: 0.80 | NS | NS | [55] | |
| Schmauch | 2020 | USA | ResNet50 | TCGA/323/NC | 224 × 224 | PCR | AUC: 0.76 | NS | NS | [54] | |
| Ovarian cancer | Zeng | 2021 | China | Random forest | TCGA/NC/229 | 1000 × 1000 | NC | AUC: 0.91 | NS | NS | [58] |
4. Discussion
4.1. Present Status of AI Models
4.1.1. Yearly, Country-Wise, and Organ-Wise Publication Trend
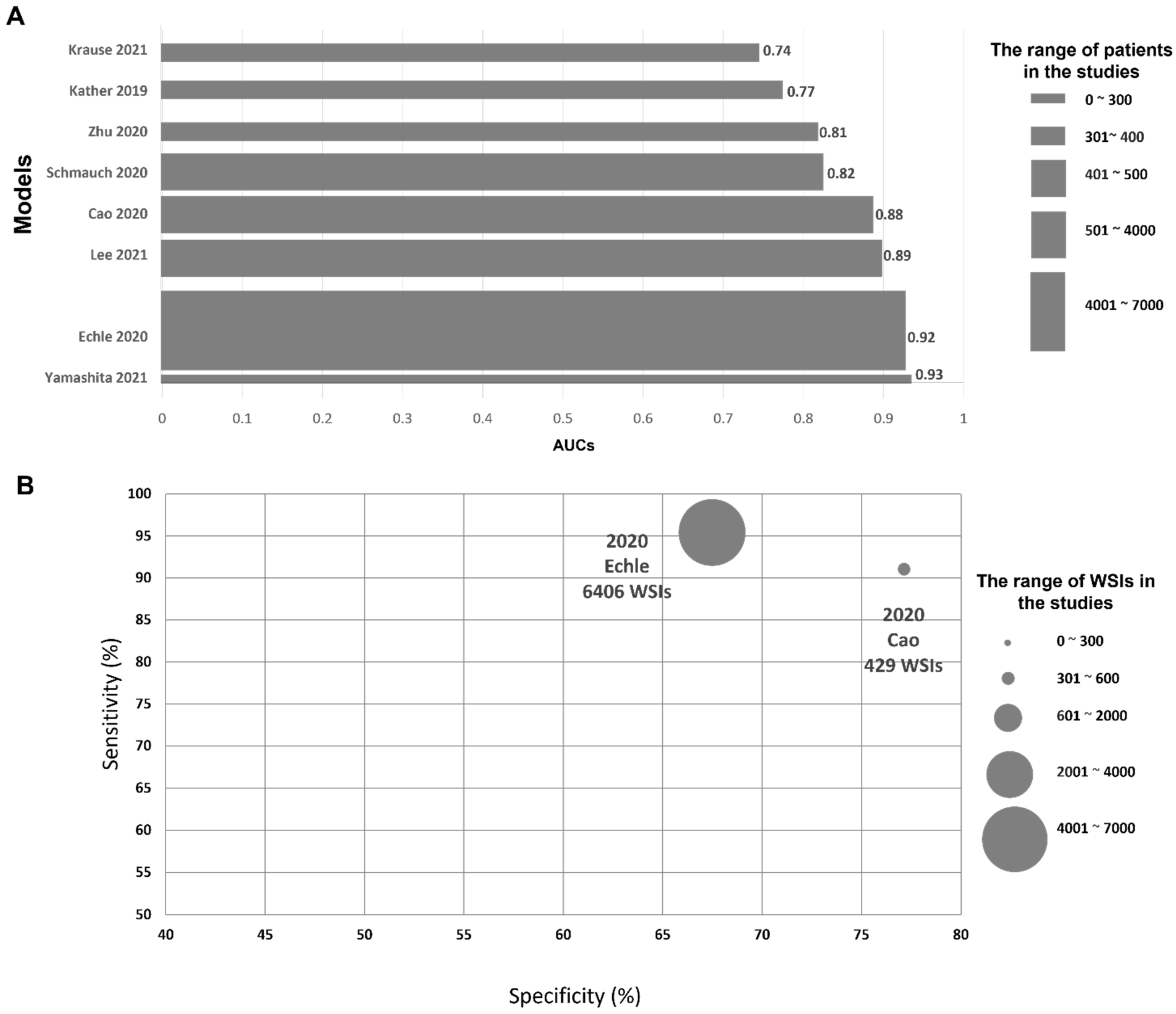
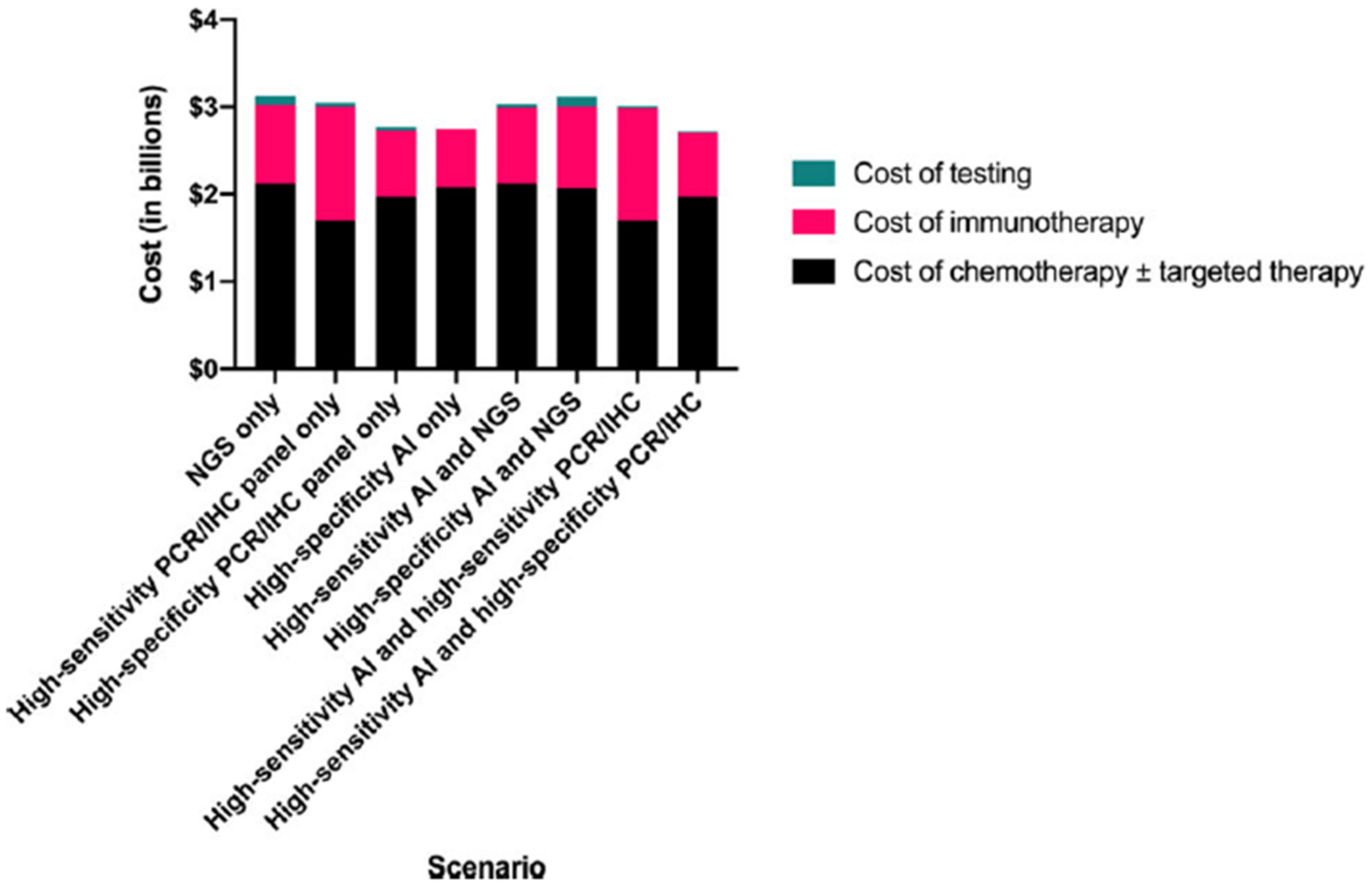
4.1.2. Performance of AI Models and Their Cost Effectiveness
4.2. Limitation and Challenge of AI Models
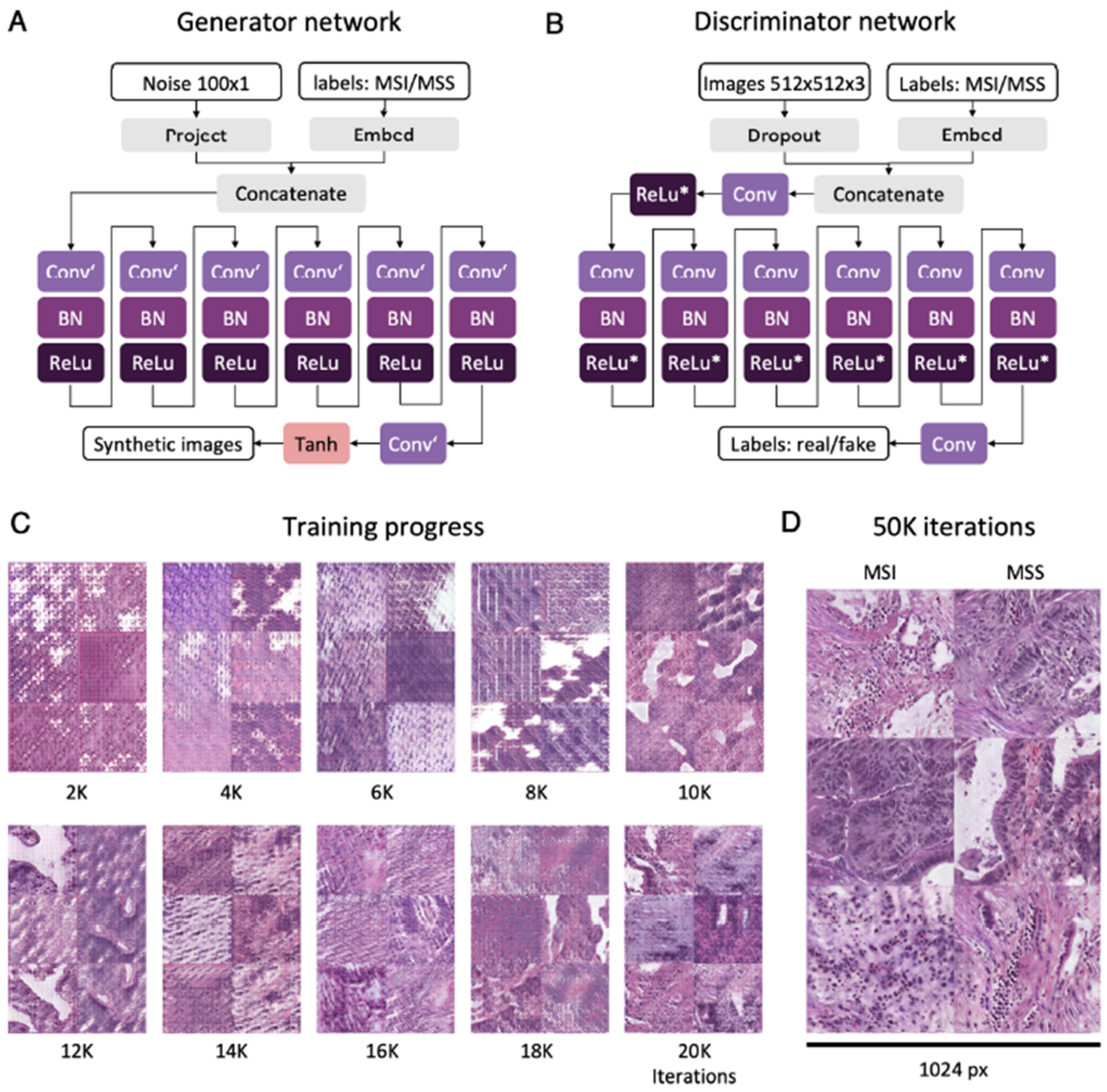
4.2.1. Data, Image Quality and CNN Architecture
4.2.2. External Validation and Multi-Institutional Study
4.2.3. MSI Prediction on Biopsy Samples
4.2.4. Establishment of Central Facility
4.3. Future Direction
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Popat, S.; Hubner, R.; Houlston, R. Systematic review of microsatellite instability and colorectal cancer prognosis. J. Clin. Oncol. 2005, 23, 609–618. [Google Scholar] [CrossRef]
- Boland, C.R.; Goel, A. Microsatellite instability in colorectal cancer. Gastroenterology 2010, 138, 2073–2087. [Google Scholar] [CrossRef]
- Le, D.T.; Uram, J.N.; Wang, H.; Bartlett, B.R.; Kemberling, H.; Eyring, A.D.; Skora, A.D.; Luber, B.S.; Azad, N.S.; Laheru, D. PD-1 blockade in tumors with mismatch-repair deficiency. N. Engl. J. Med. 2015, 372, 2509–2520. [Google Scholar] [CrossRef] [Green Version]
- Greenson, J.K.; Bonner, J.D.; Ben-Yzhak, O.; Cohen, H.I.; Miselevich, I.; Resnick, M.B.; Trougouboff, P.; Tomsho, L.D.; Kim, E.; Low, M. Phenotype of microsatellite unstable colorectal carcinomas: Well-differentiated and focally mucinous tumors and the absence of dirty necrosis correlate with microsatellite instability. Am. J. Surg. Path. 2003, 27, 563–570. [Google Scholar] [CrossRef]
- Smyrk, T.C.; Watson, P.; Kaul, K.; Lynch, H.T. Tumor-infiltrating lymphocytes are a marker for microsatellite instability in colorectal carcinoma. Cancer 2001, 91, 2417–2422. [Google Scholar] [CrossRef]
- Tariq, K.; Ghias, K. Colorectal cancer carcinogenesis: A review of mechanisms. Cancer Biol. Med. 2016, 13, 120–135. [Google Scholar] [CrossRef] [Green Version]
- Devaud, N.; Gallinger, S. Chemotherapy of MMR-deficient colorectal cancer. Fam. Cancer 2013, 12, 301–306. [Google Scholar] [CrossRef]
- Cheng, L.; Zhang, D.Y.; Eble, J.N. Molecular Genetic Pathology, 2nd ed.; Springer: New York, NY, USA, 2013. [Google Scholar]
- Hewish, M.; Lord, C.J.; Martin, S.A.; Cunningham, D.; Ashworth, A. Mismatch repair deficient colorectal cancer in the era of personalized treatment. Nat. Rev. Clin. Oncol. 2010, 7, 197–208. [Google Scholar] [CrossRef]
