Characteristics of Bacterial Communities under Different Tree Species and Their Response to Soil Physicochemical Properties
Abstract
:1. Introduction
2. Materials and Methods
2.1. Study Area Description
2.2. Plot Setup and Sample Collection
2.3. Determination of Soil Physicochemical Properties
2.4. Analysis of Soil Bacterial Community Diversity and Composition
2.5. Data Processing
3. Results
3.1. Soil Physicochemical Properties Characteristics
3.2. Bacterial Diversity
3.3. Bacterial Community Composition
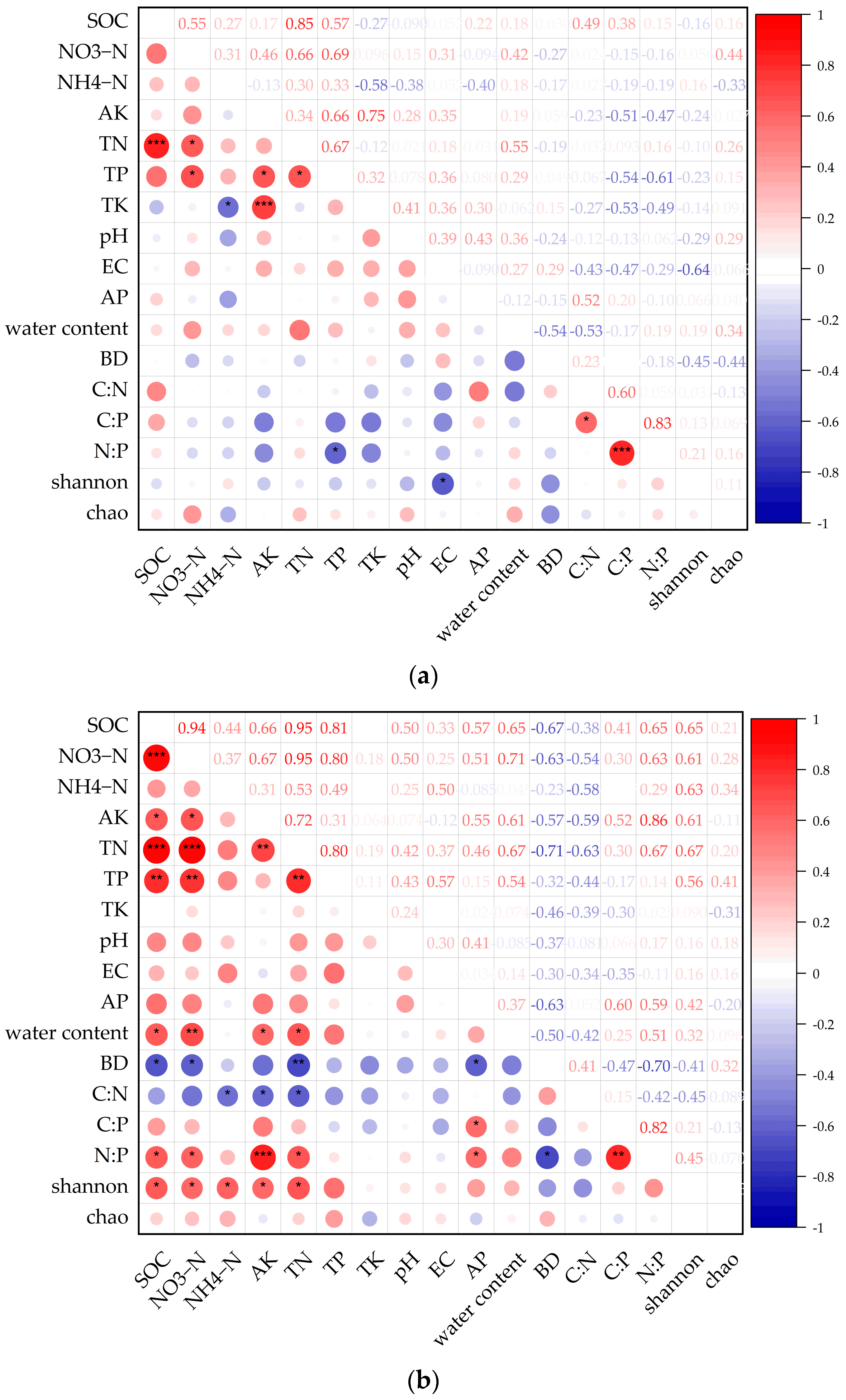
3.4. Relationship between Soil Physicochemical Properties and Bacterial Alpha Diversity
3.5. Soil Physicochemical Properties and Individual Bacterial Community Correlation Analysis
3.6. The Influence of Soil Physicochemical Properties on the Bacterial Community Structure
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Das, S.; Deb, S.; Sahoo, S.S.; Sahoo, U.K. Soil microbial biomass carbon stock and its relation with climatic and other environmental factors in forest ecosystems: A review. Acta Ecol. Sin. 2023, 43, 933–945. [Google Scholar] [CrossRef]
- Narayanan, M.; Ma, Y. Mitigation of heavy metal stress in the soil through optimized interaction between plants and microbes. J. Environ. Manag. 2023, 345, 118732. [Google Scholar] [CrossRef]
- Ji, H.; Wei, H.; Wang, R.; Zhang, J.; Liu, Z.; Abdellah, Y.A.Y.; Ren, X.; Shan, X.; Zhong, J.; He, Z. Heterogeneity and its drivers of microbial communities and diversity in six typical soils under two different land uses in tropical and subtropical southern China. Appl. Soil Ecol. 2022, 179, 104555. [Google Scholar] [CrossRef]
- Sokol, N.W.; Slessarev, E.; Marschmann, G.L.; Nicolas, A.; Blazewicz, S.J.; Brodie, E.L.; Firestone, M.K.; Foley, M.M.; Hestrin, R.; Hungate, B.A.; et al. Life and death in the soil microbiome: How ecological processes influence biogeochemistry. Nat. Rev. Microbiol. 2022, 20, 415–430. [Google Scholar] [CrossRef]
- Dunbar, J.; Barns, S.M.; Ticknor, L.O.; Kuske, C.R. Empirical and theoretical bacterial diversity in four Arizona soils. Appl. Environ. Microb. 2002, 68, 3035–3045. [Google Scholar] [CrossRef]
