Cytotoxic Potential and Molecular Pathway Analysis of Silver Nanoparticles in Human Colon Cancer Cells HCT116
Abstract
:1. Introduction
2. Results and Discussion
2.1. Synthesis and Characterization of AgNPs Using NAR
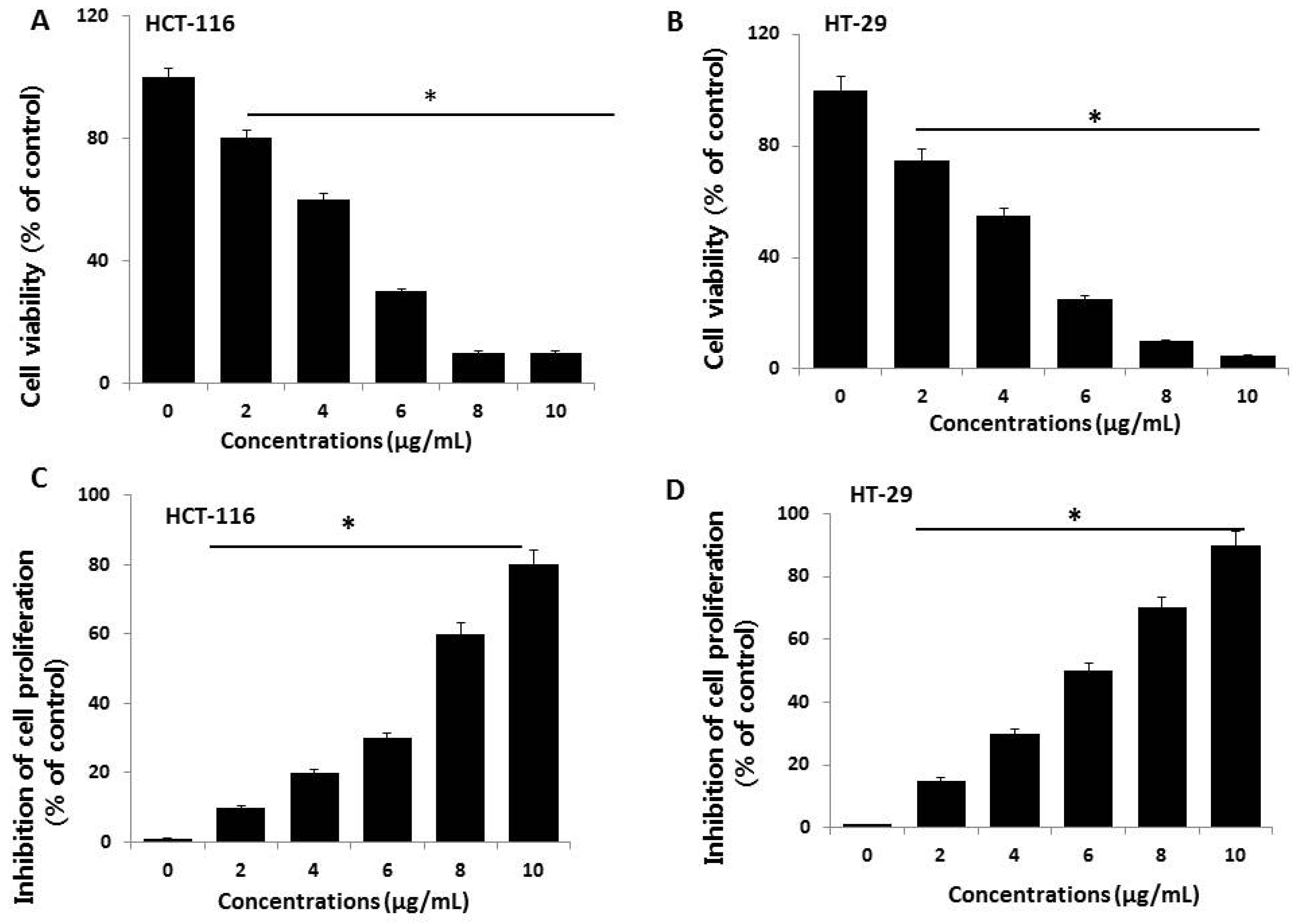
2.2. Effect of AgNPs on Cell Viability and Proliferation of HCT116 and HT-29 Cells
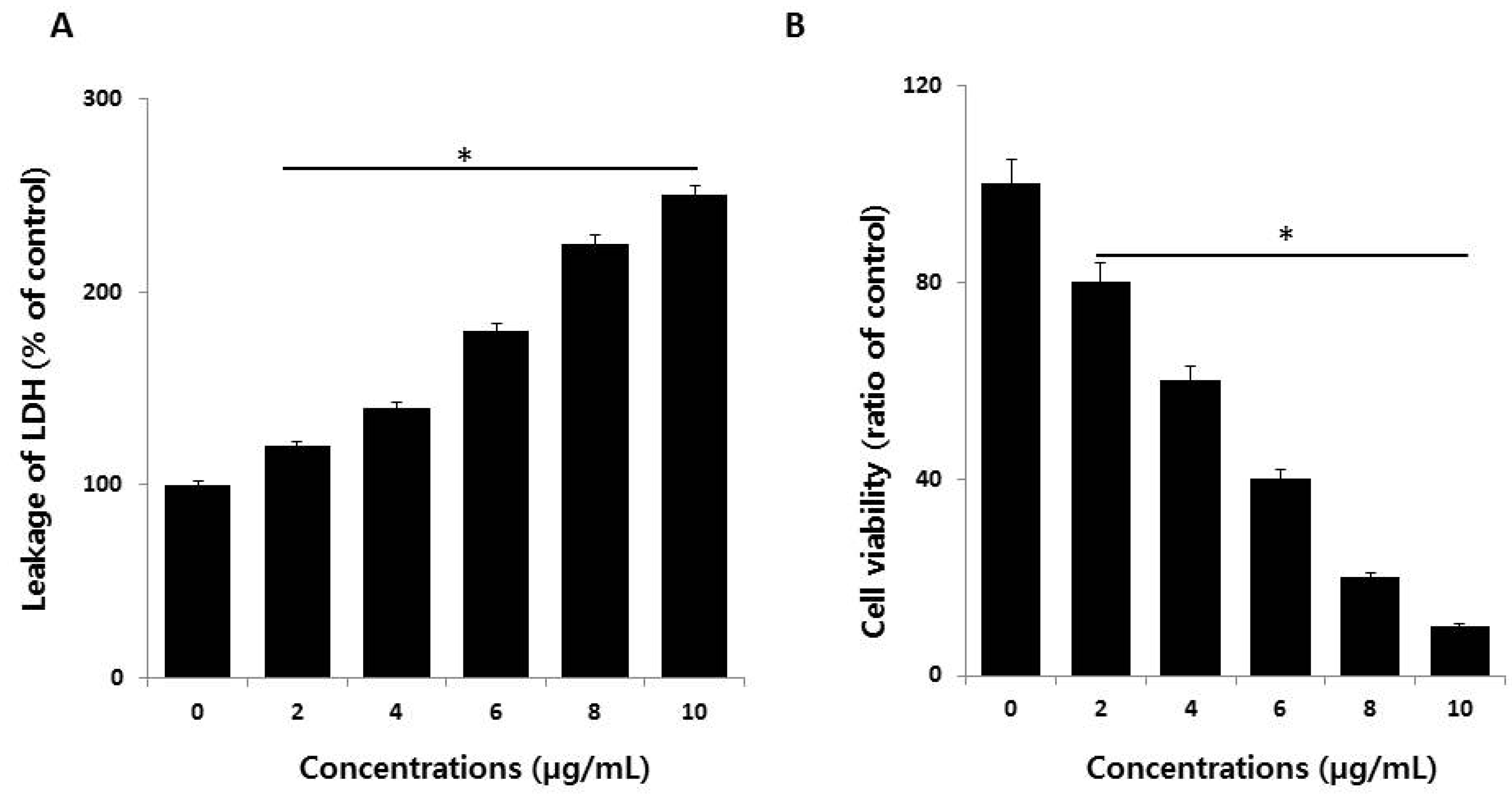
2.3. AgNPs Increase Cytotoxicity in HCT116 Cells
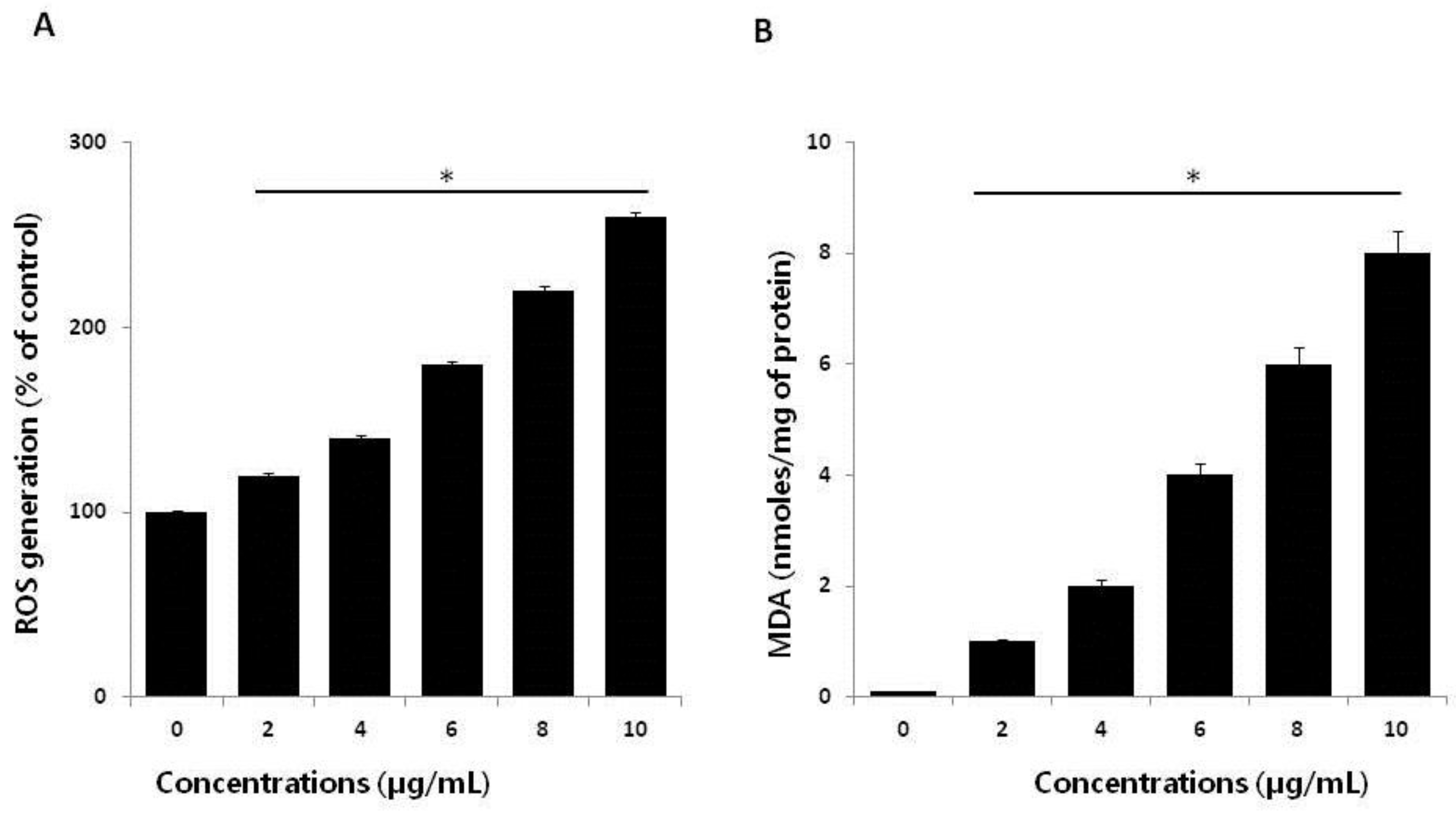
2.4. Effect of AgNPs on ROS Generation and Malondialdehyde (MDA)
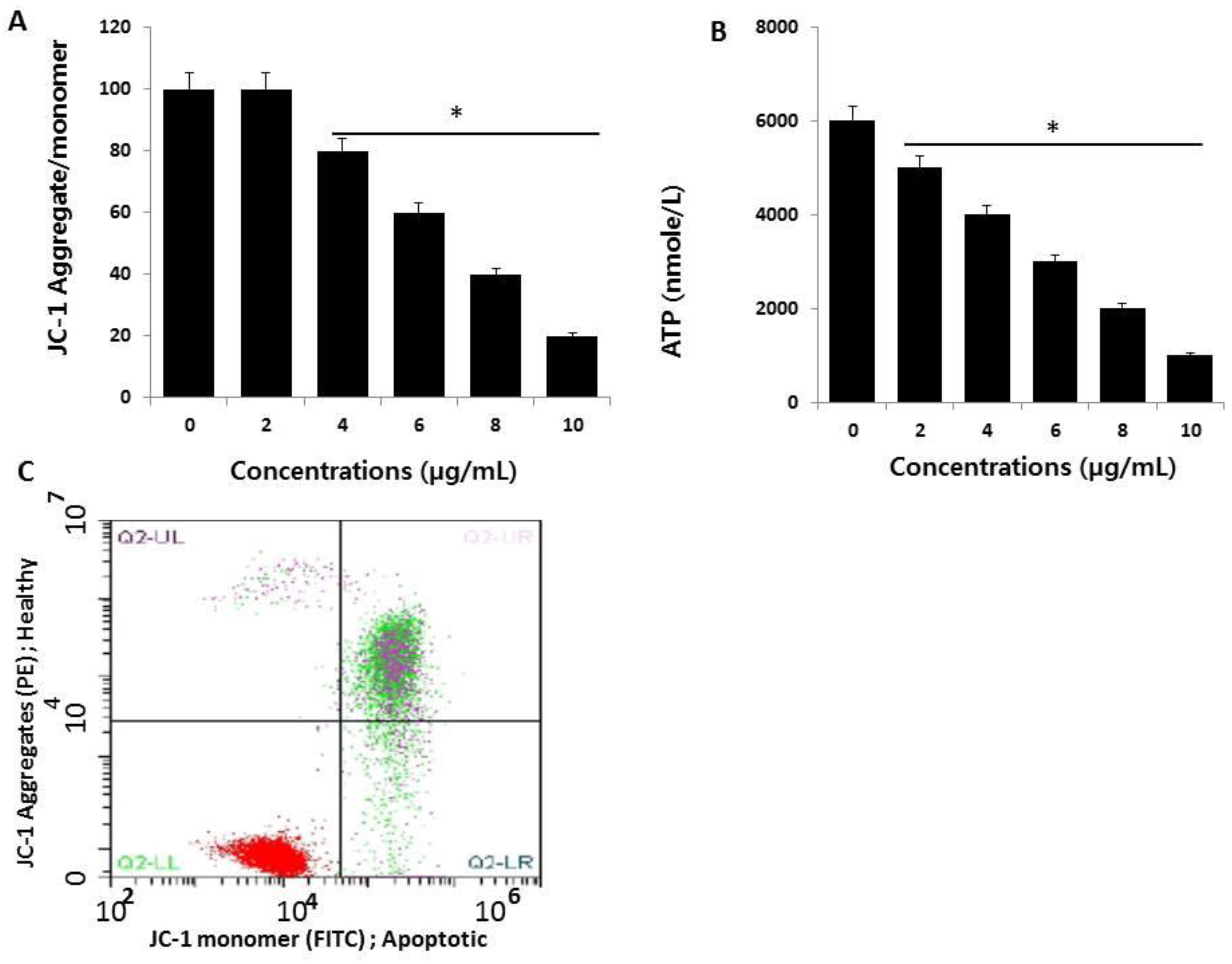
2.5. AgNPs Induce Mitochondrial Dysfunction and Reduce ATP Generation in HCT116 Cells
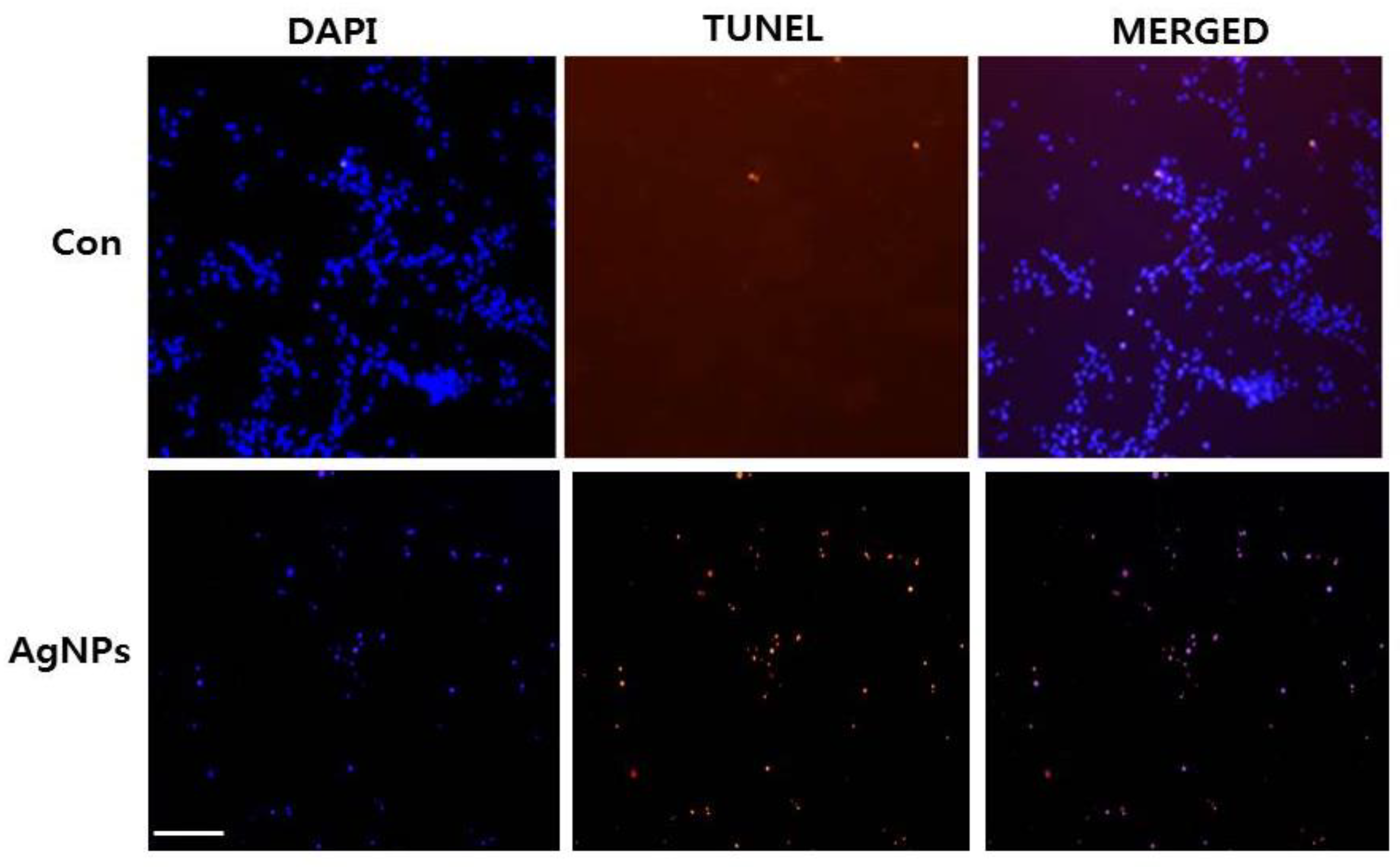
2.6. AgNPs Induce Apoptosis
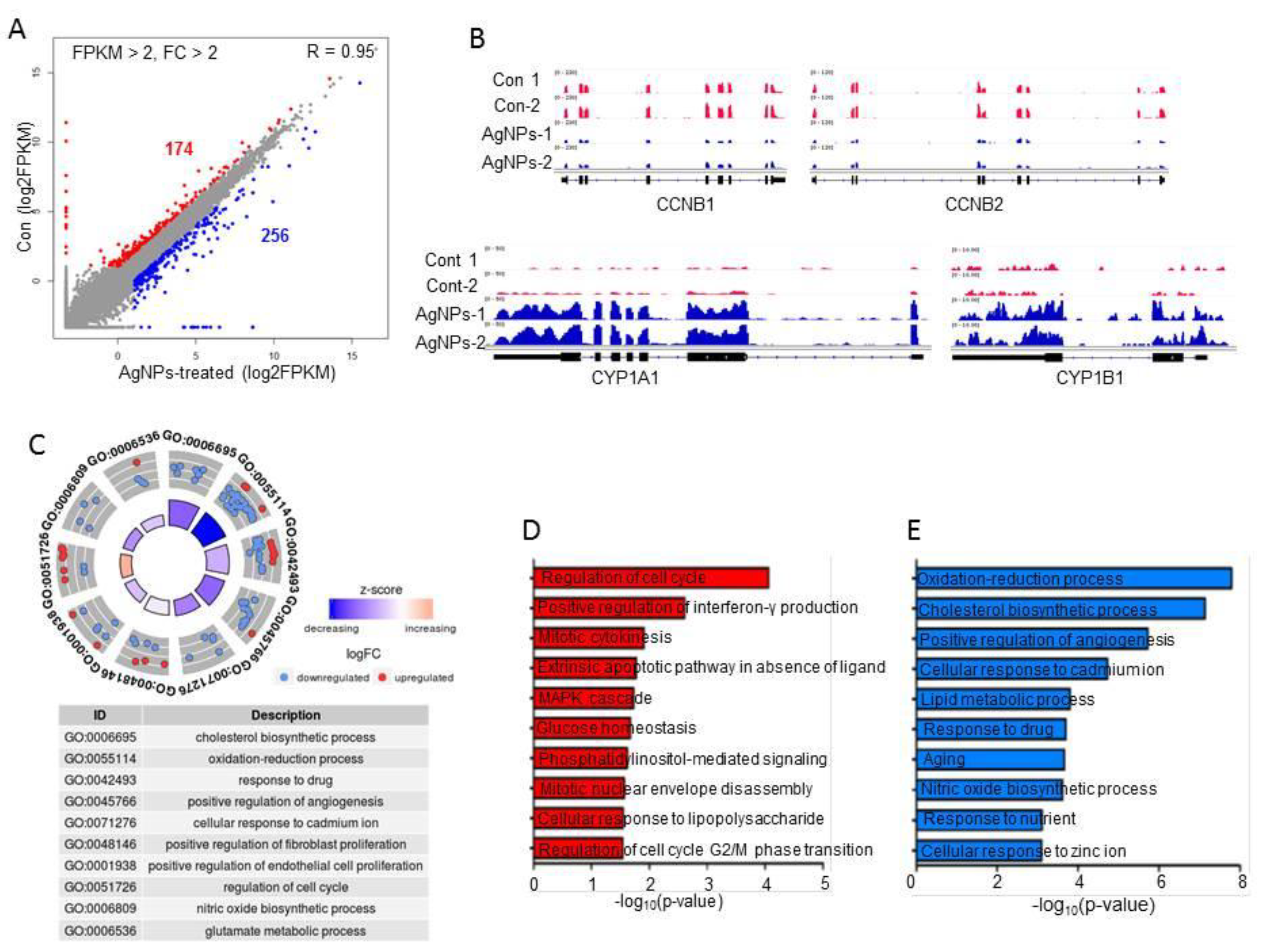
2.7. AgNP Treatment Impairs Expression of Genes Involved in Mitochondrial Function and Cell Apoptosis
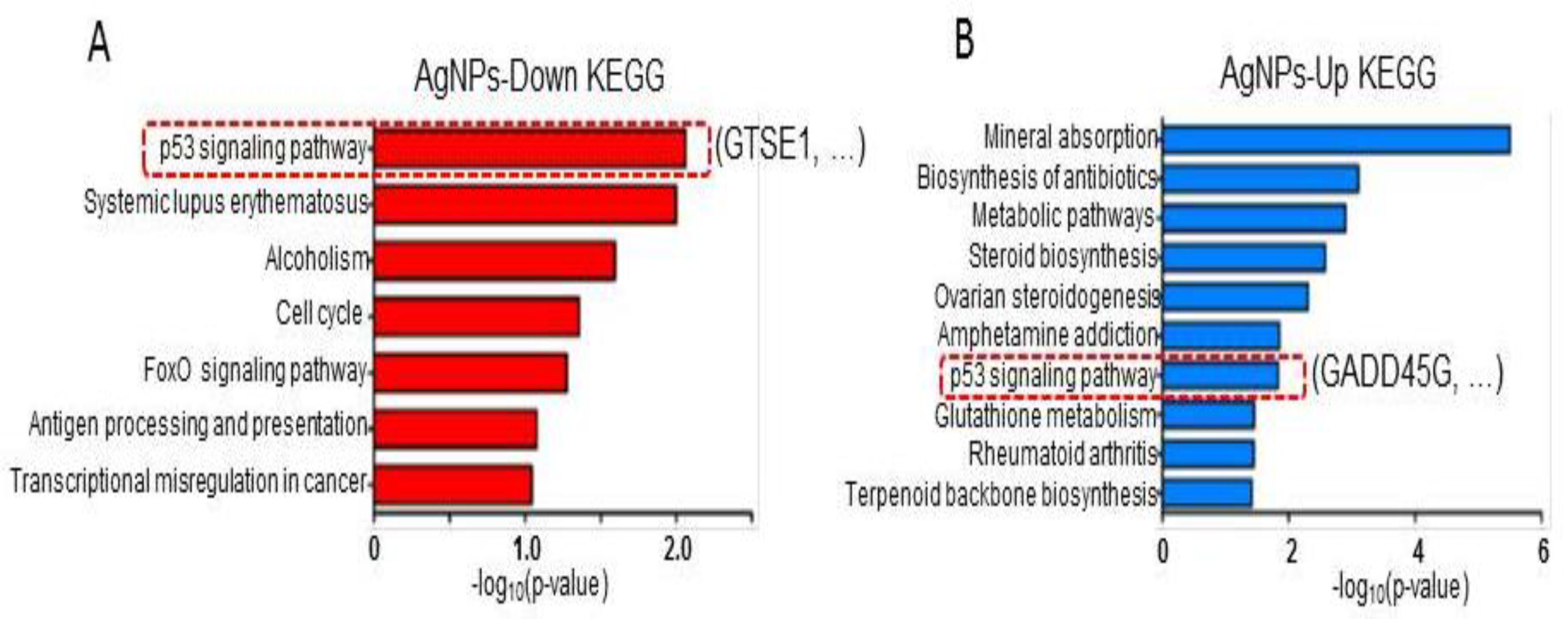
2.8. AgNP Treatment Dysregulates Multiple Biological Pathways
3. Materials and Methods
3.1. Materials
3.2. Synthesis and Characterization of AgNPs
3.3. Cell Viability and Cell Proliferation
3.4. Membrane Integrity
3.5. Assessment of Dead-Cell Protease Activity
3.6. Determination of Intracellular ROS
3.7. Measurement of MDA
3.8. JC-1 Assay
3.9. Measurement of ATP
3.10. TUNEL Analysis
3.11. RNA-Seq and Downstream Bioinformatics Analysis
3.12. Statistical Methods
4. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Arnold, M.; Sierra, M.S.; Laversanne, M.; Soerjomataram, I.; Jemal, A.; Bray, F. Global patterns and trends in colorectal cancer incidence and mortality. Gut 2017, 66, 683–691. [Google Scholar] [CrossRef] [PubMed]
- Navarro, M.; Nicolas, A.; Ferrandez, A.; Lanas, A. Colorectal cancer population screening programs worldwide in 2016: An update. World J. Gastroenterol. 2017, 23, 3632–3642. [Google Scholar] [CrossRef] [PubMed]
- Marley, A.R.; Nan, H. Epidemiology of colorectal cancer. Int. J. Mol. Epidemiol. Genet. 2016, 7, 105–114. [Google Scholar] [PubMed]
- Kelly, C.; Bhuva, N.; Harrison, M.; Buckley, A.; Saunders, M. Use of raltitrexed as an alternative to 5-fluorouracil and capecitabine in cancer patients with cardiac history. Eur. J. Cancer 2013, 49, 2303–2310. [Google Scholar] [CrossRef] [PubMed]
