ρ0 Cells Feature De-Ubiquitination of SLC Transporters and Increased Levels and Fluxes of Amino Acids
Abstract
:1. Introduction
2. Results and Discussion
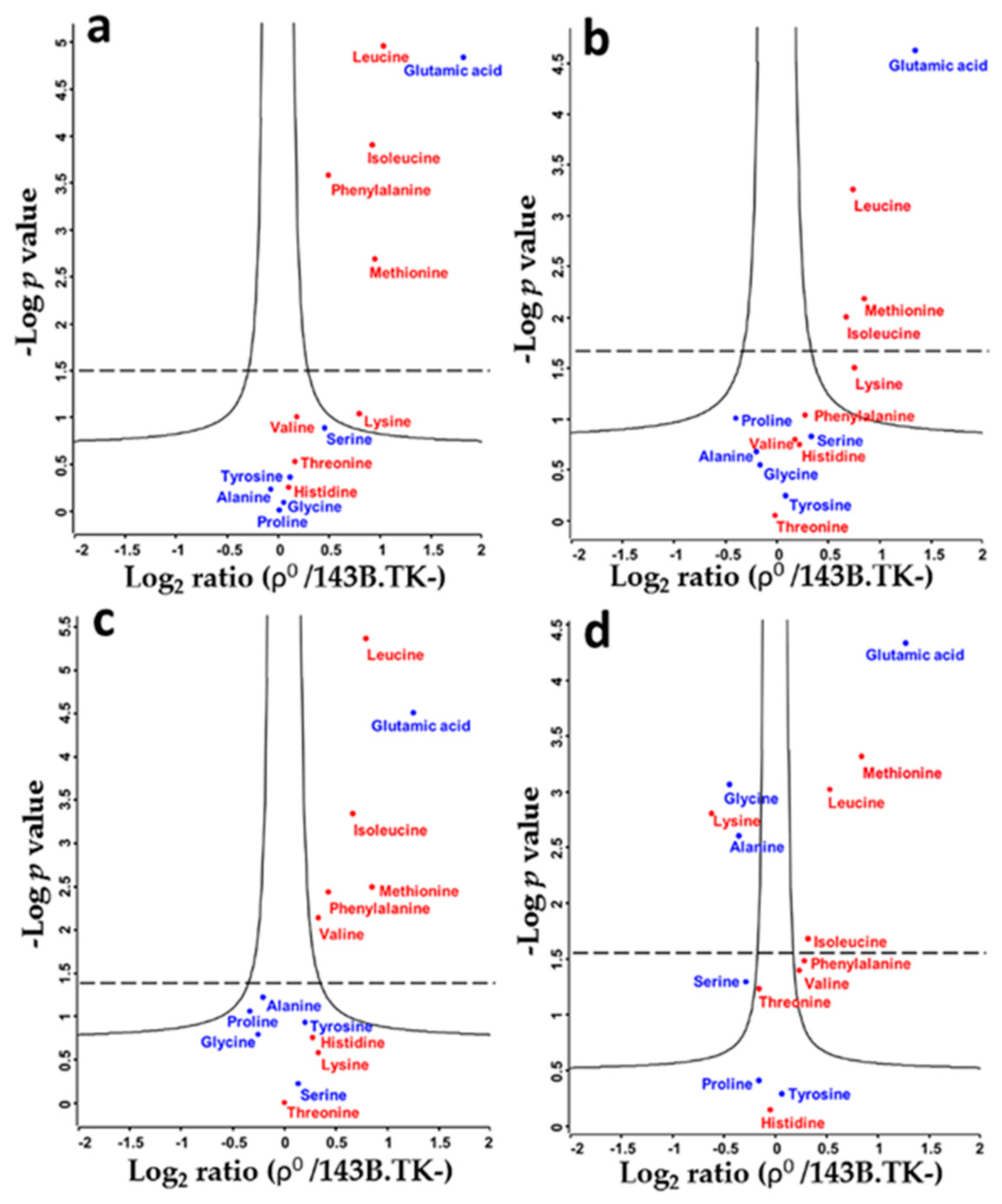
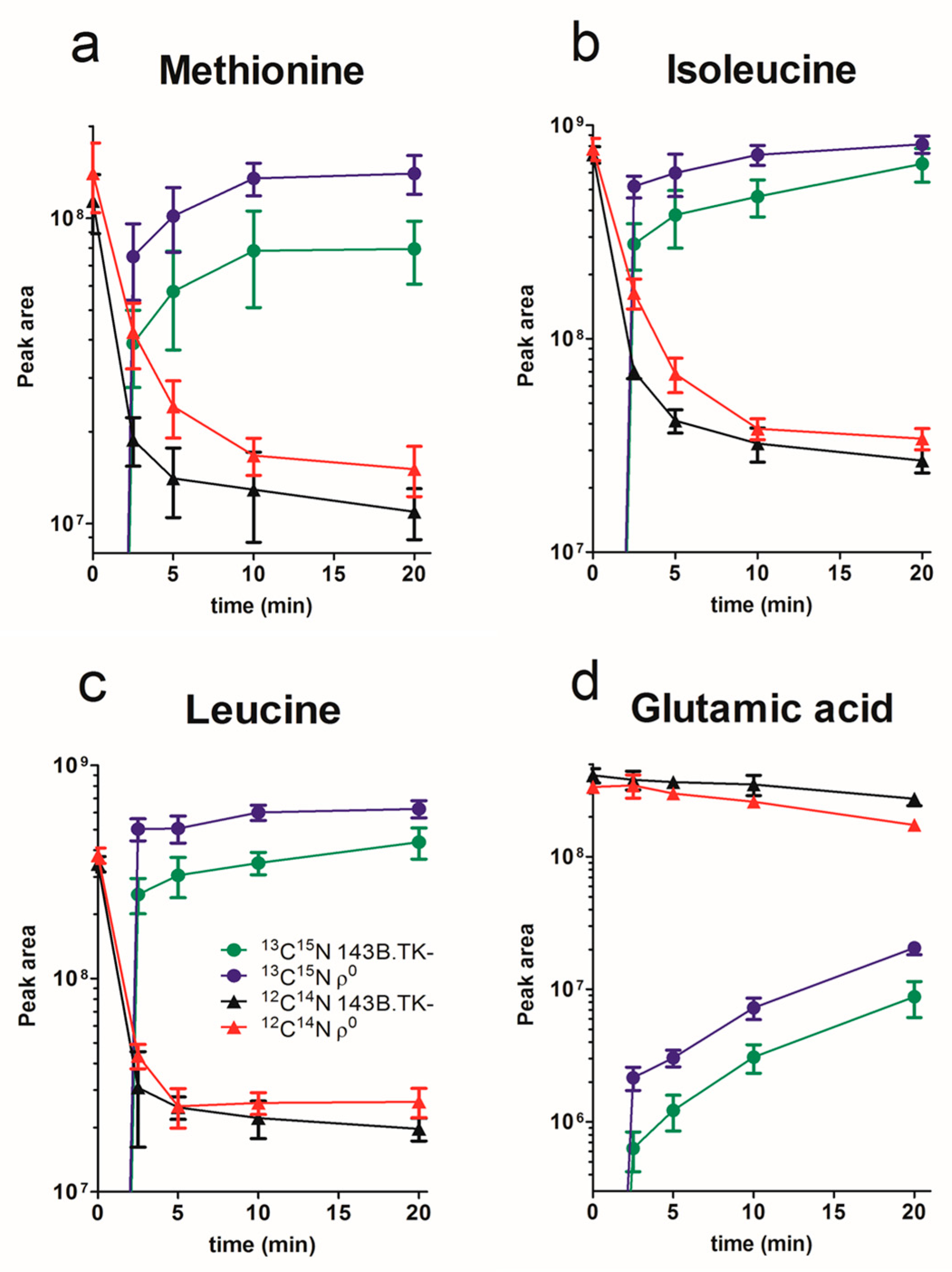
2.1. Amino Acid Flux of ρ0 and 143B.TK- Cells
2.2. Determination of Absolute Amino Acid Concentrations
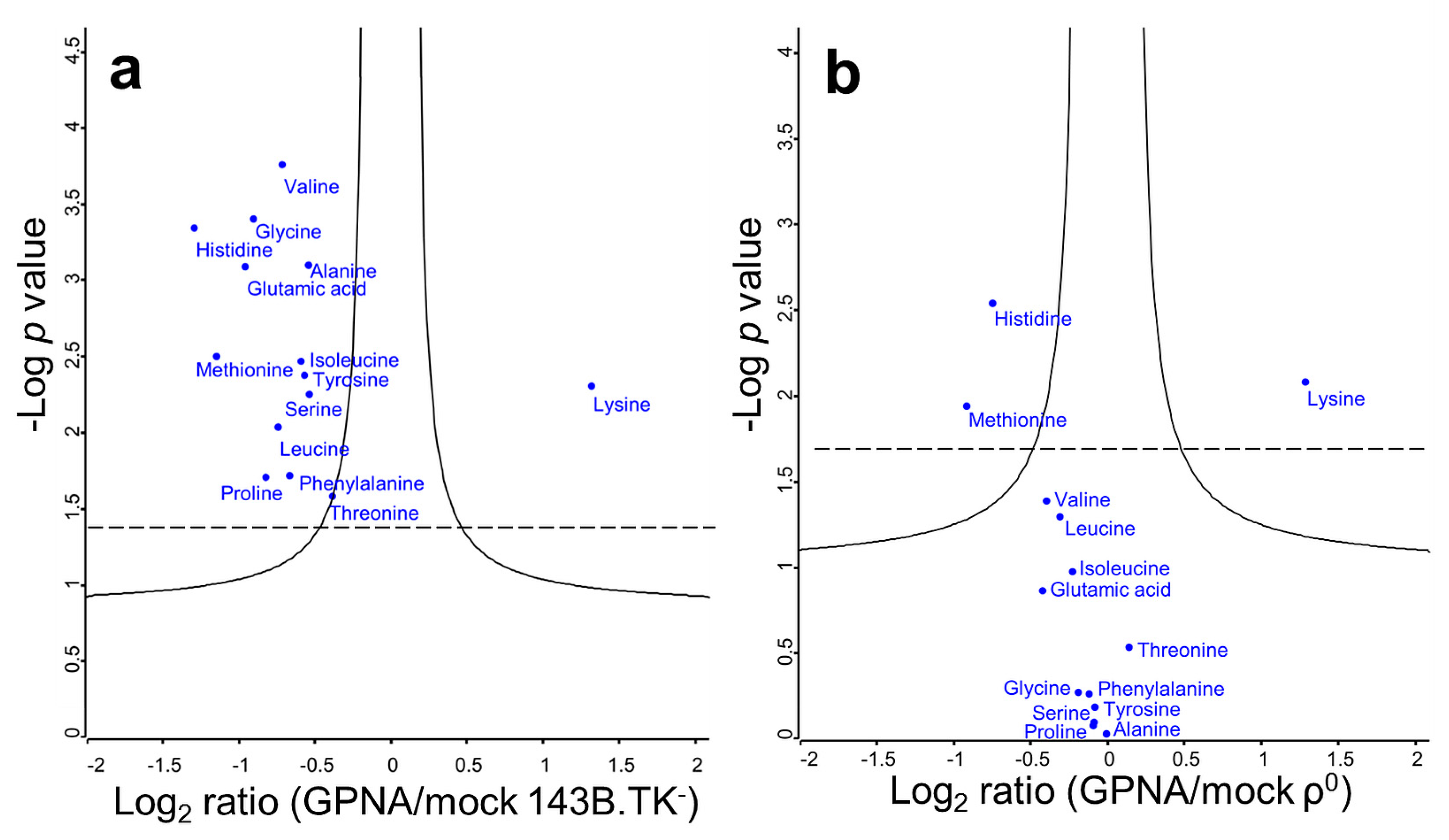
2.3. Solute Carrier Transporter Inhibition
3. Methods
3.1. Cell Culture
3.2. Amino Acid Flux Assay
3.3. SLC Transporter Inhibitor Assay
3.4. Metabolite Extraction and Profiling by Targeted Liquid Chromatography Mass Spectrometry (LC-MS)
3.5. Statistical Analyses
4. Conclusions
Supplementary Materials
Acknowledgments
Author Contributions
Conflicts of Interest
Abbreviations
| ATP | Adenosine triphosphate |
| LC-MS | Liquid chromatography mass spectrometry |
| MRM | Multiple reaction monitoring |
| SILAC | Stable isotope labeling with amino acids in cell culture |
| SLC | Solute carrier |
| PTM | Post-translational modification |
References
- Lin, L.; Yee, S.W.; Kim, R.B.; Giacomini, K.M. SLC transporters as therapeutic targets: Emerging opportunities. Nat. Rev. Drug Discov. 2015, 14, 543–560. [Google Scholar] [CrossRef] [PubMed]
- Bhutia, Y.D.; Babu, E.; Ramachandran, S.; Yang, S.; Thangaraju, M.; Ganapathy, V. SLC transporters as a novel class of tumour suppressors: Identity, function and molecular mechanisms. Biochem. J. 2016, 473, 1113–1124. [Google Scholar] [CrossRef] [PubMed]
- Li, H.; Myeroff, L.; Smiraglia, D.; Romero, M.F.; Pretlow, T.P.; Kasturi, L.; Lutterbaugh, J.; Rerko, R.M.; Casey, G.; Issa, J.P.; et al. SLC5A8, a sodium transporter, is a tumor suppressor gene silenced by methylation in human colon aberrant crypt foci and cancers. Proc. Natl. Acad. Sci. USA 2003, 100, 8412–8417. [Google Scholar] [CrossRef] [PubMed]