- Evrard, C.; Tachon, G.; Randrian, V.; Karayan-Tapon, L.; Tougeron, D. Microsatellite instability: Diagnosis, heterogeneity, discordance, and clinical impact in colorectal cancer. Cancers 2019, 11, 1567. [Google Scholar] [CrossRef] [Green Version]
- Revythis, A.; Shah, S.; Kutka, M.; Moschetta, M.; Ozturk, M.A.; Pappas-Gogos, G.; Ioannidou, E.; Sheriff, M.; Rassy, E.; Boussios, S. Unraveling the wide spectrum of melanoma biomarkers. Diagnostics 2021, 11, 1341. [Google Scholar] [CrossRef]
- Bailey, M.H.; Tokheim, C.; Porta-Pardo, E.; Sengupta, S.; Bertrand, D.; Weerasinghe, A.; Colaprico, A.; Wendl, M.C.; Kim, J.; Reardon, B. Comprehensive characterization of cancer driver genes and mutations. Cell 2018, 173, 371–385. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bonneville, R.; Krook, M.A.; Kautto, E.A.; Miya, J.; Wing, M.R.; Chen, H.-Z.; Reeser, J.W.; Yu, L.; Roychowdhury, S. Landscape of microsatellite instability across 39 cancer types. JCO Precis. Oncol. 2017, 2017, PO.17.00073. [Google Scholar] [CrossRef] [PubMed]
- Ghose, A.; Moschetta, M.; Pappas-Gogos, G.; Sheriff, M.; Boussios, S. Genetic Aberrations of DNA Repair Pathways in Prostate Cancer: Translation to the Clinic. Int. J. Mol. Sci. 2021, 22, 9783. [Google Scholar] [CrossRef] [PubMed]
- Mosele, F.; Remon, J.; Mateo, J.; Westphalen, C.; Barlesi, F.; Lolkema, M.; Normanno, N.; Scarpa, A.; Robson, M.; Meric-Bernstam, F. Recommendations for the use of next-generation sequencing (NGS) for patients with metastatic cancers: A report from the ESMO Precision Medicine Working Group. Ann. Oncol. 2020, 31, 1491–1505. [Google Scholar] [CrossRef]
- Khalil, D.N.; Smith, E.L.; Brentjens, R.J.; Wolchok, J.D. The future of cancer treatment: Immunomodulation, CARs and combination immunotherapy. Nat. Rev. Clin. Oncol. 2016, 13, 273–290. [Google Scholar] [CrossRef] [Green Version]
- Mittal, D.; Gubin, M.M.; Schreiber, R.D.; Smyth, M.J. New insights into cancer immunoediting and its three component phases—Elimination, equilibrium and escape. Curr. Opin. Immunol. 2014, 27, 16–25. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Darvin, P.; Toor, S.M.; Nair, V.S.; Elkord, E. Immune checkpoint inhibitors: Recent progress and potential biomarkers. Exp. Mol. Med. 2018, 50, 165. [Google Scholar] [CrossRef] [Green Version]
- Herbst, R.S.; Soria, J.-C.; Kowanetz, M.; Fine, G.D.; Hamid, O.; Gordon, M.S.; Sosman, J.A.; McDermott, D.F.; Powderly, J.D.; Gettinger, S.N. Predictive correlates of response to the anti-PD-L1 antibody MPDL3280A in cancer patients. Nature 2014, 515, 563–567. [Google Scholar]