- Kanterman, J.; Sade-Feldman, M.; Biton, M.; Ish-Shalom, E.; Lasry, A.; Goldshtein, A.; Hubert, A.; Baniyash, M. Adverse immunoregulatory effects of 5FU and CPT11 chemotherapy on myeloid-derived suppressor cells and colorectal cancer outcomes. Cancer Res. 2014, 74, 6022–6035. [Google Scholar] [CrossRef] [PubMed]
- Qasim, M.; Lim, D.-J.; Park, H.; Na, D. Nanotechnology for Diagnosis and Treatment of Infectious Diseases. J. Nanosci. Nanotechnol. 2014, 14, 7374–7387. [Google Scholar] [CrossRef] [PubMed]
- De Matteis, V.; Cascione, M.; Toma, C.C.; Leporatti, S. Silver Nanoparticles: Synthetic Routes, In Vitro Toxicity and Theranostic Applications for Cancer Disease. Nanomaterials 2018, 8, 319. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.-F.; Liu, Z.-G.; Shen, W.; Gurunathan, S. Silver Nanoparticles: Synthesis, Characterization, Properties, Applications, and Therapeutic Approaches. Int. J. Mol. Sci. 2016, 17, 1534. [Google Scholar] [CrossRef] [PubMed]
- AbdelRahim, K.; Mahmoud, S.Y.; Ali, A.M.; Almaary, K.S.; Mustafa, A.E.-Z.M.A.; Husseiny, S.M. Extracellular biosynthesis of silver nanoparticles using Rhizopus stolonifer. Saudi J. Biol. Sci. 2017, 24, 208–216. [Google Scholar] [CrossRef] [PubMed]
- De Lima, R.; Seabra, A.B.; Durán, N. Silver nanoparticles: A brief review of cytotoxicity and genotoxicity of chemically and biogenically synthesized nanoparticles. J. Appl. Toxicol. 2012, 32, 867–879. [Google Scholar] [CrossRef] [PubMed]
- Gurunathan, S.; Lee, K.-J.; Kalishwaralal, K.; Sheikpranbabu, S.; Vaidyanathan, R.; Eom, S.H. Antiangiogenic properties of silver nanoparticles. Biomaterials 2009, 30, 6341–6350. [Google Scholar] [CrossRef] [PubMed]
- Sriram, M.I.; Kanth, S.B.M.; Kalishwaralal, K.; Gurunathan, S. Antitumor activity of silver nanoparticles in Dalton’s lymphoma ascites tumor model. Int. J. Nanomed. 2010, 5, 753–762. [Google Scholar] [CrossRef]
- Guo, D.; Zhu, L.; Huang, Z.; Zhou, H.; Ge, Y.; Ma, W.; Wu, J.; Zhang, X.; Zhou, X.; Zhang, Y.; et al. Anti-leukemia activity of PVP-coated silver nanoparticles via generation of reactive oxygen species and release of silver ions. Biomaterials 2013, 34, 7884–7894. [Google Scholar] [CrossRef] [PubMed]
