SAK-HV Decreases the Self-Ubiquitination of MEKK1 to Promote Macrophage Proliferation via MAPK/ERK and JNK Pathways
Abstract
:1. Introduction
2. Results
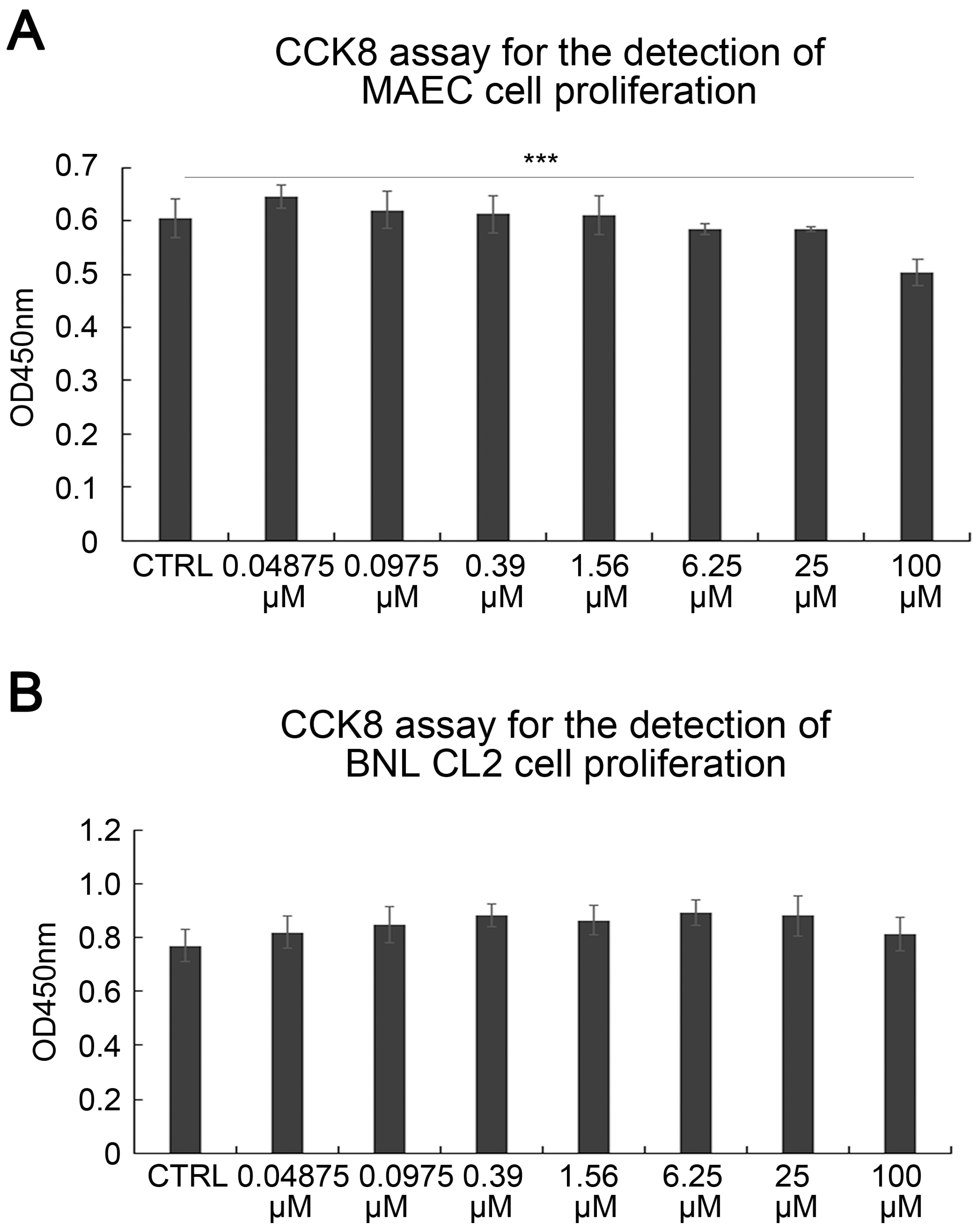
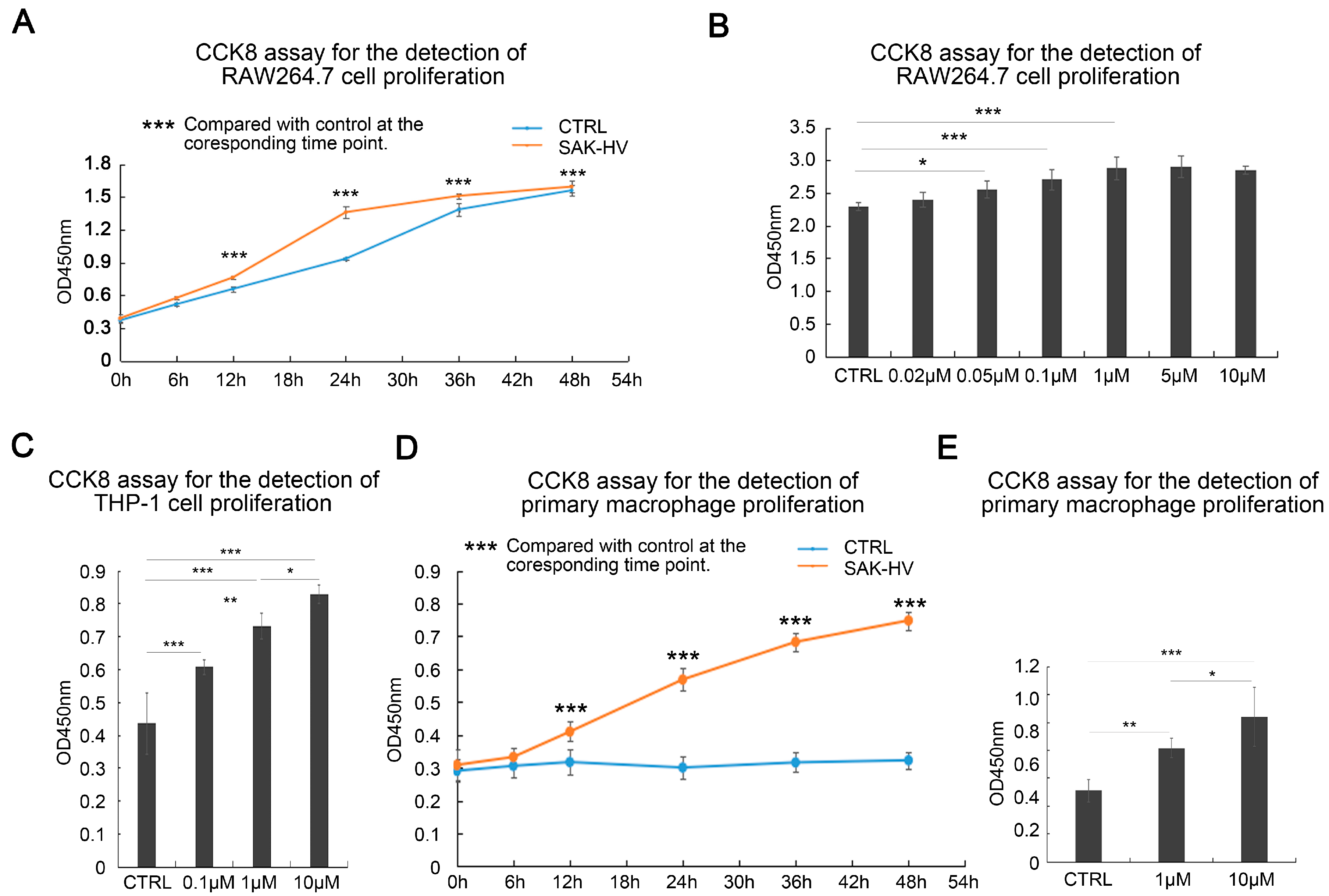
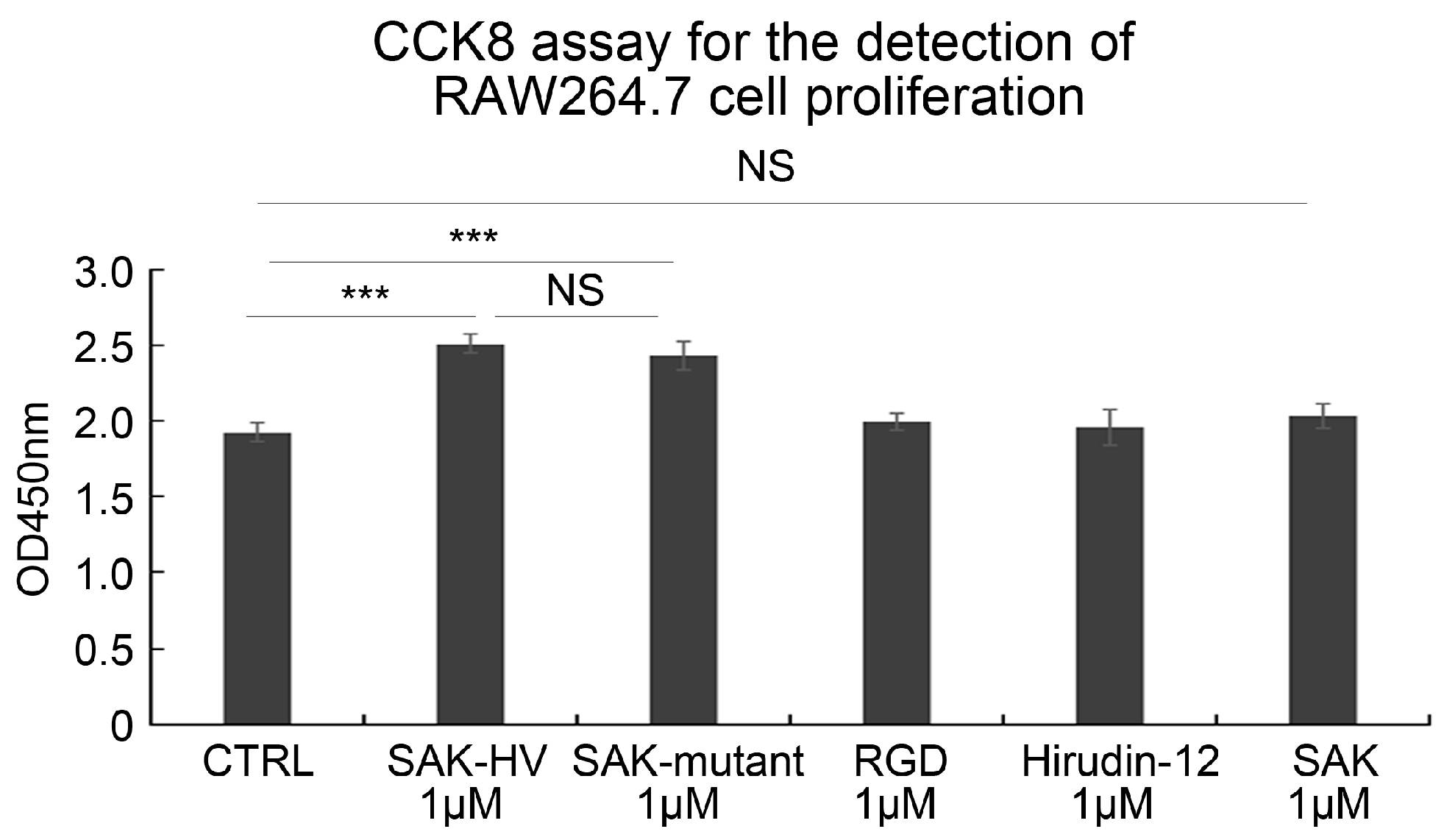
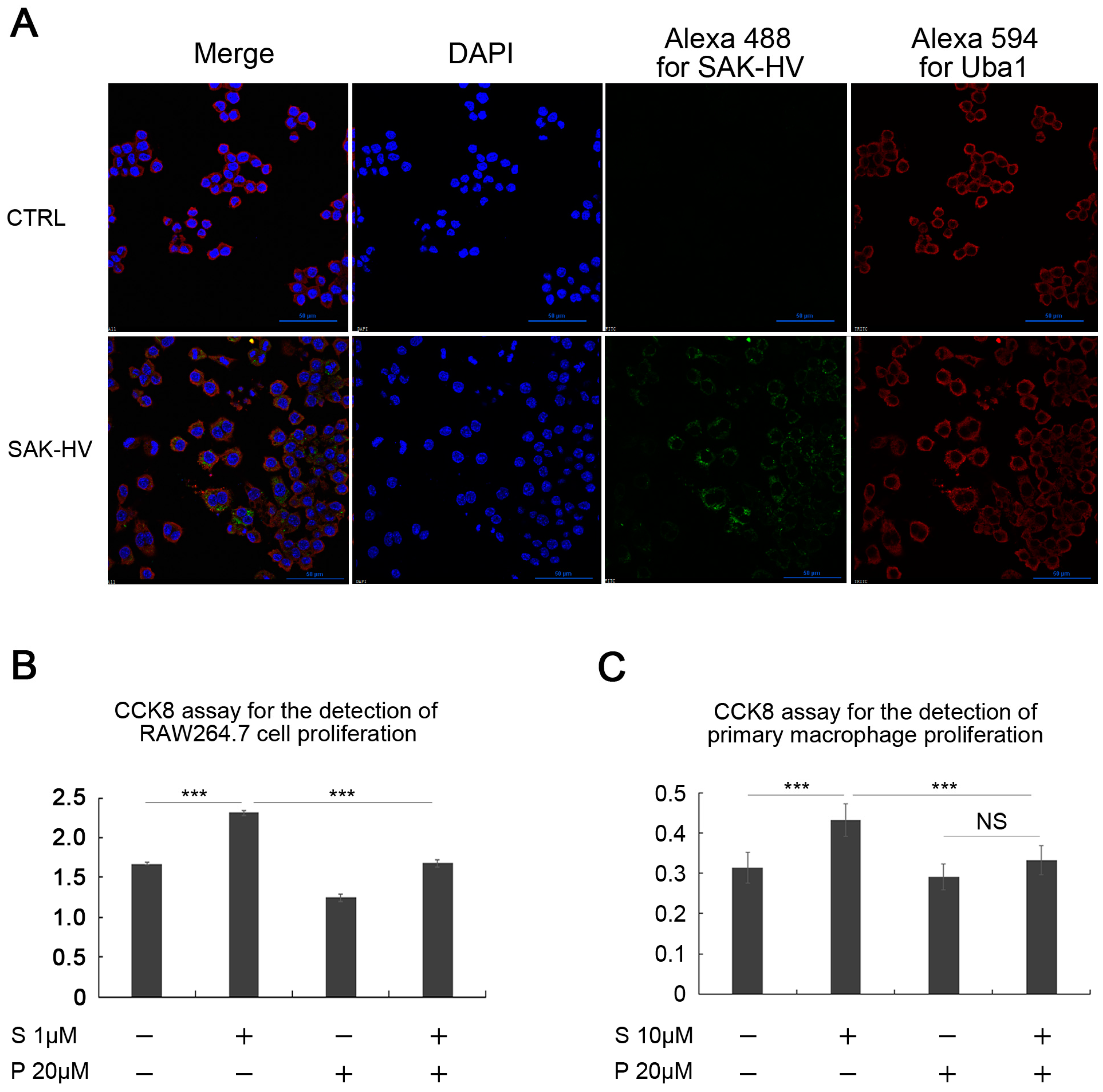
2.1. SAK-HV Promotes the Proliferation of RAW264.7 Murine Macrophages
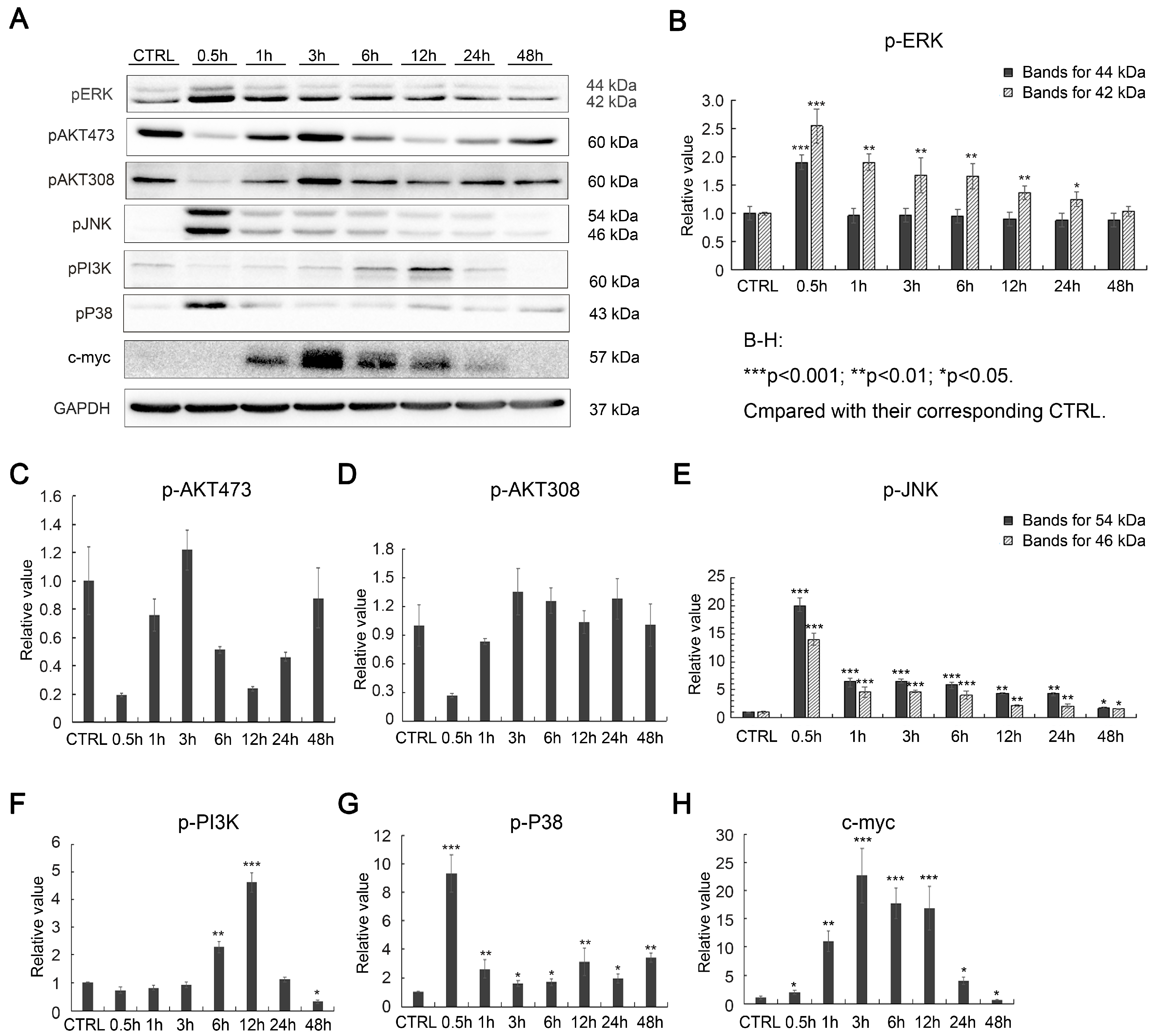
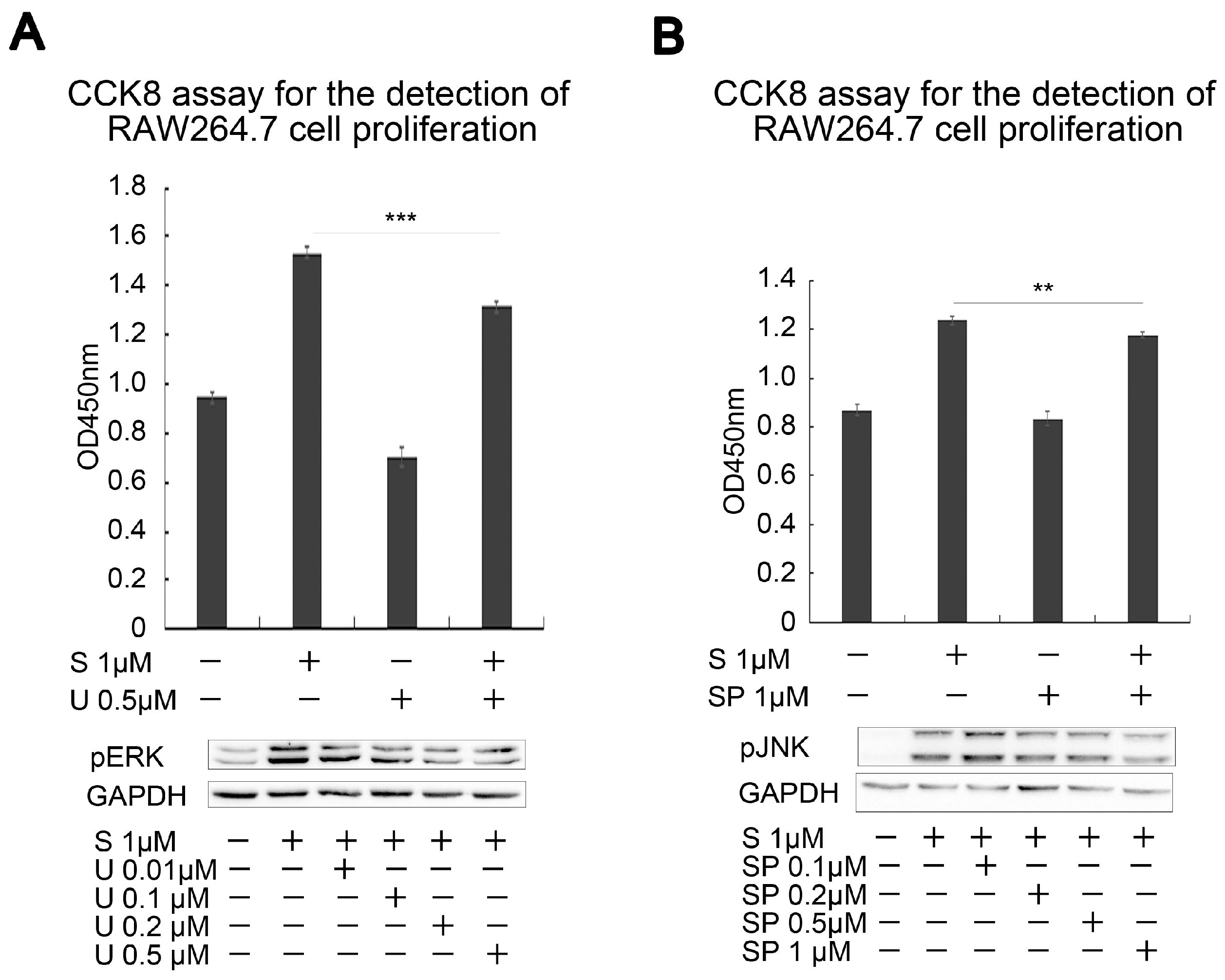
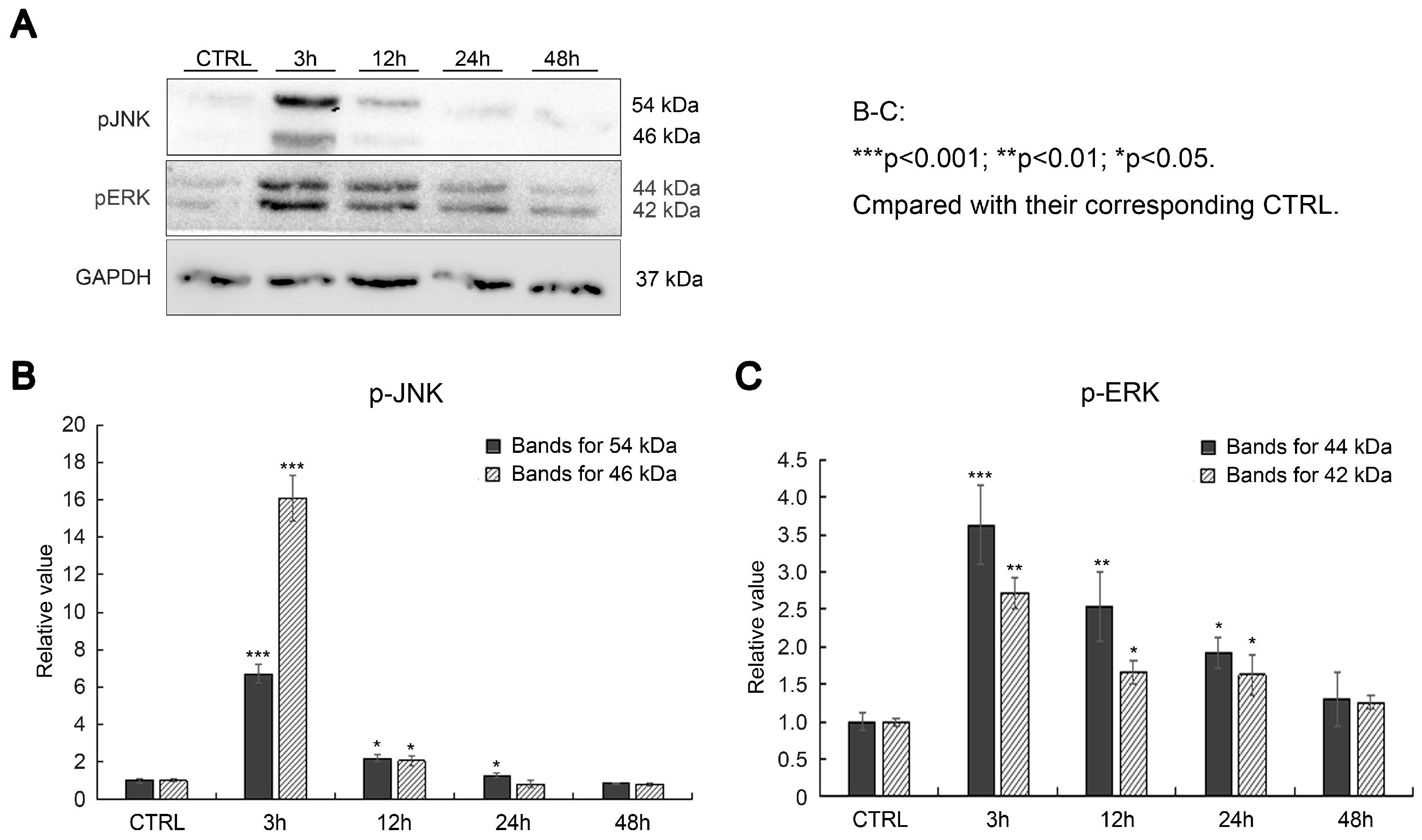
2.2. Effects of SAK-HV on Proliferation-Related Pathways in RAW264.7 Cells
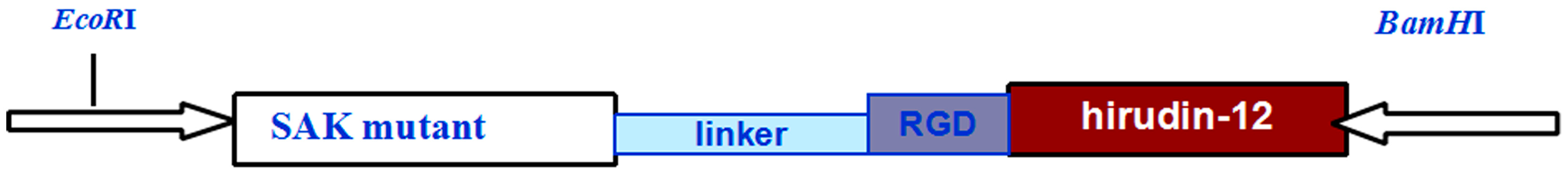
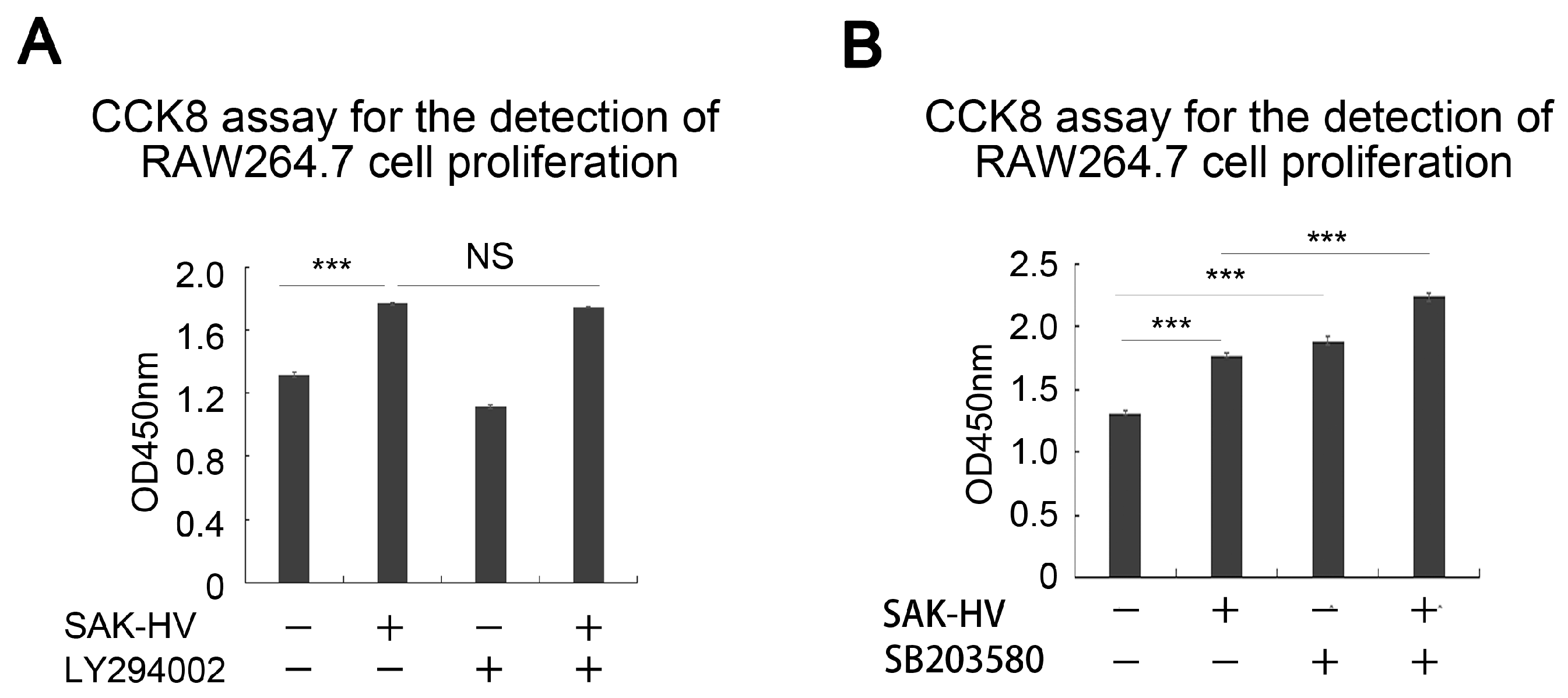
2.3. SAK-HV Promotes Proliferation through Its SAK-Mutant Functional Domain in RAW264.7 Cells
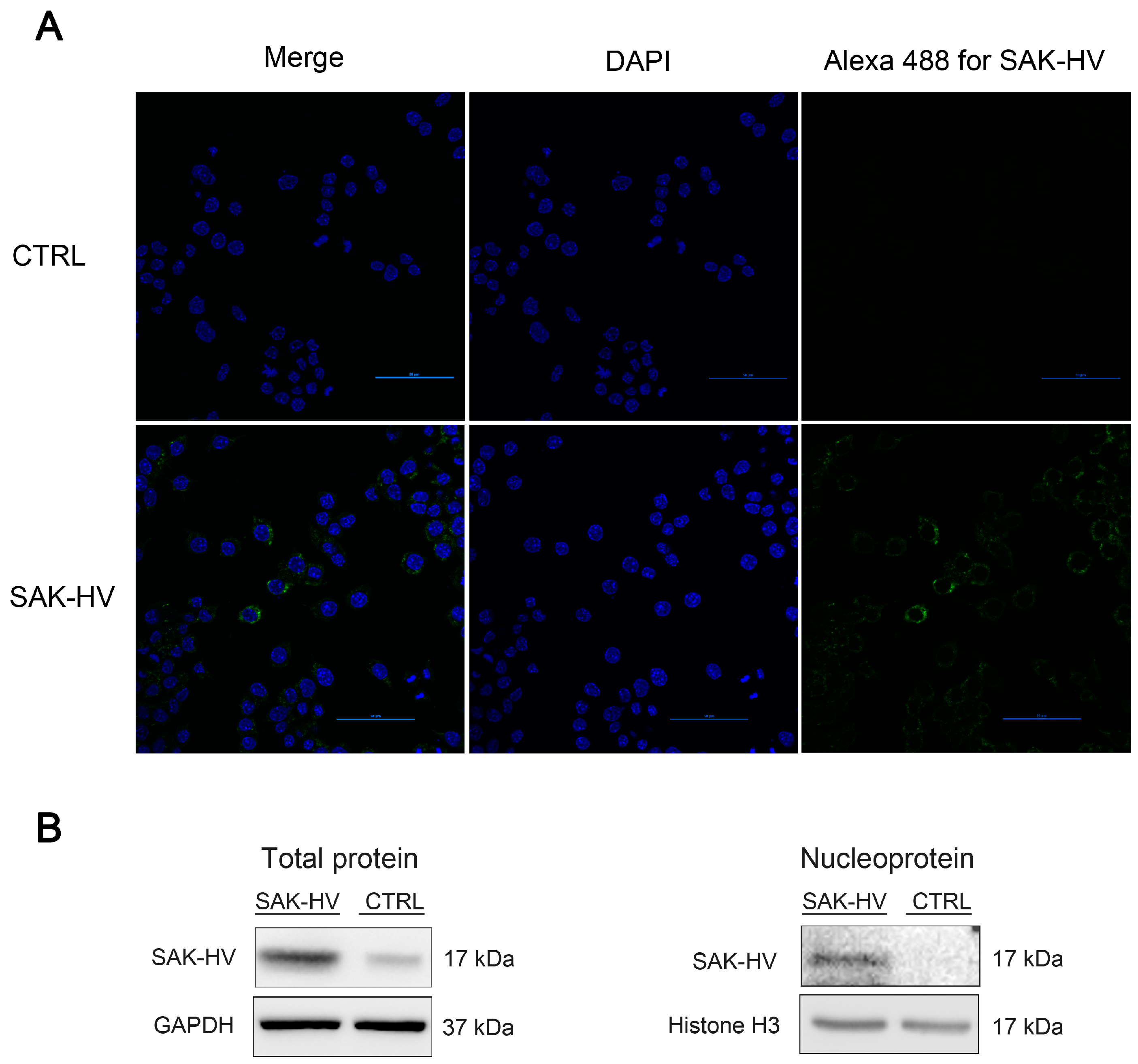
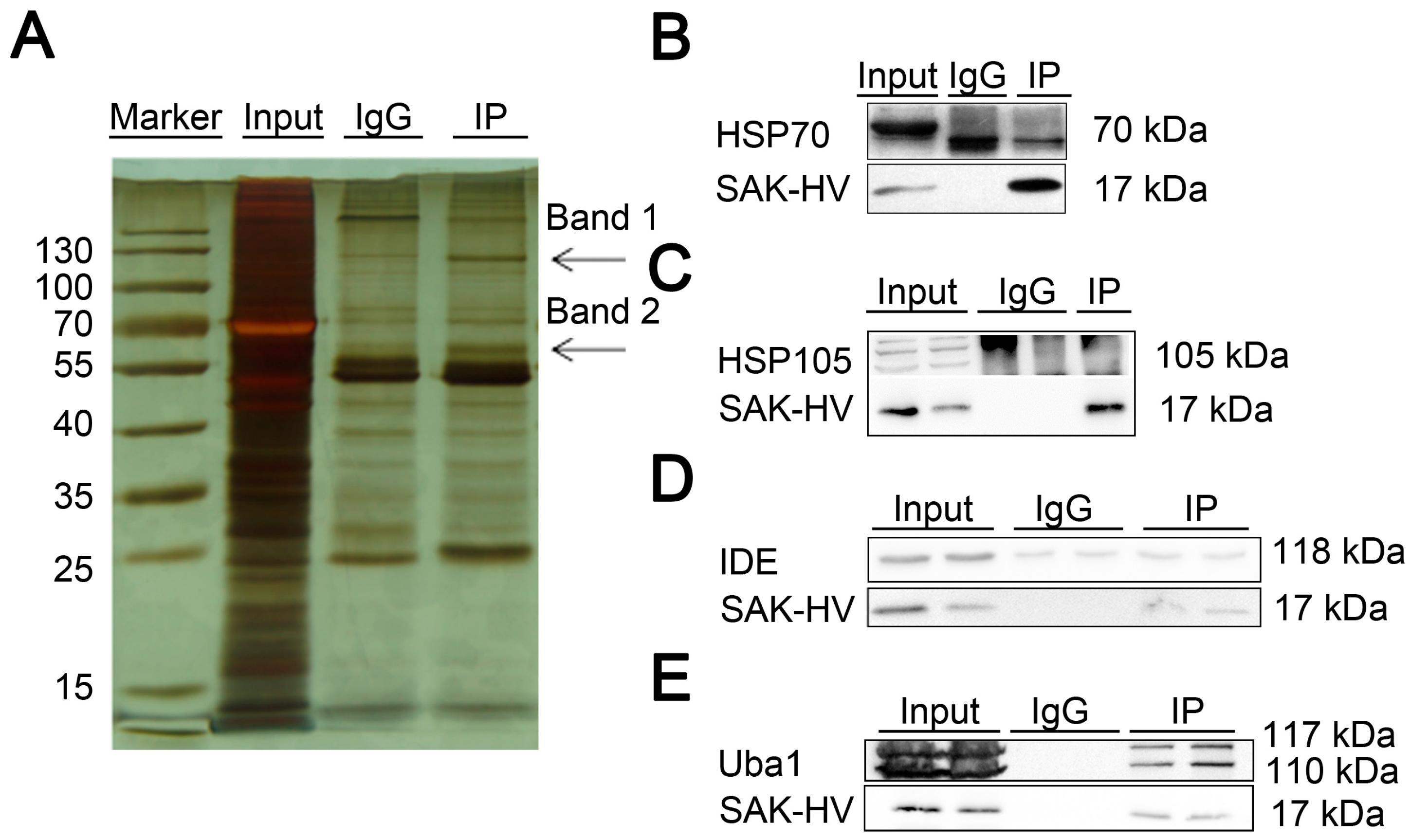
2.4. Screening and Verification of SAK-HV-Interacting Proteins in RAW264.7 Cells
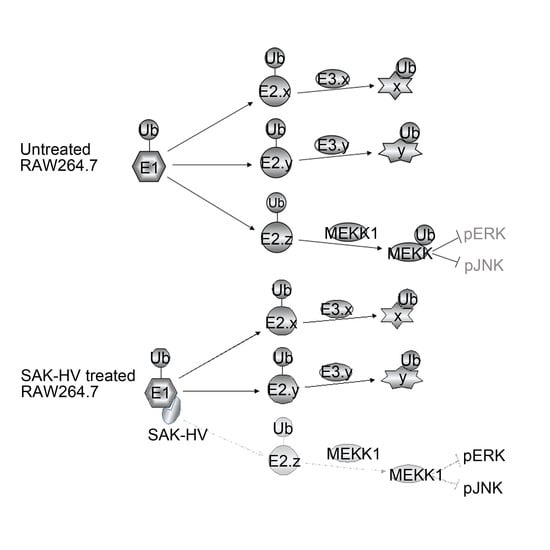
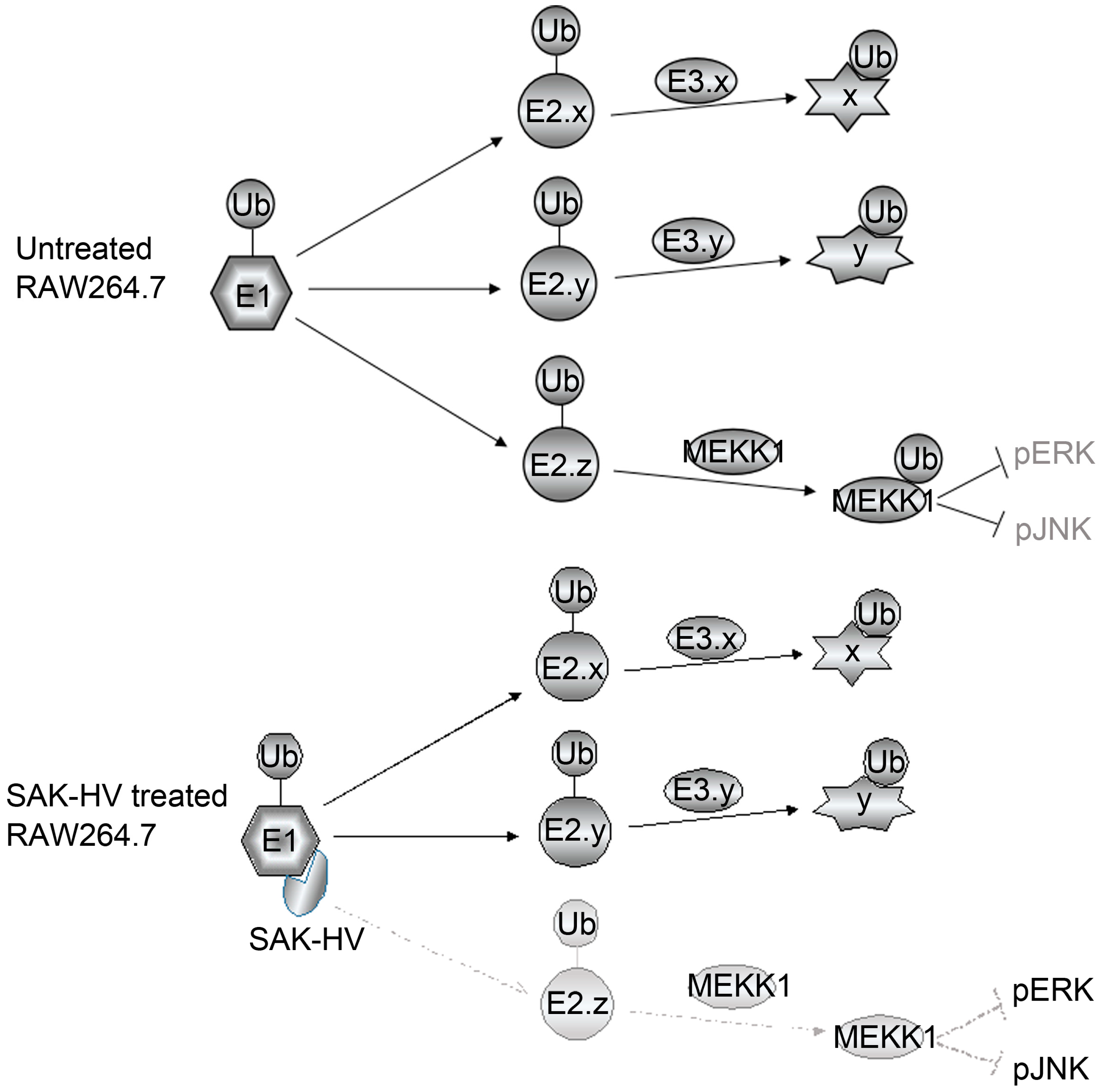
2.5. SAK-HV Inhibited the Self-Ubiquitination of the Ubiquitin Ligase MAPK/ERK Kinase Kinase 1 (MEKK1) to Activate both ERK and JNK Pathways
3. Discussion
4. Materials and Methods
4.1. Cell Culture
4.2. CCK8 Assay
4.3. Western Blot
4.4. Mass Spectrometric Analysis
4.5. Confocal Microscopy
4.6. Co-Immunoprecipitation Assay
4.7. Ubiquitination Assay
4.8. The Extraction of Primary Peritoneal Macrophage Cells from C57BL/6J Mice
4.9. Statistics
5. Conclusions
Acknowledgments
Author Contributions
Conflicts of Interest
Abbreviations
| MAPK | mitogen-activated protein kinase |
| ERKs | extracellular signal-regulated kinases |
| JNKs | c-Jun N-terminal kinases |
| LD | ubiquitin-like modifier-activating enzyme 1 |
| RGD | Arg-Gly-Asp |