- Schweinfest, C.W.; Henderson, K.W.; Suster, S.; Kondoh, N.; Papas, T.S. Identification of a colon mucosa gene that is down-regulated in colon adenomas and adenocarcinomas. Proc. Natl. Acad. Sci. USA 1993, 90, 4166–4170. [Google Scholar] [CrossRef] [PubMed]
- Costello, L.C.; Franklin, R.B. The clinical relevance of the metabolism of prostate cancer; zinc and tumor suppression: Connecting the dots. Mol. Cancer 2006, 5, 17. [Google Scholar] [CrossRef] [PubMed]
- Hopkins, A.L.; Groom, C.R. The druggable genome. Nat. Rev. Drug Discov. 2002, 1, 727–730. [Google Scholar] [CrossRef] [PubMed]
- Polillo, M.; Galimberti, S.; Barate, C.; Petrini, M.; Danesi, R.; Di Paolo, A. Pharmacogenetics of BCR/ABL inhibitors in chronic myeloid leukemia. Int. J. Mol. Sci. 2015, 16, 22811–22829. [Google Scholar] [CrossRef] [PubMed]
- Franke, R.M.; Gardner, E.R.; Sparreboom, A. Pharmacogenetics of drug transporters. Curr. Pharm. Des. 2010, 16, 220–230. [Google Scholar] [CrossRef] [PubMed]
- Wang, Q.; Beaumont, K.A.; Otte, N.J.; Font, J.; Bailey, C.G.; van Geldermalsen, M.; Sharp, D.M.; Tiffen, J.C.; Ryan, R.M.; Jormakka, M.; et al. Targeting glutamine transport to suppress melanoma cell growth. Int. J. Cancer 2014, 135, 1060–1071. [Google Scholar] [CrossRef] [PubMed]
- Dall’Igna, O.P.; Bobermin, L.D.; Souza, D.O.; Quincozes-Santos, A. Riluzole increases glutamate uptake by cultured c6 astroglial cells. Int. J. Dev. Neurosci. 2013, 31, 482–486. [Google Scholar] [CrossRef] [PubMed]