- Gurunathan, S.; Han, J.W.; Eppakayala, V.; Jeyaraj, M.; Kim, J.-H. Cytotoxicity of biologically synthesized silver nanoparticles in MDA-MB-231 human breast cancer cells. Biomed Res. Int. 2013, 2013, 535796. [Google Scholar] [CrossRef] [PubMed]
- Gurunathan, S.; Jeong, J.-K.; Han, J.W.; Zhang, X.-F.; Park, J.H.; Kim, J.-H. Multidimensional effects of biologically synthesized silver nanoparticles in Helicobacter pylori, Helicobacter felis, and human lung (L132) and lung carcinoma A549 cells. Nanoscale Res. Lett. 2015, 10, 35. [Google Scholar] [CrossRef] [PubMed]
- Gurunathan, S.; Han, J.W.; Park, J.H.; Kim, E.; Choi, Y.-J.; Kwon, D.-N.; Kim, J.-H. Reduced graphene oxide-silver nanoparticle nanocomposite: A potential anticancer nanotherapy. Int. J. Nanomed. 2015, 10, 6257–6276. [Google Scholar] [CrossRef] [PubMed]
- Han, J.; Gurunathan, S.; Jeong, J.-K.; Choi, Y.-J.; Kwon, D.-N.; Park, J.-K.; Kim, J.-H. Oxidative stress mediated cytotoxicity of biologically synthesized silver nanoparticles in human lung epithelial adenocarcinoma cell line. Nanoscale Res. Lett. 2014, 9, 459. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Online, V.A.; Sahu, S.; Sinha, N.; Bhutia, S.K.; Majhi, M.; Mohapatra, S. Luminescent magnetic hollow mesoporous silica nanotheranostics for camptothecin delivery and multimodal imaging. J. Mater. Chem. B 2014, 3799–3808. [Google Scholar] [CrossRef]
- Jagannath, C.V.; Radheshyam, B.K. Potentiation of antiepileptic activity of phenytoin using β carotene against maximal electroshock induced convulsions in mice. World J. Pharm. Pharm. Sci. 2017, 1574–1585. [Google Scholar] [CrossRef]
- Look, M.; Bandyopadhyay, A.; Blum, J.S.; Fahmy, T.M. Application of nanotechnologies for improved immune response against infectious diseases in the developing world. Adv. Drug Deliv. Rev. 2010, 62, 378–393. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Feliu, N.; Kohonen, P.; Ji, J.; Zhang, Y.; Karlsson, H.L.; Palmberg, L.; Nyström, A.; Fadeel, B. Next-Generation Sequencing Reveals Low-Dose Effects of Cationic Dendrimers in Primary Human Bronchial Epithelial Cells. ACS Nano 2015, 9, 146–163. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Guo, Y.; Ma, C.; Zhang, D.; Wang, C.; Yang, Q. Transcriptome analysis of maize resistance to Fusarium graminearum. BMC Genom. 2016, 17, 477. [Google Scholar] [CrossRef]