- Zou, W.; Wolchok, J.D.; Chen, L. PD-L1 (B7-H1) and PD-1 pathway blockade for cancer therapy: Mechanisms, response biomarkers, and combinations. Sci. Transl. Med. 2016, 8, 328rv324. [Google Scholar] [CrossRef] [Green Version]
- Jenkins, M.A.; Hayashi, S.; O’shea, A.-M.; Burgart, L.J.; Smyrk, T.C.; Shimizu, D.; Waring, P.M.; Ruszkiewicz, A.R.; Pollett, A.F.; Redston, M. Pathology features in Bethesda guidelines predict colorectal cancer microsatellite instability: A population-based study. Gastroenterology 2007, 133, 48–56. [Google Scholar] [CrossRef] [Green Version]
- Alexander, J.; Watanabe, T.; Wu, T.-T.; Rashid, A.; Li, S.; Hamilton, S.R. Histopathological identification of colon cancer with microsatellite instability. Am. J. Pathol. 2001, 158, 527–535. [Google Scholar] [CrossRef] [Green Version]
- Benson, A.B.; Venook, A.P.; Al-Hawary, M.M.; Arain, M.A.; Chen, Y.-J.; Ciombor, K.K.; Cohen, S.A.; Cooper, H.S.; Deming, D.A.; Garrido-Laguna, I. Small bowel adenocarcinoma, version 1.2020, NCCN clinical practice guidelines in oncology. J. Natl. Compr. Canc. Netw. 2019, 17, 1109–1133. [Google Scholar] [PubMed]
- Koh, W.-J.; Abu-Rustum, N.R.; Bean, S.; Bradley, K.; Campos, S.M.; Cho, K.R.; Chon, H.S.; Chu, C.; Clark, R.; Cohn, D. Cervical cancer, version 3.2019, NCCN clinical practice guidelines in oncology. J. Natl. Compr. Cancer Netw. 2019, 17, 64–84. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sepulveda, A.R.; Hamilton, S.R.; Allegra, C.J.; Grody, W.; Cushman-Vokoun, A.M.; Funkhouser, W.K.; Kopetz, S.E.; Lieu, C.; Lindor, N.M.; Minsky, B.D. Molecular Biomarkers for the Evaluation of Colorectal Cancer: Guideline From the American Society for Clinical Pathology, College of American Pathologists, Association for Molecular Pathology, and American Society of Clinical Oncology. J. Mol. Diagn. 2017, 19, 187–225. [Google Scholar] [CrossRef] [Green Version]
- Percesepe, A.; Borghi, F.; Menigatti, M.; Losi, L.; Foroni, M.; Di Gregorio, C.; Rossi, G.; Pedroni, M.; Sala, E.; Vaccina, F. Molecular screening for hereditary nonpolyposis colorectal cancer: A prospective, population-based study. J. Clin. Oncol. 2001, 19, 3944–3950. [Google Scholar] [CrossRef] [PubMed]
- Aaltonen, L.A.; Salovaara, R.; Kristo, P.; Canzian, F.; Hemminki, A.; Peltomäki, P.; Chadwick, R.B.; Kääriäinen, H.; Eskelinen, M.; Järvinen, H. Incidence of hereditary nonpolyposis colorectal cancer and the feasibility of molecular screening for the disease. N. Engl. J. Med. 1998, 338, 1481–1487. [Google Scholar] [PubMed]
- Singh, M.P.; Rai, S.; Pandey, A.; Singh, N.K.; Srivastava, S. Molecular subtypes of colorectal cancer: An emerging therapeutic opportunity for personalized medicine. Genes Dis. 2021, 8, 133–145. [Google Scholar] [CrossRef]