- Daniel, B.; Green, O.; Viskind, O.; Gruzman, A. Riluzole increases the rate of glucose transport in L6 myotubes and NSC-34 motor neuron-like cells via AMPK pathway activation. Amyotroph. Lateral Scler. Frontotemporal Degener. 2013, 14, 434–443. [Google Scholar] [CrossRef] [PubMed]
- Fumagalli, E.; Funicello, M.; Rauen, T.; Gobbi, M.; Mennini, T. Riluzole enhances the activity of glutamate transporters GLAST, GLT1 and EAAC1. Eur. J. Pharmacol. 2008, 578, 171–176. [Google Scholar] [CrossRef] [PubMed]
- Carbone, M.; Duty, S.; Rattray, M. Riluzole elevates GLT-1 activity and levels in striatal astrocytes. Neurochem. Int. 2012, 60, 31–38. [Google Scholar] [CrossRef] [PubMed]
- Komander, D.; Rape, M. The ubiquitin code. Annu. Rev. Biochem. 2012, 81, 203–229. [Google Scholar] [CrossRef] [PubMed]
- Yau, R.; Rape, M. The increasing complexity of the ubiquitin code. Nat. Cell Biol. 2016, 18, 579–586. [Google Scholar] [CrossRef] [PubMed]
- Rajalingam, K.; Dikic, I. Expanding the ubiquitin code. Cell 2016, 164, 1074. [Google Scholar] [CrossRef] [PubMed]
- Herhaus, L.; Dikic, I. Expanding the ubiquitin code through post-translational modification. EMBO Rep. 2015, 16, 1071–1083. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; Smits, A.H.; van Tilburg, G.B.; Jansen, P.W.; Makowski, M.M.; Ovaa, H.; Vermeulen, M. An interaction landscape of ubiquitin signaling. Mol. Cell 2017, 65, 941–955. [Google Scholar] [CrossRef] [PubMed]
- Sane, S.; Rezvani, K. Essential roles of E3 ubiquitin ligases in p53 regulation. Int. J. Mol. Sci. 2017, 18, 442. [Google Scholar] [CrossRef] [PubMed]
- Broer, S. Amino acid transport across mammalian intestinal and renal epithelia. Physiol. Rev. 2008, 88, 249–286. [Google Scholar] [CrossRef] [PubMed]