- Sun, Q.-L.; Zhao, C.-P.; Wang, T.-Y.; Hao, X.-B.; Wang, X.-Y.; Zhang, X.; Li, Y.-C. Expression profile analysis of long non-coding RNA associated with vincristine resistance in colon cancer cells by next-generation sequencing. Gene 2015, 572, 79–86. [Google Scholar] [CrossRef] [PubMed]
- Morrissy, A.S.; Garzia, L.; Shih, D.J.H.; Zuyderduyn, S.; Huang, X.; Skowron, P.; Remke, M.; Cavalli, F.M.G.; Ramaswamy, V.; Lindsay, P.E.; et al. Divergent clonal selection dominates medulloblastoma at recurrence. Nature 2016, 529, 351–357. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cong, L.; Ran, F.A.; Cox, D.; Lin, S.; Barretto, R.; Habib, N.; Hsu, P.D.; Wu, X.; Jiang, W.; Marraffini, L.A.; et al. Multiplex Genome Engineering Using CRISPR/Cas Systems. Science 2013, 339, 819–823. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kanno, S.; Shouji, A.; Hirata, R.; Asou, K.; Ishikawa, M. Effects of naringin on cytosine arabinoside (Ara-C)-induced cytotoxicity and apoptosis in P388 cells. Life Sci. 2004, 75, 353–365. [Google Scholar] [CrossRef] [PubMed]
- Song, H.M.; Park, G.H.; Eo, H.J.; Lee, J.W.; Kim, M.K.; Lee, J.R.; Lee, M.H.; Koo, J.S.; Jeong, J.B. Anti-Proliferative Effect of Naringenin through p38-Dependent Downregulation of Cyclin D1 in Human Colorectal Cancer Cells. Biomol. Ther. (Seoul) 2015, 23, 339–344. [Google Scholar] [CrossRef] [PubMed]
- Sahu, N.; Soni, D.; Chandrashekhar, B.; Satpute, D.B.; Saravanadevi, S.; Sarangi, B.K.; Pandey, R.A. Synthesis of silver nanoparticles using flavonoids: Hesperidin, naringin and diosmin, and their antibacterial effects and cytotoxicity. Int. Nano Lett. 2016, 6, 173–181. [Google Scholar] [CrossRef]
- Shankar, S.S.; Rai, A.; Ahmad, A.; Sastry, M. Rapid synthesis of Au, Ag, and bimetallic Au core–Ag shell nanoparticles using Neem (Azadirachta indica) leaf broth. J. Colloid Interface Sci. 2004, 275, 496–502. [Google Scholar] [CrossRef] [PubMed]
- Jain, S.; Mehata, M.S. Medicinal Plant Leaf Extract and Pure Flavonoid Mediated Green Synthesis of Silver Nanoparticles and their Enhanced Antibacterial Property. Sci. Rep. 2017, 7, 15867. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Singhal, G.; Bhavesh, R.; Kasariya, K.; Sharma, A.R.; Singh, R.P. Biosynthesis of silver nanoparticles using Ocimum sanctum (Tulsi) leaf extract and screening its antimicrobial activity. J. Nanopart. Res. 2011, 13, 2981–2988. [Google Scholar] [CrossRef]