- Raza, T.; Qadir, M.F.; Khan, K.S.; Eash, N.S.; Yousuf, M.; Chatterjee, S.; Manzoor, R.; Rehman, S.U.; Oetting, J.N. Unrevealing the potential of microbes in decomposition of organic matter and release of carbon in the ecosystem. J. Environ. Manag. 2023, 344, 118529. [Google Scholar] [CrossRef]
- Uroz, S.; Calvaruso, C.; Turpault, M.P.; Pierrat, J.C.; Mustin, C.; Frey-Klett, P. Effect of the mycorrhizosphere on the genotypic and metabolic diversity of the bacterial communities involved in mineral weathering in a forest soil. Appl. Environ. Microb. 2007, 73, 3019–3027. [Google Scholar] [CrossRef] [PubMed]
- Lladó, S.; Lopez-Mondejar, R.; Baldrian, P. Forest Soil Bacteria: Diversity, Involvement in Ecosystem Processes, and Response to Global Change. Microbiol. Mol. Biol. Rev. 2017, 81, e00063-16. [Google Scholar] [CrossRef]
- Uroz, S.; Picard, L.; Turpault, M. Recent progress in understanding the ecology and molecular genetics of soil mineral weathering bacteria. Trends Microbiol. 2022, 30, 882–897. [Google Scholar] [CrossRef]
- Zhang, L.; Fan, J.; Feng, G.; Declerck, S. The arbuscular mycorrhizal fungus Rhizophagus irregularis MUCL 43194 induces the gene expression of citrate synthase in the tricarboxylic acid cycle of the phosphate-solubilizing bacterium Rahnella aquatilis HX2. Mycorrhiza 2019, 29, 69–75. [Google Scholar] [CrossRef]
- Xue, J.; Li, Z.; Feng, Q.; Li, Z.; Gui, J.; Li, Y. Ecological conservation pattern based on ecosystem services in the Qilian Mountains, northwest China. Environ. Dev. 2023, 46, 100834. [Google Scholar] [CrossRef]
- Jiang, S.; Chen, X.; Huang, R.; Wang, T.; Smettem, K. Effect of the altitudinal climate change on growing season length for deciduous broadleaved forest in southwest China. Sci. Total Environ. 2022, 828, 154306. [Google Scholar] [CrossRef] [PubMed]
- Lian, Z.; Wang, J.; Fan, C.; von Gadow, K. Structure complexity is the primary driver of functional diversity in the temperate forests of northeastern China. For. Ecosyst. 2022, 9, 100048. [Google Scholar] [CrossRef]