- Kather, J.N.; Pearson, A.T.; Halama, N.; Jäger, D.; Krause, J.; Loosen, S.H.; Marx, A.; Boor, P.; Tacke, F.; Neumann, U.P. Deep learning can predict microsatellite instability directly from histology in gastrointestinal cancer. Nat. Med. 2019, 25, 1054–1056. [Google Scholar] [CrossRef]
- Echle, A.; Grabsch, H.I.; Quirke, P.; van den Brandt, P.A.; West, N.P.; Hutchins, G.G.; Heij, L.R.; Tan, X.; Richman, S.D.; Krause, J. Clinical-grade detection of microsatellite instability in colorectal tumors by deep learning. Gastroenterology 2020, 159, 1406–1416. [Google Scholar] [CrossRef]
- Coelho, H.; Jones-Hughes, T.; Snowsill, T.; Briscoe, S.; Huxley, N.; Frayling, I.M.; Hyde, C. A Systematic Review of Test Accuracy Studies Evaluating Molecular Micro-Satellite Instability Testing for the Detection of Individuals With Lynch Syndrome. BMC Cancer 2017, 17, 836. [Google Scholar] [CrossRef]
- Snowsill, T.; Coelho, H.; Huxley, N.; Jones-Hughes, T.; Briscoe, S.; Frayling, I.M.; Hyde, C. Molecular testing for Lynch syndrome in people with colorectal cancer: Systematic reviews and economic evaluation. Health Technol Assess 2017, 21, 1–238. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, X.; Li, J. Era of universal testing of microsatellite instability in colorectal cancer. World. J. Gastrointest. Oncol. 2013, 5, 12–19. [Google Scholar] [CrossRef] [PubMed]
- Cohen, R.; Hain, E.; Buhard, O.; Guilloux, A.; Bardier, A.; Kaci, R.; Bertheau, P.; Renaud, F.; Bibeau, F.; Fléjou, J.-F. Association of primary resistance to immune checkpoint inhibitors in metastatic colorectal cancer with misdiagnosis of microsatellite instability or mismatch repair deficiency status. JAMA Oncol. 2019, 5, 551–555. [Google Scholar] [CrossRef] [PubMed]
- Andre, T.; Shiu, K.-K.; Kim, T.W.; Jensen, B.V.; Jensen, L.H.; Punt, C.J.; Smith, D.M.; Garcia-Carbonero, R.; Benavides, M.; Gibbs, P. Pembrolizumab versus chemotherapy for microsatellite instability-high/mismatch repair deficient metastatic colorectal cancer: The phase 3 KEYNOTE-177 Study. J. Clin. Oncol. 2020, 38, LBA4. [Google Scholar] [CrossRef]
- Ardila, D.; Kiraly, A.P.; Bharadwaj, S.; Choi, B.; Reicher, J.J.; Peng, L.; Tse, D.; Etemadi, M.; Ye, W.; Corrado, G. End-to-end lung cancer screening with three-dimensional deep learning on low-dose chest computed tomography. Nat. Med. 2019, 25, 954–961. [Google Scholar] [CrossRef]
- Bejnordi, B.E.; Veta, M.; Van Diest, P.J.; Van Ginneken, B.; Karssemeijer, N.; Litjens, G.; Van Der Laak, J.A.; Hermsen, M.; Manson, Q.F.; Balkenhol, M. Diagnostic assessment of deep learning algorithms for detection of lymph node metastases in women with breast cancer. JAMA 2017, 318, 2199–2210. [Google Scholar] [CrossRef]