- Verma, A.; Mehata, M.S. Controllable synthesis of silver nanoparticles using Neem leaves and their antimicrobial activity. J. Radiat. Res. Appl. Sci. 2016, 9, 109–115. [Google Scholar] [CrossRef]
- Prathna, T.C.; Chandrasekaran, N.; Raichur, A.M.; Mukherjee, A. Biomimetic synthesis of silver nanoparticles by Citrus limon (lemon) aqueous extract and theoretical prediction of particle size. Colloids Surf. B Biointerfaces 2011, 82, 152–159. [Google Scholar] [CrossRef] [PubMed]
- Miethling-Graff, R.; Rumpker, R.; Richter, M.; Verano-Braga, T.; Kjeldsen, F.; Brewer, J.; Hoyland, J.; Rubahn, H.-G.; Erdmann, H. Exposure to silver nanoparticles induces size- and dose-dependent oxidative stress and cytotoxicity in human colon carcinoma cells. Toxicol. In Vitro 2014, 28, 1280–1289. [Google Scholar] [CrossRef] [PubMed]
- Durai, P.; Chinnasamy, A.; Gajendran, B.; Ramar, M.; Pappu, S.; Kasivelu, G.; Thirunavukkarasu, A. Synthesis and characterization of silver nanoparticles using crystal compound of sodium para-hydroxybenzoate tetrahydrate isolated from Vitex negundo L. leaves and its apoptotic effect on human colon cancer cell lines. Eur. J. Med. Chem. 2014, 84, 90–99. [Google Scholar] [CrossRef] [PubMed]
- Satapathy, S.R.; Mohapatra, P.; Preet, R.; Das, D.; Sarkar, B.; Choudhuri, T.; Wyatt, M.D.; Kundu, C.N. Silver-based nanoparticles induce apoptosis in human colon cancer cells mediated through p53. Nanomedicine 2013, 8, 1307–1322. [Google Scholar] [CrossRef] [PubMed]
- Mata, R.; Nakkala, J.R.; Sadras, S.R. Biogenic silver nanoparticles from Abutilon indicum: Their antioxidant, antibacterial and cytotoxic effects in vitro. Colloids Surf. B Biointerfaces 2015, 128, 276–286. [Google Scholar] [CrossRef] [PubMed]
- Azizi, M.; Ghourchian, H.; Yazdian, F.; Bagherifam, S.; Bekhradnia, S.; Nyström, B. Anti-cancerous effect of albumin coated silver nanoparticles on MDA-MB 231 human breast cancer cell line. Sci. Rep. 2017, 7, 5178. [Google Scholar] [CrossRef] [PubMed]
- Yuan, Y.-G.; Wang, Y.-H.; Xing, H.-H.; Gurunathan, S. Quercetin-mediated synthesis of graphene oxide–silver nanoparticle nanocomposites: A suitable alternative nanotherapy for neuroblastoma. Int. J. Nanomed. 2017, 12, 5819–5839. [Google Scholar] [CrossRef] [PubMed]