- He, R.; Hu, M.; Shi, H.; Zhou, Q.; Shu, X.; Zhang, K.; Zhang, Q.; Dang, H. Patterns of species diversity and its determinants in a temperate deciduous broad-leaved forest. For. Ecosyst. 2022, 9, 100062. [Google Scholar] [CrossRef]
- Gu, H.; Li, J.; Qi, G.; Wang, S. Species spatial distributions in a warm-temperate deciduous broad-leaved forest in China. J. For. Res. 2020, 31, 1187–1194. [Google Scholar] [CrossRef]
- Morreale, L.L.; Thompson, J.R.; Tang, X.; Reinmann, A.B.; Hutyra, L.R. Elevated growth and biomass along temperate forest edges. Nat. Commun. 2021, 12, 7181. [Google Scholar] [CrossRef] [PubMed]
- Cubino, J.P.; Lenoir, J.; Li, D.; Montano-Centellas, F.A.; Retana, J.; Baeten, L.; Bernhardt-Roemermann, M.; Chudomelova, M.; Closset, D.; Decocq, G.; et al. Evaluating plant lineage losses and gains in temperate forest understories: A phylogenetic perspective on climate change and nitrogen deposition. New Phytol. 2023, 241, 2287–2299. [Google Scholar] [CrossRef]
- Rillig, M.C.; Ryo, M.; Lehmann, A.; Aguilar-Trigueros, C.A.; Buchert, S.; Wulf, A.; Iwasaki, A.; Roy, J.; Yang, G. The role of multiple global change factors in driving soil functions and microbial biodiversity. Science 2019, 366, 886–890. [Google Scholar] [CrossRef] [PubMed]
- Keyimu, M.; Wei, J.; Zhang, Y.; Zhang, S.; Li, Z.; Ma, K.; Fu, B. Climate signal shift under the influence of prevailing climate warming–Evidence from Quercus liaotungensis on Dongling Mountain, Beijing, China. Dendrochronologia 2020, 60, 125683. [Google Scholar] [CrossRef]
- Zhang, Q.; Lyu, L.; Wang, Y. Patterns of daily stem growth in different tree species in a warm-temperate forest in northern China. Dendrochronologia 2022, 72, 125934. [Google Scholar] [CrossRef]
- Bao, S.D. Agricultural Chemical Analysis of Soil; Agriculture Press: Beijing, China, 2000. [Google Scholar]
- National Soil Survey Office. China Soil Survey Technology; Agricultural Press: Beijing, China, 1992. [Google Scholar]
- Li, T.; Liang, J.; Chen, X.; Wang, H.; Zhang, S.; Pu, Y.; Xu, X.; Li, H.; Xu, J.; Wu, X.; et al. The interacting roles and relative importance of climate, topography, soil properties and mineralogical composition on soil potassium variations at a national scale in China. Catena 2021, 196, 104875. [Google Scholar] [CrossRef]