| PI3K | Phosphoinositide 3-kinase |
| MAEC | mouse aortic endothelial cells |
| hirudin-12 | 12-amino acid peptide from the C-terminus of hirudin |
| MEKK1 | MAPK/ERK kinase kinase 1 |
| NLS | nuclear localization signal |
References
- Wynn, T.A.; Chawla, A.; Pollard, J.W. Macrophage biology in development, homeostasis and disease. Nature 2013, 496, 445–455. [Google Scholar] [CrossRef] [PubMed]
- Van Furth, R.; Cohn, Z.A. The origin and kinetics of mononuclear phagocytes. J. Exp. Med. 1968, 128, 415–435. [Google Scholar] [CrossRef] [PubMed]
- Ginhoux, F.; Greter, M.; Leboeuf, M.; Nandi, S.; See, P.; Gokhan, S.; Mehler, M.F.; Conway, S.J.; Ng, L.G.; Stanley, E.R.; et al. Fate mapping analysis reveals that adult microglia derive from primitive macrophages. Science 2010, 330, 841–845. [Google Scholar] [CrossRef] [PubMed]
- Schulz, C.; Gomez, P.E.; Chorro, L.; Szabo-Rogers, H.; Cagnard, N.; Kierdorf, K.; Prinz, M.; Wu, B.; Jacobsen, S.E.; Pollard, J.W.; et al. A lineage of myeloid cells independent of Myb and hematopoietic stem cells. Science 2012, 336, 86–90. [Google Scholar] [CrossRef] [PubMed]
- Hashimoto, D.; Chow, A.; Noizat, C.; Teo, P.; Beasley, M.B.; Leboeuf, M.; Becker, C.D.; See, P.; Price, J.; Lucas, D.; et al. Tissue-resident macrophages self-maintain locally throughout adult life with minimal contribution from circulating monocytes. Immunity 2013, 38, 792–804. [Google Scholar] [CrossRef] [PubMed]
- Amano, S.U.; Cohen, J.L.; Vangala, P.; Tencerova, M.; Nicoloro, S.M.; Yawe, J.C.; Shen, Y.; Czech, M.P.; Aouadi, M. Local proliferation of macrophages contributes to obesity-associated adipose tissue inflammation. Cell Metab. 2014, 19, 162–171. [Google Scholar] [CrossRef] [PubMed]