- Burd, J.F.; Usategui-Gomez, M. A colorimetric assay for serum lactate dehydrogenase. Clin. Chim. Acta 1973, 46, 223–227. [Google Scholar] [CrossRef]
- Chan, F.K.-M.; Moriwaki, K.; De Rosa, M.J. Detection of necrosis by release of lactate dehydrogenase activity. Methods Mol. Biol. 2013, 979, 65–70. [Google Scholar] [CrossRef] [PubMed]
- Castiglioni, S.; Caspani, C.; Cazzaniga, A.; Maier, J.A. Short- and long-term effects of silver nanoparticles on human microvascular endothelial cells. World J. Biol. Chem. 2014, 5, 457–464. [Google Scholar] [CrossRef] [PubMed]
- Brayner, R. The toxicological impact of nanoparticles. Nano Today 2008, 3, 48–55. [Google Scholar] [CrossRef]
- Yuan, Y.-G.; Peng, Q.-L.; Gurunathan, S. Silver nanoparticles enhance the apoptotic potential of gemcitabine in human ovarian cancer cells: Combination therapy for effective cancer treatment. Int. J. Nanomed. 2017, 12, 6487–6502. [Google Scholar] [CrossRef] [PubMed]
- Han, J.W.; Gurunathan, S.; Choi, Y.-J.; Kim, J.-H. Dual functions of silver nanoparticles in F9 teratocarcinoma stem cells, a suitable model for evaluating cytotoxicity- and differentiation-mediated cancer therapy. Int. J. Nanomed. 2017, 12, 7529–7549. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Han, J.W.; Jeong, J.-K.; Gurunathan, S.; Choi, Y.-J.; Das, J.; Kwon, D.-N.; Cho, S.-G.; Park, C.; Seo, H.G.; Park, J.-K.; et al. Male- and female-derived somatic and germ cell-specific toxicity of silver nanoparticles in mouse. Nanotoxicology 2016, 10, 361–373. [Google Scholar] [CrossRef] [PubMed]
- Buttacavoli, M.; Albanese, N.N.; di Cara, G.; Alduina, R.; Faleri, C.; Gallo, M.; Pizzolanti, G.; Gallo, G.; Feo, S.; Baldi, F.; Cancemi, P. Anticancer activity of biogenerated silver nanoparticles: An integrated proteomic investigation. Oncotarget 2018, 9, 9685–9705. [Google Scholar] [CrossRef] [PubMed]
- Travan, A.; Pelillo, C.; Donati, I.; Marsich, E.; Benincasa, M.; Scarpa, T.; Semeraro, S.; Turco, G.; Gennaro, R.; Paoletti, S. Non-cytotoxic Silver Nanoparticle-Polysaccharide Nanocomposites with Antimicrobial Activity. Biomacromolecules 2009, 10, 1429–1435. [Google Scholar] [CrossRef] [PubMed]