- Chen, T.; Duan, L.; Cheng, S.; Jiang, S.; Yan, B. The preparation of paddy soil amendment using granite and marble waste: Performance and mechanisms. J. Environ. Sci. 2023, 127, 564–576. [Google Scholar] [CrossRef] [PubMed]
- Giweta, M. Role of litter production and its decomposition, and factors affecting the processes in a tropical forest ecosystem: A review. J. Ecol. Environ. 2020, 44, 11. [Google Scholar] [CrossRef]
- Ribeiro, I.D.A.; Volpiano, C.G.; Vargas, L.K.; Granada, C.E.; Lisboa, B.B.; Passaglia, L.M.P. Use of Mineral Weathering Bacteria to Enhance Nutrient Availability in Crops: A Review. Front. Plant Sci. 2020, 11, 590774. [Google Scholar] [CrossRef] [PubMed]
- Zhao, W.; Huang, L. Changes in soil nutrients and stoichiometric ratios reveal increasing phosphorus deficiency along a tropical soil chronosequence. Catena 2023, 222, 106893. [Google Scholar] [CrossRef]
- Li, M.; Dai, G.; Mu, L. Composition and diversity of soil bacterial communities under identical vegetation along an elevational gradient in Changbai Mountains, China. Front. Microbiol. 2022, 13, 1065412. [Google Scholar] [CrossRef] [PubMed]
- Ren, C.; Zhao, F.; Kang, D.; Yang, G.; Han, X.; Tong, X.; Feng, Y.; Ren, G. Linkages of C:N:P stoichiometry and bacterial community in soil following afforestation of former farmland. For. Ecol. Manag. 2016, 376, 59–66. [Google Scholar] [CrossRef]
- Cleveland, C.C.; Liptzin, D. C:N:P stoichiometry in soil: Is there a “Redfield ratio“ for the microbial biomass? Biogeochemistry 2007, 85, 235–252. [Google Scholar] [CrossRef]
- Tian, H.; Chen, G.; Zhang, C.; Melillo, J.M.; Hall, C.A.S. Pattern and variation of C:N:P ratios in China’s soils: A synthesis of observational data. Biogeochemistry 2010, 98, 139–151. [Google Scholar] [CrossRef]
- Tessier, J.T.; Raynal, D.J. Use of nitrogen to phosphorus ratios in plant tissue as an indicator of nutrient limitation and nitrogen saturation. J. Appl. Ecol. 2003, 40, 523–534. [Google Scholar] [CrossRef]
