1. Introduction
Self-incompatibility (SI) is a common mechanism that promotes outcrossing in angiosperms. Based on floral morphology, SI can be classified as homomorphic or heteromorphic. Homomorphic SI systems include the gametophytic SI (GSI) and sporophytic SI (SSI) systems, and plants with homomorphic SI systems have no distinct floral morphologies [
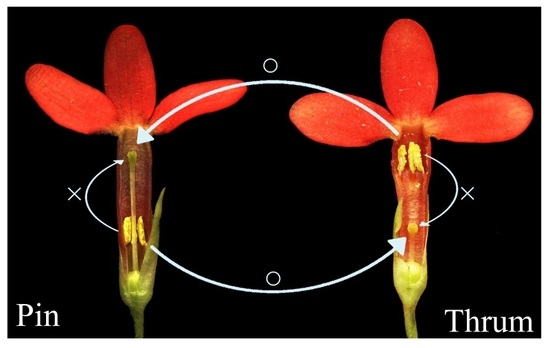
1]. Heteromorphic SI is usually considered sporophytic and is present in distylous and tristylous populations that possess long and short styles [
2].
Heteromorphic SI has been reported in at least 28 families scattered throughout the angiosperm lineage [
3]. In the Primulaceae family, floral morphs with long styles are termed pin morphs and those with short styles are termed thrum morphs. Dimorphic heteromorphic SI has been observed in 91% of
Primula species, and this species is often considered to be a model system for heterostyly [
4], with the two types of flowers varying in style length, anther position, and pollen size [
5]. Classical theories proposed that heteromorphic SI systems are controlled by the S locus, with the thrum morph being heterozygous (
S/
s) and the pin being homozygous recessive (
s/
s) [
6,
7,
8,
9]. In contrast, Li et al. proposed that the S locus is actually hemizygous, not heterozygous, in thrum morphs [
10]. Further studies indicated that three genes are present at the S locus as a co-adapted linkage group:
G (style length and incompatibility),
P (pollen size and incompatibility), and
A (anther position) [
2]. The first molecular marker for the
Primula S locus was identified in
Primula vulgaris [
11], and two S locus-linked genes of
Primula were identified using differential display technology [
12]. The
Primula CYP734A50 gene was identified at the S locus and is expressed specifically in thrum styles [
13], while the
GLOT,
CYPT,
PUMT,
KFBT, and
CCMT genes were shown to be present at the S locus only in the thrum morph [
10]. Moreover,
PvGLO2 was shown to be adjacent to
CYP734A50 which lies within the S locus [
14]. However, the key genes that regulate heteromorphic SI in
Primula remain unknown.
As mentioned above, heteromorphic SI is generally regarded as sporophytic. However, Shivanna et al. found that the thrum and pin morphs exhibited different self-pollen tube inhibition behaviors, with thrum self-pollen tube inhibition occurring on the stigma and pin self-pollen tube inhibition occurring frequently in the style [
15]. Similarly, in the SI system of
P. obconica, the thrum morph incompatibility barrier appears to be present on or in the stigma, while in the pin morph, the site of self-pollen tube inhibition could be the stigma or the style [
16]. In two distylous
Turnera species, obvious ultrastructural differences were observed between the pin and thrum self-pollen tubes [
17]. Lewis and Jones proposed that the SI systems of thrum morphs may be sporophytic and those of pin morphs may be both sporophytic and gametophytic [
2]. Thus, the SI mechanisms of pin and thrum morphs may be different.
Some researchers have proposed that SI systems originated from pathogen defense mechanisms [
18,
19,
20,
21]. The female S determinant of SSI in
Brassica is the S-locus receptor kinase (SRK), a member of a large gene family that controls host–pathogen defense [
22,
23]. Similarly, the female S determinants of many GSI systems are S-locus ribonucleases (S-RNases), which are also involved in pathogen defense responses [
24]. Some symptoms of programmed cell death (PCD), e.g., swollen mitochondria and nuclear DNA degradation, were observed in the self-pollen tubes of pear after incompatible pollination [
25]. Similarly, PCD was observed in self-pollen tubes after interactions between the female and male S determinants in the
Papaver SI system [
26]. These findings demonstrate the close association between pathogen defense mechanisms and SI systems.