- Murano, I.; Barbatelli, G.; Parisani, V.; Latini, C.; Muzzonigro, G.; Castellucci, M.; Cinti, S. Dead adipocytes, detected as crown-like structures, are prevalent in visceral fat depots of genetically obese mice. J. Lipid Res. 2008, 49, 1562–1568. [Google Scholar] [CrossRef] [PubMed]
- Robbins, C.S.; Hilgendorf, I.; Weber, G.F.; Theurl, I.; Iwamoto, Y.; Figueiredo, J.L.; Gorbatov, R.; Sukhova, G.K.; Gerhardt, L.M.; Smyth, D.; et al. Local proliferation dominates lesional macrophage accumulation in atherosclerosis. Nat. Med. 2013, 19, 1166–1172. [Google Scholar] [CrossRef] [PubMed]
- Randolph, G.J. Proliferating macrophages prevail in atherosclerosis. Nat. Med. 2013, 19, 1094–1095. [Google Scholar] [CrossRef] [PubMed]
- Saba-El-Leil, M.K.; Frémin, C.; Meloche, S. Redundancy in the world of MAP kinases: All for one. Front. Cell Dev. Biol. 2016, 4, 67. [Google Scholar] [CrossRef] [PubMed]
- Sun, Y.; Liu, W.Z.; Liu, T.; Feng, X.; Yang, N.; Zhou, H.F. Signaling pathway of MAPK/ERK in cell proliferation, differentiation, migration, senescence and apoptosis. J. Recept. Signal Transduct. Res. 2015, 35, 600–604. [Google Scholar] [CrossRef] [PubMed]
- Torres, M.; Forman, H.J. Redox signaling and the MAP kinase pathways. Biofactors 2003, 17, 287–296. [Google Scholar] [CrossRef] [PubMed]
- Zhang, L.; Wang, Y.; Xiao, F.; Wang, S.; Xing, G.; Li, Y.; Yin, X.; Lu, K.; Wei, R.; Fan, J.; et al. CKIP-1 regulates macrophage proliferation by inhibiting TRAF6-mediated Akt activation. Cell Res. 2014, 24, 742–761. [Google Scholar] [CrossRef] [PubMed]
- Pavlidou, A.; Vlahos, N.F. Molecular alterations of PI3K/Akt/mTOR pathway: A therapeutic target in endometrial cancer. Molecular alterations of PI3K/Akt/mTOR pathway: A therapeutic target in endometrial cancer. Sci. World J. 2014, 2014, 709736. [Google Scholar] [CrossRef] [PubMed]