- Pelicano, H.; Carney, D.; Huang, P. ROS stress in cancer cells and therapeutic implications. Drug Resist. Updat. 2004, 7, 97–110. [Google Scholar] [CrossRef] [PubMed]
- Gliga, A.R.; di Bucchianico, S.; Lindvall, J.; Fadeel, B.; Karlsson, H.L. RNA-sequencing reveals long-term effects of silver nanoparticles on human lung cells. Sci. Rep. 2018, 8, 6668. [Google Scholar] [CrossRef] [PubMed]
- AshaRani, P.V.; Low Kah Mun, G.; Hande, M.P.; Valiyaveettil, S. Cytotoxicity and Genotoxicity of Silver Nanoparticles in Human Cells. ACS Nano 2009, 3, 279–290. [Google Scholar] [CrossRef] [PubMed]
- Choi, Y.-J.; Park, J.-H.; Han, J.W.; Kim, E.; Jae-Wook, O.; Lee, S.Y.; Kim, J.-H.; Gurunathan, S. Differential Cytotoxic Potential of Silver Nanoparticles in Human Ovarian Cancer Cells and Ovarian Cancer Stem Cells. Int. J. Mol. Sci. 2016, 17, 2077. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.-F.; Shen, W.; Gurunathan, S. Silver Nanoparticle-Mediated Cellular Responses in Various Cell Lines: An in Vitro Model. Int. J. Mol. Sci. 2016, 17, 1603. [Google Scholar] [CrossRef] [PubMed]
- Orlowski, P.; Krzyzowska, M.; Zdanowski, R.; Winnicka, A.; Nowakowska, J.; Stankiewicz, W.; Tomaszewska, E.; Celichowski, G.; Grobelny, J. Assessment of in vitro cellular responses of monocytes and keratinocytes to tannic acid modified silver nanoparticles. Toxicol. In Vitro. 2013, 27, 1798–1808. [Google Scholar] [CrossRef] [PubMed]
- Maurer, L.L.; Meyer, J.N. A systematic review of evidence for silver nanoparticle-induced mitochondrial toxicity. Environ. Sci. Nano 2016, 3, 311–322. [Google Scholar] [CrossRef] [Green Version]
- Neustadt, J.; Pieczenik, S.R. Medication-induced mitochondrial damage and disease. Mol. Nutr. Food Res. 2008, 52, 780–788. [Google Scholar] [CrossRef] [PubMed]
- Navarro, E.; Piccapietra, F.; Wagner, B.; Marconi, F.; Kaegi, R.; Odzak, N.; Sigg, L.; Behra, R. Toxicity of silver nanoparticles to Chlamydomonas reinhardtii. Environ. Sci. Technol. 2008, 42, 8959–8964. [Google Scholar] [CrossRef] [PubMed]
- Schieber, M.; Chandel, N.S. ROS function in redox signaling and oxidative stress. Curr. Biol. 2014, 24, R453–R462. [Google Scholar] [CrossRef] [PubMed]
- Elmore, S. Apoptosis: A Review of Programmed Cell Death. Toxicol. Pathol. 2007, 35, 495–516. [Google Scholar] [CrossRef] [PubMed]