- Esteva, A.; Kuprel, B.; Novoa, R.A.; Ko, J.; Swetter, S.M.; Blau, H.M.; Thrun, S. Dermatologist-level classification of skin cancer with deep neural networks. Nature 2017, 542, 115–118. [Google Scholar] [CrossRef]
- Nam, S.; Chong, Y.; Jung, C.K.; Kwak, T.-Y.; Lee, J.Y.; Park, J.; Rho, M.J.; Go, H. Introduction to digital pathology and computer-aided pathology. J. Pathol. Transl. Med. 2020, 54, 125–134. [Google Scholar] [CrossRef] [Green Version]
- De Fauw, J.; Ledsam, J.R.; Romera-Paredes, B.; Nikolov, S.; Tomasev, N.; Blackwell, S.; Askham, H.; Glorot, X.; O’Donoghue, B.; Visentin, D. Clinically applicable deep learning for diagnosis and referral in retinal disease. Nat. Med. 2018, 24, 1342–1350. [Google Scholar] [CrossRef]
- Diao, J.A.; Wang, J.K.; Chui, W.F.; Mountain, V.; Gullapally, S.C.; Srinivasan, R.; Mitchell, R.N.; Glass, B.; Hoffman, S.; Rao, S.K. Human-interpretable image features derived from densely mapped cancer pathology slides predict diverse molecular phenotypes. Nat. Commun. 2021, 12, 1613. [Google Scholar] [CrossRef]
- Sirinukunwattana, K.; Domingo, E.; Richman, S.D.; Redmond, K.L.; Blake, A.; Verrill, C.; Leedham, S.J.; Chatzipli, A.; Hardy, C.; Whalley, C.M. Image-based consensus molecular subtype (imCMS) classification of colorectal cancer using deep learning. Gut 2021, 70, 544–554. [Google Scholar] [CrossRef] [PubMed]
- Skrede, O.-J.; De Raedt, S.; Kleppe, A.; Hveem, T.S.; Liestøl, K.; Maddison, J.; Askautrud, H.A.; Pradhan, M.; Nesheim, J.A.; Albregtsen, F. Deep learning for prediction of colorectal cancer outcome: A discovery and validation study. Lancet 2020, 395, 350–360. [Google Scholar] [CrossRef]
- Chong, Y.; Kim, D.C.; Jung, C.K.; Kim, D.-c.; Song, S.Y.; Joo, H.J.; Yi, S.-Y. Recommendations for pathologic practice using digital pathology: Consensus report of the Korean Society of Pathologists. J. Pathol. Transl. Med. 2020, 54, 437–452. [Google Scholar] [CrossRef] [PubMed]
- Kim, H.; Yoon, H.; Thakur, N.; Hwang, G.; Lee, E.J.; Kim, C.; Chong, Y. Deep learning-based histopathological segmentation for whole slide images of colorectal cancer in a compressed domain. Sci. Rep. 2021, 11, 22520. [Google Scholar] [CrossRef]
- Tizhoosh, H.R.; Pantanowitz, L. Artificial intelligence and digital pathology: Challenges and opportunities. J. Pathol. Inform. 2018, 9, 38. [Google Scholar] [CrossRef]
- Greenson, J.K.; Huang, S.-C.; Herron, C.; Moreno, V.; Bonner, J.D.; Tomsho, L.P.; Ben-Izhak, O.; Cohen, H.I.; Trougouboff, P.; Bejhar, J. Pathologic predictors of microsatellite instability in colorectal cancer. Am. J. Surg. Path. 2009, 33, 126–133. [Google Scholar] [CrossRef] [Green Version]