- Yao, Y.; Yang, D.; Han, Y.; Wang, W.; Wang, N.; Yang, J.; Zeng, C. Dopamine D1-like receptors suppress the proliferation of macrophages induced by Ox-LDL. Cell Physiol. Biochem. 2016, 38, 415–426. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Zheng, Y.; Li, T.; Wang, Q.; Qian, J.; Lu, Y.; Zhang, M.; Bi, E.; Yang, M.; Reu, F.; et al. Chemokines CCL2, 3, 14 stimulate macrophage bone marrow homing, proliferation, and polarization in multiple myeloma. Oncotarget 2015, 6, 24218–24229. [Google Scholar] [CrossRef] [PubMed]
- Nagai, Y.; Kaneda, S.; Nomura, K.; Yasuda, H.; Seno, T.; Yamao, F. Ubiquitin-activating enzyme, E1, is phosphorylated in mammalian cells by the protein kinase Cdc2. J. Cell Sci. 1995, 108, 2145–2152. [Google Scholar] [PubMed]
- Nguyen, L.K.; Kolch, W.; Kholodenko, B.N. When ubiquitination meets phosphorylation a systems biology perspective of EGFR-MAPK signalling. Cell Commun. Signal. 2013, 11, 52. [Google Scholar] [CrossRef] [PubMed]
- Groen, E.J.; Gillingwater, T.H. UBA1: At the crossroads of ubiquitin homeostasis and neurodegeneration. Trends Mol. Med. 2015, 21, 622–632. [Google Scholar] [CrossRef] [PubMed]
- Lee, I.; Schindelin, H. Structural insights into E1-catalyzed ubiquitin activation and transfer to conjugating enzymes. Cell 2008, 134, 268–278. [Google Scholar] [CrossRef] [PubMed]
- Schulman, B.A.; Harper, J.W. Ubiquitin-like protein activation by E1 enzymes: The apex for downstream signalling pathways. Nat. Rev. Mol. Cell Biol. 2009, 10, 319–331. [Google Scholar] [CrossRef] [PubMed]
- Sugaya, K.; Ishihara, Y.; Inoue, S. Analysis of a temperature-sensitive mutation in Uba1: Effects of the click reaction on subsequent immunolabeling of proteins involved in DNA replication. FEBS Open Bio 2015, 5, 167–174. [Google Scholar] [CrossRef] [PubMed]