- Mundra, S.; Kauserud, H.; Okland, T.; Nordbakken, J.; Ransedokken, Y.; Kjonaas, O.J. Shift in tree species changes the belowground biota of boreal forests. New Phytol. 2022, 234, 2073–2087. [Google Scholar] [CrossRef] [PubMed]
- Gillespie, L.M.; Hattenschwiler, S.; Milcu, A.; Wambsganss, J.; Shihan, A.; Fromin, N. Tree species mixing affects soil microbial functioning indirectly via root and litter traits and soil parameters in European forests. Funct. Ecol. 2021, 35, 2190–2204. [Google Scholar] [CrossRef]
- Kerfahi, D.; Guo, Y.; Dong, K.; Wang, Q.; Adams, J.M. pH is the major predictor of soil microbial network complexity in Chinese forests along a latitudinal gradient. Catena 2024, 234, 107595. [Google Scholar] [CrossRef]
- Fu, X.; Cheng, Z.; Ni, H.; Zhang, R. Latitude variations of soil bacterial community diversity and composition in three typical forests of temperate, northeastern of China. Front. Earth Sc-Switz. 2023, 10, 1096931. [Google Scholar] [CrossRef]
- Landesman, W.J.; Freedman, Z.B.; Nelson, D.M. Seasonal, sub-seasonal and diurnal variation of soil bacterial community composition in a temperate deciduous forest. Fems Microbiol. Ecol. 2019, 95, fiz002. [Google Scholar] [CrossRef]
- Yang, Y.; Wu, P. Soil bacterial community varies but fungal community stabilizes along five vertical climate zones. Catena 2020, 195, 104841. [Google Scholar] [CrossRef]
- Zhang, Q.; Wang, X.; Zhang, Z.; Liu, H.; Liu, Y.; Feng, Y.; Yang, G.; Ren, C.; Han, X. Linking soil bacterial community assembly with the composition of organic carbon during forest succession. Soil Biol. Biochem. 2022, 173, 108790. [Google Scholar] [CrossRef]
- Brust, G.E. Management strategies for organic vegetable fertility. In Safety and Practice for Organic Food; Elsevier: Amsterdam, The Netherlands, 2019; pp. 193–212. [Google Scholar]
- Mitra, D.; Mondal, R.; Khoshru, B.; Senapati, A.; Radha, T.K.; Mahakur, B.; Uniyal, N.; Myo, E.M.; Boutaj, H.; Sierra, B.E.G.; et al. Actinobacteria-enhanced plant growth, nutrient acquisition, and crop protection: Advances in soil, plant, and microbial multifactorial interactions. Pedosphere 2022, 32, 149–170. [Google Scholar] [CrossRef]