- Lee, S.H.; Song, I.H.; Jang, H.J. Feasibility of deep learning-based fully automated classification of microsatellite instability in tissue slides of colorectal cancer. Int. J. Cancer 2021, 149, 728–740. [Google Scholar] [CrossRef]
- Yamashita, R.; Long, J.; Longacre, T.; Peng, L.; Berry, G.; Martin, B.; Higgins, J.; Rubin, D.L.; Shen, J. Deep learning model for the prediction of microsatellite instability in colorectal cancer: A diagnostic study. Lancet Oncol. 2021, 22, 132–141. [Google Scholar] [CrossRef]
- Cao, R.; Yang, F.; Ma, S.-C.; Liu, L.; Zhao, Y.; Li, Y.; Wu, D.-H.; Wang, T.; Lu, W.-J.; Cai, W.-J. Development and interpretation of a pathomics-based model for the prediction of microsatellite instability in Colorectal Cancer. Theranostics 2020, 10, 11080. [Google Scholar] [CrossRef]
- Zhang, R.; Osinski, B.L.; Taxter, T.J.; Perera, J.; Lau, D.J.; Khan, A.A. Adversarial deep learning for microsatellite instability prediction from histopathology slides. In Proceedings of the 1st Conference on Medical Imaging with Deep Learning (MIDL 2018), Amsterdam, The Netherlands, 4–6 July 2018; pp. 4–6. [Google Scholar]
- Ke, J.; Shen, Y.; Guo, Y.; Wright, J.D.; Liang, X. A prediction model of microsatellite status from histology images. In Proceedings of the 2020 10th International Conference on Biomedical Engineering and Technology, Tokyo, Japan, 15–18 September 2020; pp. 334–338. [Google Scholar]
- Kather, J.N.; Heij, L.R.; Grabsch, H.I.; Loeffler, C.; Echle, A.; Muti, H.S.; Krause, J.; Niehues, J.M.; Sommer, K.A.; Bankhead, P. Pan-cancer image-based detection of clinically actionable genetic alterations. Nat. Cancer 2020, 1, 789–799. [Google Scholar] [CrossRef]
- Schmauch, B.; Romagnoni, A.; Pronier, E.; Saillard, C.; Maillé, P.; Calderaro, J.; Kamoun, A.; Sefta, M.; Toldo, S.; Zaslavskiy, M. A deep learning model to predict RNA-Seq expression of tumours from whole slide images. Nat. Commun. 2020, 11, 3877. [Google Scholar] [CrossRef] [PubMed]
- Zhu, J.; Wu, W.; Zhang, Y.; Lin, S.; Jiang, Y.; Liu, R.; Wang, X. Computational analysis of pathological image enables interpretable prediction for microsatellite instability. arXiv 2020, arXiv:2010.03130. [Google Scholar]
- Krause, J.; Grabsch, H.I.; Kloor, M.; Jendrusch, M.; Echle, A.; Buelow, R.D.; Boor, P.; Luedde, T.; Brinker, T.J.; Trautwein, C. Deep learning detects genetic alterations in cancer histology generated by adversarial networks. J. Pathol. 2021, 254, 70–79. [Google Scholar] [CrossRef] [PubMed]
- Hong, R.; Liu, W.; DeLair, D.; Razavian, N.; Fenyö, D. Predicting endometrial cancer subtypes and molecular features from histopathology images using multi-resolution deep learning models. Cell Rep. Med. 2021, 2, 100400. [Google Scholar] [CrossRef]