- Sugaya, K.; Ishihara, Y.; Inoue, S. Nuclear localization of ubiquitin-activating enzyme Uba1 is characterized in its mammalian temperature-sensitive mutant. Genes Cells 2015, 20, 659–666. [Google Scholar] [CrossRef] [PubMed]
- Wang, M.; Wang, Y.; Wang, J.; Zou, M.; Liu, S.; Xu, T.; Cai, X.; Wu, C.; Wang, J.; Xu, D. Construction and characterization of a novel staphylokinase variant with thrombin-inhibitory activity. Biotechnol. Lett. 2009, 31, 1923–1927. [Google Scholar] [CrossRef] [PubMed]
- Zhang, C.; Huang, Z.; Jing, H.; Fu, W.; Yuan, M.; Xia, W.; Cai, L.; Gan, X.; Chen, Y.; Zou, M.; et al. SAK-HV triggered a short-period lipid-lowering biotherapy based on the energy model of liver proliferation via a novel pathway. Theranostics 2017. accepted. [Google Scholar]
- Wang, M.; Fu, W.; Cai, X.; Xing, W.; Xu, T.; Wang, Y.; Li, R.; Jing, H.; Yuan, M.; Zhang, C.; et al. The fusion protein (SAK-HV) attenuates atherosclerosis in Apoe−/− mouse via an anti-inflammation mechanism mediated by PAR1 and and GPIIb/IIIa. Blood 2017. under review. [Google Scholar]
- Henklova, P.; Vrzal, R.; Papouskova, B.; Bednar, P.; Jancova, P.; Anzenbacherova, E.; Ulrichova, J.; Maurel, P.; Pavek, P.; Dvorak, Z. SB203580, a pharmacological inhibitor of p38 MAP kinase transduction pathway activates ERK and JNK MAP kinases in primary cultures of human hepatocytes. Eur. J. Pharmacol. 2008, 593, 16–23. [Google Scholar] [CrossRef] [PubMed]
- Zacksenhaus, E.; Sheinin, R. Molecular cloning, primary structure and expression of the human X linked AlS9 gene cDNA which complements the ts A1S9 mouse L cell defect in DNA replication. EMBO J. 1990, 9, 2923–2929. [Google Scholar] [PubMed]