In recent years, most studies of the
Primula SI system have focused on differences in pollination behaviors between morphs, such as the roles of bumblebee nectar robbers and syrphid flies in
P. secundiflora pollination, the strength of SI and distylous syndrome in
P. veris, the effect of anther–stigma distance on stigma pollen grains in two
Primula species, and the relationship between flower display and SI in
P. tibetica [
27,
28,
29,
30]. However, little is known about the biological events that occur during the process of
Primula SI. In addition, while extensive molecular studies on
Primula have been carried out, such as genomics analyses of
P. veris and
P. vulgaris [
10,
31], the regulatory factors involved in heteromorphic SI have not been identified. In the current study, we used
P. maximowiczii, a perennial species native to Northern China, to investigate the pollen germination time, pollen tube elongation, and fruiting rates of self- and cross-pollinations of pin and thrum morphs, and we collected the pistils after four types of pollination to perform a transcriptomics analysis. Our results provide the first transcriptomics analysis of the heteromorphic SI of
Primula and offer new data for future genetic and genomic studies on
Primula SI.
3. Discussion
Compared to GSI and SSI, the molecular mechanisms of heteromorphic SI are poorly understood. The SI mating types of
Primula are thought to be controlled by a diallelic S locus [
8]. Some genes in the S locus of
Primula have been reported recently [
10,
18]; however, the female and male determinants of heteromorphic SI, the signaling components activated after the interaction of determinants, and the events involved in the process of heteromorphic SI remain unknown.
SSI systems usually take the form of stigmatic inhibition of self-pollen, and GSI systems frequently show pollen inhibition in the style [
37,
38]. Therefore, although heteromorphic SI has been traditionally classified as sporophytic, it has been hypothesized that the SI mechanism of the thrum morph is sporophytic while that of the pin morph may be both sporophytic and gametophytic [
2]. In our study, self-pollen tubes of the thrum morph rarely reached the style, while the self-pollen tubes of the pin morph occasionally, though rarely, entered the style. Therefore, our results support this hypothesis. Furthermore, based on our transcriptomics analyses, about 90% of the 2135 DEGs identified between the self- and cross-pollinated pistils were specifically expressed in either the pin (PS vs. PT) or thrum (TS vs. TP) morphs. Moreover, the GO and KEGG enrichment analyses demonstrated that the genes involved in the pin SI process were distinct from those involved in thrum SI. Based on these results, we hypothesize that the pin and thrum SI responses differ and have different underlying molecular mechanisms.
Carbohydrate metabolism participates in many plant biological processes. Our transcriptomics data showed that the DEGs identified in the heteromorphic SI process of
P. maximowiczii were significantly enriched in those involved in carbohydrate metabolism. A total of 17 downregulated DEGs and 24 upregulated DEGs were associated with carbohydrate metabolism (
Figures S11 and S13). Furthermore, the results of the GO and KEGG enrichment analyses for all DEGs highlighted the responses to stimuli and environmental adaptation categories as important in the processes of compatibility and incompatibility, and the analyses of upregulated and downregulated DEGs returned similar results. Notably, the most significantly enriched KEGG pathway in the PS vs. PT comparison was plant-pathogen interactions which were enriched with 29 DEGs from the PS vs. PT comparison and nine DEGs from the TS vs. TP comparison. Based on our analysis, TFs, ERF, C2H2, and WRKY proteins were identified as instrumental in the SI process and are known to be involved in plant defense and other stress responses. In conclusion, the bioinformatics analysis of the transcriptomics data suggested that the pathways associated with environmental adaptation are involved in heteromorphic SI, especially those involved in pathogen defense. Thus, the heteromorphic SI response of
P. maximowiczii may be associated with both environmental adaptation and carbohydrate metabolism.
In the plant-pathogen interaction pathway, we observed that three KO nodes enriched for DEGs act across the cell membrane, namely, CNGCs, BAK1/SERK4, and FLSs (
Figure 8). It is known that the female S determinants, SRK in SSI and S-RNase in GSI, in several plant species (Brassicaceae, Solanaceae, and Papaveraceae) are transmembrane proteins containing transmembrane domains [
39,
40,
41,
42,
43]. The genes encoding these transmembrane proteins may participate in the recognition of the female and male S determinants of
P. maximowiczii SI. In the FLSs node, there were enriched with the DEGs of both PS vs. PT and TS vs. TP, but the DEGs were different and showed different expression patterns. In the PS vs. PT comparison, two FLS DEGs, both putative polygalacturonase inhibitors (PGIPs), were implicated, while in the TS vs. TP comparison, one FLS DEG, a putative endoglucanase (EG), was implicated. It has been reported that PGIPs play important roles in the defense against pathogenic fungi and pollen development [
44,
45] and that EGs are associated with the degradation of the cell wall and pollen tube growth [
46,
47]. Thus, the putative PGIP and EG proteins may participate in the processes of pin and thrum SI, respectively. In the future, we plan to investigate the function of these DEGs in more detail and to identify the key genes involved in the recognition processes of pin and thrum SI.