- Zeng, H.; Chen, L.; Zhang, M.; Luo, Y.; Ma, X. Integration of histopathological images and multi-dimensional omics analyses predicts molecular features and prognosis in high-grade serous ovarian cancer. Gynecol. Oncol. 2021, 163, 171–180. [Google Scholar] [CrossRef]
- Wang, T.; Lu, W.; Yang, F.; Liu, L.; Dong, Z.; Tang, W.; Chang, J.; Huan, W.; Huang, K.; Yao, J. Microsatellite instability prediction of uterine corpus endometrial carcinoma based on H&E histology whole-slide imaging. In Proceedings of the 2020 IEEE 17th international symposium on biomedical imaging (ISBI), Iowa City, IA, USA, 3–7 April 2020; pp. 1289–1292. [Google Scholar]
- Musa, I.H.; Zamit, I.; Okeke, M.; Akintunde, T.Y.; Musa, T.H. Artificial Intelligence and Machine Learning in Oncology: Historical Overview of Documents Indexed in the Web of Science Database. EJMO 2021, 5, 239–248. [Google Scholar] [CrossRef]
- Tran, B.X.; Vu, G.T.; Ha, G.H.; Vuong, Q.-H.; Ho, M.-T.; Vuong, T.-T.; La, V.-P.; Ho, M.-T.; Nghiem, K.-C.P.; Nguyen, H.L.T. Global evolution of research in artificial intelligence in health and medicine: A bibliometric study. J. Clin. Med. 2019, 8, 360. [Google Scholar] [CrossRef] [Green Version]
- Yang, G.; Zheng, R.-Y.; Jin, Z.-S. Correlations between microsatellite instability and the biological behaviour of tumours. J. Cancer Res. Clin. Oncol. 2019, 145, 2891–2899. [Google Scholar] [CrossRef] [Green Version]
- Carethers, J.M.; Jung, B.H. Genetics and genetic biomarkers in sporadic colorectal cancer. Gastroenterology 2015, 149, 1177–1190. [Google Scholar] [CrossRef] [Green Version]
- Kloor, M.; Doeberitz, M.V.K. The immune biology of microsatellite-unstable cancer. Trends Cancer 2016, 2, 121–133. [Google Scholar] [CrossRef] [Green Version]
- Chang, L.; Chang, M.; Chang, H.M.; Chang, F. Microsatellite instability: A predictive biomarker for cancer immunotherapy. Appl. Immunohistochem. Mol. Morphol. 2018, 26, e15–e21. [Google Scholar] [CrossRef] [PubMed]
- Le, D.T.; Durham, J.N.; Smith, K.N.; Wang, H.; Bartlett, B.R.; Aulakh, L.K.; Lu, S.; Kemberling, H.; Wilt, C.; Luber, B.S. Mismatch repair deficiency predicts response of solid tumors to PD-1 blockade. Science 2017, 357, 409–413. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kacew, A.J.; Strohbehn, G.W.; Saulsberry, L.; Laiteerapong, N.; Cipriani, N.A.; Kather, J.N.; Pearson, A.T. Artificial intelligence can cut costs while maintaining accuracy in colorectal cancer genotyping. Front. Oncol. 2021, 11, 630953. [Google Scholar] [CrossRef]
- Boussios, S.; Mikropoulos, C.; Samartzis, E.; Karihtala, P.; Moschetta, M.; Sheriff, M.; Karathanasi, A.; Sadauskaite, A.; Rassy, E.; Pavlidis, N. Wise management of ovarian cancer: On the cutting edge. J. Pers. Med. 2020, 10, 41. [Google Scholar] [CrossRef] [PubMed]