- Witowsky, J.A.; Johnson, G.L. Ubiquitylation of MEKK1 inhibits its phosphorylation of MKK1 and MKK4 and activation of the ERK1/2 and JNK pathways. J. Biol. Chem. 2003, 278, 1403–1406. [Google Scholar] [CrossRef] [PubMed]
- Van Gassen, N.; van Overmeire, E.; Leuckx, G.; Heremans, Y.; de Groef, S.; Cai, Y.; Elkrim, Y.; Gysemans, C.; Stijlemans, B.; van de Casteele, M.; et al. Macrophage dynamics are regulated by local macrophage proliferation and monocyte recruitment in injured pancreas. Eur. J. Immunol. 2015, 45, 1482–1493. [Google Scholar] [CrossRef] [PubMed]
- Côté, C.H.; Bouchard, P.; van Rooijen, N.; Marsolais, D.; Duchesne, E. Monocyte depletion increases local proliferation of macrophage subsets after skeletal muscle injury. BMC Musculoskelet. Disord. 2013, 14, 359. [Google Scholar] [CrossRef] [PubMed]
- Jenkins, S.J. Local macrophage proliferation, rather than recruitment from the blood, is a signature of Th2 inflammation. Science 2011, 332, 1284–1288. [Google Scholar] [CrossRef] [PubMed]
- Tsuji, H.; Matsudo, Y.; Tsuji, S.; Hanaoka, F.; Hyodo, M.; Hori, T. Isolation of temperature-sensitive CHO-K1 cell mutants exhibiting chromosomal instability and reduced DNA synthesis at nonpermissive temperature. Somat. Cell Mol. Genet. 1990, 16, 461–476. [Google Scholar] [CrossRef] [PubMed]
- Sugaya, K.; Ishihara, Y.; Inoue, S.; Tsuji, H. Characterization of ubiquitin-activating enzyme Uba1 in the nucleus by its mammalian temperature-sensitive mutant. PLoS ONE 2014, 9, e96666. [Google Scholar] [CrossRef] [PubMed]
- Thompson, L.H.; Mankovitz, R.; Baker, R.M.; Till, J.E.; Siminovitch, L.; Whitmore, G.F. Isolation of temperature-sensitive mutants of L-cells. Proc. Natl. Acad. Sci. USA 1970, 66, 377–384. [Google Scholar] [CrossRef] [PubMed]