- Aretz, I.; Hardt, C.; Wittig, I.; Meierhofer, D. An impaired respiratory electron chain triggers down-regulation of the energy metabolism and de-ubiquitination of solute carrier amino acid transporters. Mol. Cell. Proteom. 2016, 15, 1526–1538. [Google Scholar] [CrossRef] [PubMed]
- Schlessinger, A.; Khuri, N.; Giacomini, K.M.; Sali, A. Molecular modeling and ligand docking for solute carrier (SLC) transporters. Curr. Top. Med. Chem. 2013, 13, 843–856. [Google Scholar] [CrossRef] [PubMed]
- Esslinger, C.S.; Cybulski, K.A.; Rhoderick, J.F. Nγ-Aryl glutamine analogues as probes of the ASCT2 neutral amino acid transporter binding site. Bioorg. Med. Chem. 2005, 13, 1111–1118. [Google Scholar] [CrossRef] [PubMed]
- Chiu, M.; Sabino, C.; Allegri, M.; Bianchi, M.G.; Bussolati, O. Multiple glutamine transporters sustain the growth of glutamine synthetase-negative human oligodedroglioma cells. FASEB J. 2016, 30, 1099–1116. [Google Scholar]
- Udeshi, N.D.; Mani, D.R.; Eisenhaure, T.; Mertins, P.; Jaffe, J.D.; Clauser, K.R.; Hacohen, N.; Carr, S.A. Methods for quantification of in vivo changes in protein ubiquitination following proteasome and deubiquitinase inhibition. Mol. Cell. Proteom. 2012, 11, 148–159. [Google Scholar] [CrossRef] [PubMed]
- Amann, T.; Hellerbrand, C. GLUT1 as a therapeutic target in hepatocellular carcinoma. Expert Opin. Ther. Targets 2009, 13, 1411–1427. [Google Scholar] [CrossRef] [PubMed]
- Pinheiro, C.; Longatto-Filho, A.; Azevedo-Silva, J.; Casal, M.; Schmitt, F.C.; Baltazar, F. Role of monocarboxylate transporters in human cancers: State of the art. J. Bioenerg. Biomembr. 2012, 44, 127–139. [Google Scholar] [CrossRef] [PubMed]