- He, K.; Zhang, X.; Ren, S.; Sun, J. Deep residual learning for image recognition. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, Las Vegas, NV, USA, 27–30 June 2016; pp. 770–778. [Google Scholar]
- Djuric, U.; Zadeh, G.; Aldape, K.; Diamandis, P. Precision histology: How deep learning is poised to revitalize histomorphology for personalized cancer care. NPJ Precis. Oncol. 2017, 1, 22. [Google Scholar] [CrossRef]
- Serag, A.; Ion-Margineanu, A.; Qureshi, H.; McMillan, R.; Saint Martin, M.-J.; Diamond, J.; O’Reilly, P.; Hamilton, P. Translational AI and deep learning in diagnostic pathology. Front. Med. 2019, 6, 185. [Google Scholar] [CrossRef] [Green Version]
- Ailia, M.J.; Thakur, N.; Abdul-Ghafar, J.; Jung, C.K.; Yim, K.; Chong, Y. Current Trend of Artificial Intelligence Patents in Digital Pathology: A Systematic Evaluation of the Patent Landscape. Cancers 2022, 14, 2400. [Google Scholar] [CrossRef]
- Chen, J.; Bai, G.; Liang, S.; Li, Z. Automatic image cropping: A computational complexity study. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, Las Vegas, NV, USA, 27–30 June 2016; pp. 507–515. [Google Scholar]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Alam, M.R.; Abdul-Ghafar, J.; Yim, K.; Thakur, N.; Lee, S.H.; Jang, H.-J.; Jung, C.K.; Chong, Y. Recent Applications of Artificial Intelligence from Histopathologic Image-Based Prediction of Microsatellite Instability in Solid Cancers: A Systematic Review. Cancers 2022, 14, 2590. https://doi.org/10.3390/cancers14112590
Alam MR, Abdul-Ghafar J, Yim K, Thakur N, Lee SH, Jang H-J, Jung CK, Chong Y. Recent Applications of Artificial Intelligence from Histopathologic Image-Based Prediction of Microsatellite Instability in Solid Cancers: A Systematic Review. Cancers. 2022; 14(11):2590. https://doi.org/10.3390/cancers14112590
Chicago/Turabian StyleAlam, Mohammad Rizwan, Jamshid Abdul-Ghafar, Kwangil Yim, Nishant Thakur, Sung Hak Lee, Hyun-Jong Jang, Chan Kwon Jung, and Yosep Chong. 2022. "Recent Applications of Artificial Intelligence from Histopathologic Image-Based Prediction of Microsatellite Instability in Solid Cancers: A Systematic Review" Cancers 14, no. 11: 2590. https://doi.org/10.3390/cancers14112590
APA StyleAlam, M. R., Abdul-Ghafar, J., Yim, K., Thakur, N., Lee, S. H., Jang, H.-J., Jung, C. K., & Chong, Y. (2022). Recent Applications of Artificial Intelligence from Histopathologic Image-Based Prediction of Microsatellite Instability in Solid Cancers: A Systematic Review. Cancers, 14(11), 2590. https://doi.org/10.3390/cancers14112590