- Finley, D.; Ciechanover, A.; Varshavsky, A. Thermolability of ubiquitin-activating enzyme from the mammalian cell cycle mutant ts 85. Cell 1984, 37, 43–55. [Google Scholar] [CrossRef]
- Kulka, R.G.; Raboy, B.; Schuster, R.; Parag, H.A.; Diamond, G.; Ciechanover, A.; Marcus, M. A Chinese hamster cell cycle mutant arrested at G2 phase has a temperature-sensitive ubiquitin-activating enzyme, E1. J. Biol. Chem. 1988, 263, 15726–15731. [Google Scholar] [PubMed]
- Stephen, A.G.; Trausch-Azar, J.S.; Ciechanover, A.; Schwartz, A.L. The ubiquitin-activating enzyme E1 is phosphorylated and localized to the nucleus in a cell cycle-dependent Manner. J. Biol. Chem. 1996, 271, 15608–15614. [Google Scholar] [PubMed]
- Stephen, A.G.; Trausch-Azar, J.S.; Handley-Gearhart, P.M.; Ciechanover, A.; Schwartz, A.L. Identification of a region within the ubiquitin-activating enzyme required for nuclear targeting and phosphorylation. J. Biol. Chem. 1997, 272, 10895–10903. [Google Scholar] [PubMed]
- Qin, Z.; Cui, B.; Jin, J.; Song, M.; Zhou, B.; Guo, H.; Qian, D.; He, Y.; Huang, L. The ubiquitin-activating enzyme E1 as a novel therapeutic target for the treatment of restenosis. Atherosclerosis 2016, 247, 142–153. [Google Scholar] [CrossRef] [PubMed]
- Xu, G.W.; Ali, M.; Wood, T.E.; Wong, D.; Maclean, N.; Wang, X.; Gronda, M.; Skrtic, M.; Li, X.; Hurren, R.; et al. The ubiquitin-activating enzyme E1 as a therapeutic target for the treatment of leukemia and multiple myeloma. Blood 2010, 115, 2251–2259. [Google Scholar] [CrossRef] [PubMed]
- Schlabach, M.R.; Luo, J.; Solimini, N.L.; Hu, G.; Xu, Q.; Li, M.Z.; Zhao, Z.; Smogorzewska, A.; Sowa, M.E.; Ang, X.L.; et al. Cancer proliferation gene discovery through functional genomics. Science 2008, 319, 620–624. [Google Scholar] [CrossRef] [PubMed]