- Liu, F.; Mahmood, M.; Xu, Y.; Watanabe, F.; Biris, A.S.; Hansen, D.K.; Inselman, A.; Casciano, D.; Patterson, T.A.; Paule, M.G.; et al. Effects of silver nanoparticles on human and rat embryonic neural stem cells. Front. Neurosci. 2015, 9, 115. [Google Scholar] [CrossRef] [PubMed]
- Aşık, E.; Aslan, T.N.; Volkan, M.; Güray, N.T. 2-Amino-2-deoxy-glucose conjugated cobalt ferrite magnetic nanoparticle (2DG-MNP) as a targeting agent for breast cancer cells. Environ. Toxicol. Pharmacol. 2016, 41, 272–278. [Google Scholar] [CrossRef] [PubMed]
- Hsu, Y.-C.; Chen, M.-J.; Huang, T.-Y. Inducement of mitosis delay by cucurbitacin E, a novel tetracyclic triterpene from climbing stem of Cucumis melo L., through GADD45γ in human brain malignant glioma (GBM) 8401 cells. Cell Death Dis. 2014, 5, e1087. [Google Scholar] [CrossRef] [PubMed]
- Hsu, Y.-C.; Huang, T.-Y.; Chen, M.-J. Therapeutic ROS targeting of GADD45γ in the induction of G2/M arrest in primary human colorectal cancer cell lines by cucurbitacin E. Cell Death Dis. 2014, 5, e1198. [Google Scholar] [CrossRef] [PubMed]

© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Gurunathan, S.; Qasim, M.; Park, C.; Yoo, H.; Kim, J.-H.; Hong, K. Cytotoxic Potential and Molecular Pathway Analysis of Silver Nanoparticles in Human Colon Cancer Cells HCT116. Int. J. Mol. Sci. 2018, 19, 2269. https://doi.org/10.3390/ijms19082269
Gurunathan S, Qasim M, Park C, Yoo H, Kim J-H, Hong K. Cytotoxic Potential and Molecular Pathway Analysis of Silver Nanoparticles in Human Colon Cancer Cells HCT116. International Journal of Molecular Sciences. 2018; 19(8):2269. https://doi.org/10.3390/ijms19082269
Chicago/Turabian StyleGurunathan, Sangiliyandi, Muhammad Qasim, Chanhyeok Park, Hyunjin Yoo, Jin-Hoi Kim, and Kwonho Hong. 2018. "Cytotoxic Potential and Molecular Pathway Analysis of Silver Nanoparticles in Human Colon Cancer Cells HCT116" International Journal of Molecular Sciences 19, no. 8: 2269. https://doi.org/10.3390/ijms19082269
APA StyleGurunathan, S., Qasim, M., Park, C., Yoo, H., Kim, J.-H., & Hong, K. (2018). Cytotoxic Potential and Molecular Pathway Analysis of Silver Nanoparticles in Human Colon Cancer Cells HCT116. International Journal of Molecular Sciences, 19(8), 2269. https://doi.org/10.3390/ijms19082269