- King, M.P.; Attardi, G. Isolation of human cell lines lacking mitochondrial DNA. Methods Enzymol. 1996, 264, 304–313. [Google Scholar] [PubMed]
- Meierhofer, D.; Halbach, M.; Sen, N.E.; Gispert, S.; Auburger, G. Atxn2-knock-out mice show branched chain amino acids and fatty acids pathway alterations. Mol. Cell. Proteomics 2016, 15, 1728–1739. [Google Scholar] [CrossRef] [PubMed]
- Gielisch, I.; Meierhofer, D. Metabolome and proteome profiling of complex I deficiency induced by rotenone. J. Proteome Res. 2015, 14, 224–235. [Google Scholar] [CrossRef] [PubMed]
- Tyanova, S.; Temu, T.; Sinitcyn, P.; Carlson, A.; Hein, M.Y.; Geiger, T.; Mann, M.; Cox, J. The perseus computational platform for comprehensive analysis of (prote)omics data. Nat. Methods 2016, 13, 731–740. [Google Scholar] [CrossRef] [PubMed]

© 2017 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Medina, A.B.; Banaszczak, M.; Ni, Y.; Aretz, I.; Meierhofer, D. ρ0 Cells Feature De-Ubiquitination of SLC Transporters and Increased Levels and Fluxes of Amino Acids. Int. J. Mol. Sci. 2017, 18, 879. https://doi.org/10.3390/ijms18040879
Medina AB, Banaszczak M, Ni Y, Aretz I, Meierhofer D. ρ0 Cells Feature De-Ubiquitination of SLC Transporters and Increased Levels and Fluxes of Amino Acids. International Journal of Molecular Sciences. 2017; 18(4):879. https://doi.org/10.3390/ijms18040879
Chicago/Turabian StyleMedina, André Bordinassi, Marcin Banaszczak, Yang Ni, Ina Aretz, and David Meierhofer. 2017. "ρ0 Cells Feature De-Ubiquitination of SLC Transporters and Increased Levels and Fluxes of Amino Acids" International Journal of Molecular Sciences 18, no. 4: 879. https://doi.org/10.3390/ijms18040879