- Nouspikel, T.; Hanawalt, P.C. Impaired nucleotide excision repair upon macrophage differentiation is corrected by E1 ubiquitin-activating enzyme. Proc. Natl. Acad. Sci. USA 2006, 103, 16188–16193. [Google Scholar] [CrossRef] [PubMed]
- Shevchenko, A.; Wilm, M.; Vorm, O.; Mann, M. Mass spectrometric sequencing of proteins from silver-stained polyacrylamide gels. Anal. Chem. 1996, 68, 850–858. [Google Scholar] [CrossRef] [PubMed]


| No. | Name | Band | Score |
|---|---|---|---|
| gi|6678483 | ubiquitin-like modifier-activating enzyme 1 isoform 1 (Uba1) | Band 1 1 | 341 |
| gi|114145505 | heat shock protein 105 kDa (HSP105) | Band 1 1 | 167 |
| gi|14548072 | nsulin-degrading enzyme (IDE) | Band 1 1 | 148 |
| gi|435839 | heat shock protein 70 kDa (HSP70) | Band 2 1 | 51 |
© 2017 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhang, C.; Chen, Y.; Gan, X.; Huang, Z.; Zou, M.; Fu, W.; Xing, W.; Xu, D. SAK-HV Decreases the Self-Ubiquitination of MEKK1 to Promote Macrophage Proliferation via MAPK/ERK and JNK Pathways. Int. J. Mol. Sci. 2017, 18, 835. https://doi.org/10.3390/ijms18040835
Zhang C, Chen Y, Gan X, Huang Z, Zou M, Fu W, Xing W, Xu D. SAK-HV Decreases the Self-Ubiquitination of MEKK1 to Promote Macrophage Proliferation via MAPK/ERK and JNK Pathways. International Journal of Molecular Sciences. 2017; 18(4):835. https://doi.org/10.3390/ijms18040835
Chicago/Turabian StyleZhang, Chao, Yao Chen, Xiangdong Gan, Zhiguang Huang, Minji Zou, Wenliang Fu, Weiwei Xing, and Donggang Xu. 2017. "SAK-HV Decreases the Self-Ubiquitination of MEKK1 to Promote Macrophage Proliferation via MAPK/ERK and JNK Pathways" International Journal of Molecular Sciences 18, no. 4: 835. https://doi.org/10.3390/ijms18040835