In our study, we found that 89 (4.17%) of the 2135 DEGs showed inverse expression patterns in the two comparisons and may therefore be associated with the pollen genotype. Therefore, this raises the question of whether these DEGs are involved in SI or whether they are simply associated with pollen morphology. For example, one unigene, predicted to be a putative RALF-like gene (
CL2572-3), was found to exhibit inverse expression patterns between PS vs. PT and TS vs. TP comparisons. It was downregulated in PS vs. PT, with a fold change >9 and upregulated in TS vs. TP with a fold change of seven (
Table S13). We have confirmed this gene is specifically expressed in thrum pollen (
Figure 7B). The RALF-like gene has been previously reported in
P. vulgaris as a pollen-expressed gene and was expressed earlier in development in thrum pollen than in pin pollen [
48]. Moreover, RALF-like genes can alter the pH of the cell and play a role in regulating plant development [
49]; cytosolic pH acidification has been confirmed as an integral and essential event for the SI response in
Papaver rhoeas [
50]. This raises the question of whether this putative RALF-like gene is involved in the
Primula SI response. One putative PME gene (
CL8881-2) was differentially expressed in the PS vs. PT and TS vs. TP comparisons and is specifically expressed in pin pollen (
Figure 7A). PMEs are involved in the degradation of the cell wall, pollen tube growth, and plant defense mechanisms against pathogens [
51,
52,
53,
54]. In addition, there are known male S determinant genes of SI systems that are specifically expressed in the anther or pollen, such as
SP11/SCR in
Brassica and
SLF in
Antirrhinum and
Prunus [
40,
55,
56,
57]. However, determining whether these genes that are associated with pollen genotype participate in the
Primula SI response or interact with the male S determinant gene requires additional research.
After removing the 89 DEGs with inverse expression patterns in the pin and thrum comparisons, there were 2046 DEGs (95.83%), including the upregulated and downregulated DEGs specific to the TS vs. TP and PS vs. PT comparisons and the upregulated and downregulated DEGs that were present in both the TS vs. TP and PS vs. PT comparisons. Nine hundred and eleven were upregulated and 1135 were downregulated in the self-pollinations vs. the cross-pollinations (
Figure 3C). The genes with different expression patterns were specific to the different pollination combinations. For example, among the genes tested, the upregulated DEGs specific to the PS vs. PT comparison were expressed specifically in the PS treatment. They showed a higher expression level in the PS pollination than in the other pollinations and no specific expression in the pistils or pollen. This result suggests that the genes are associated with the incompatible response of pin morph. Similarly, the upregulated DEGs specific to the TS vs. TP comparison were expressed specifically in the TS pollination, and the downregulated DEGs specific to the PS vs. PT comparison were expressed specifically in the PT pollination and so on. In the cross-pollinated pistils, the pollen germinated and grew into the ovary, while in the self-pollinated plants, most pollen grains or pollen tubes were inhibited on the stigma surface or in the stigma. As the downregulated DEGs were more highly expressed in compatible combinations than in incompatible combinations, downregulated DEGs may be associated with the compatibility response. As described above, downregulated DEGs involved in carbohydrate metabolism may be involved in supplying the energy for pollen tube growth. In contrast, upregulated DEGs were more highly expressed in the incompatible combinations. Some of these DEGs were also involved in carbohydrate metabolism and energy metabolism (
Figure S13). However, even if the downregulated DEGs play a role in supplying energy to compatible pollen tubes, the functions of the upregulated DEGs associated with carbohydrate metabolism and energy metabolism in the SI response remain unknown. Moreover, how do these genes regulate the compatible or incompatible responses? Does this mean that silencing of the highly expressed genes would inhibit the compatible or incompatible responses? Alternatively, if the lowly expressed genes were overexpressed, would the compatible or incompatible response be promoted? Our study provides interesting data that may help to answer these questions and is a basis for further investigations into the molecular mechanisms of
Primula SI.