- Liu, T.; Wu, X.; Li, H.; Alharbi, H.; Wang, J.; Dang, P.; Chen, X.; Kuzyakov, Y.; Yan, W. Soil organic matter, nitrogen and pH driven change in bacterial community following forest conversion. For. Ecol. Manag. 2020, 477, 118473. [Google Scholar] [CrossRef]
- Fan, W.; Wu, J. Short-term effects of returning granulated straw on soil microbial community and organic carbon fractions in dryland farming. J. Microbiol. 2020, 58, 657–667. [Google Scholar] [CrossRef]
- Ling, N.; Wang, T.; Kuzyakov, Y. Rhizosphere bacteriome structure and functions. Nat. Commun. 2022, 13, 836. [Google Scholar] [CrossRef] [PubMed]
- Rao, M.P.N.; Luo, Z.; Dong, Z.; Li, Q.; Liu, B.; Guo, S.; Nie, G.; Li, W.J. Metagenomic analysis further extends the role of Chloroflexi in fundamental biogeochemical cycles. Environ. Res. 2022, 209, 112888. [Google Scholar]
- Ho, A.; Di Lonardo, D.P.; Bodelier, P.L. Revisiting life strategy concepts in environmental microbial ecology. Fems. Microbiol. Ecol. 2017, 93, x6. [Google Scholar] [CrossRef] [PubMed]
- Kielak, A.M.; Barreto, C.C.; Kowalchuk, G.A.; van Veen, J.A.; Kuramae, E.E. The Ecology of Acidobacteria: Moving beyond Genes and Genomes. Front. Microbials. 2016, 7, 744. [Google Scholar] [CrossRef] [PubMed]
- Uroz, S.; Ioannidis, P.; Lengelle, J.; Cebron, A.; Morin, E.; Buee, M.; Martin, F. Functional assays and metagenomic analyses reveals differences between the microbial communities inhabiting the soil horizons of a Norway spruce plantation. PLoS ONE 2013, 8, e55929. [Google Scholar] [CrossRef] [PubMed]
- Hemkemeyer, M.; Schwalb, S.A.; Heinze, S.; Joergensen, R.G.; Wichern, F. Functions of elements in soil microorganisms. Microbiol. Res. 2021, 252, 126832. [Google Scholar] [CrossRef]
- Fallah, N.; Tayyab, M.; Yang, Z.; Pang, Z.; Zhang, C.; Lin, Z.; Stewart, L.J.; Ntambo, M.S.; Abubakar, A.Y.; Lin, W.; et al. Free-living bacteria stimulate sugarcane growth traits and edaphic factors along soil depth gradients under contrasting fertilization. Sci. Rep. 2023, 13, 6288. [Google Scholar] [CrossRef]

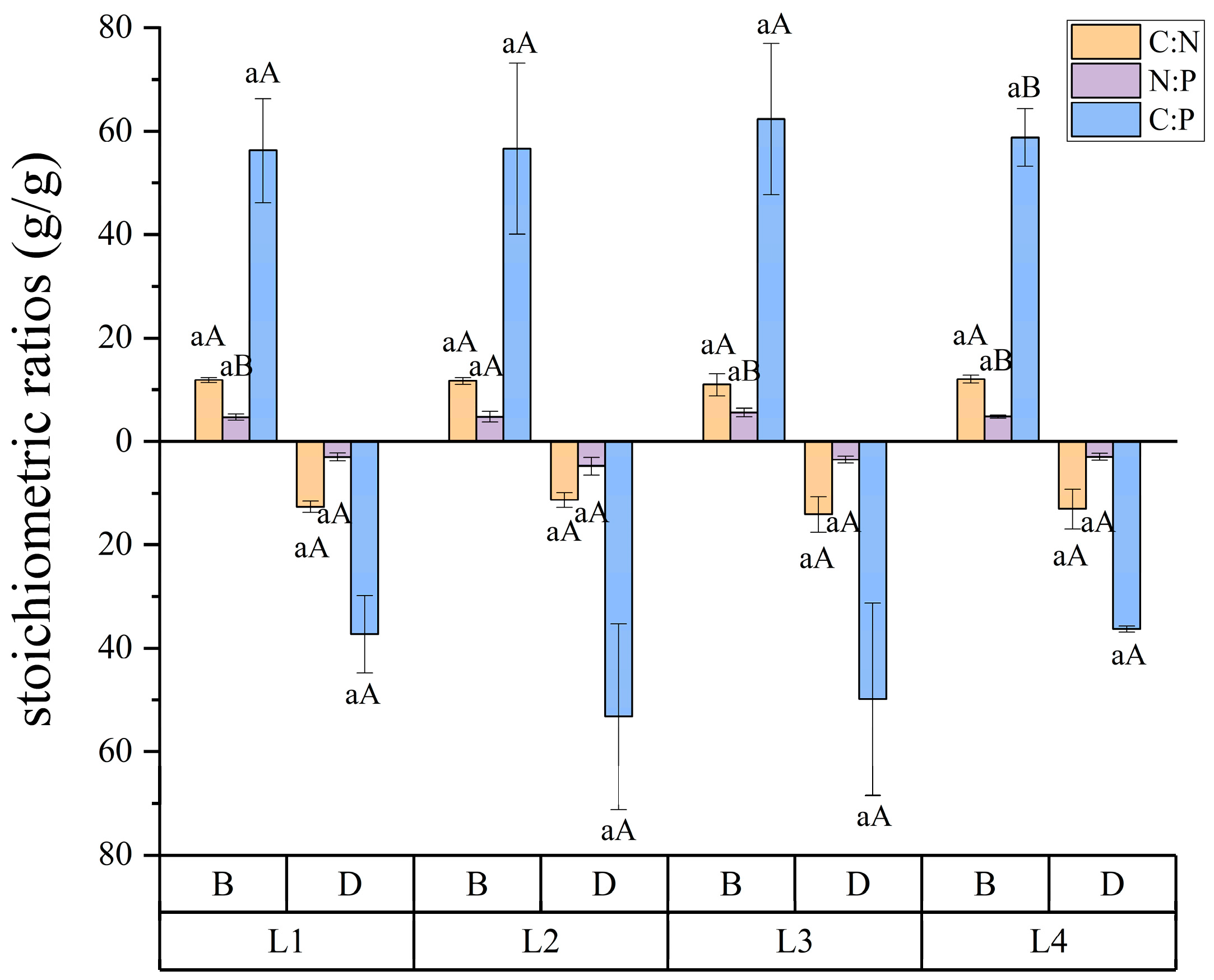
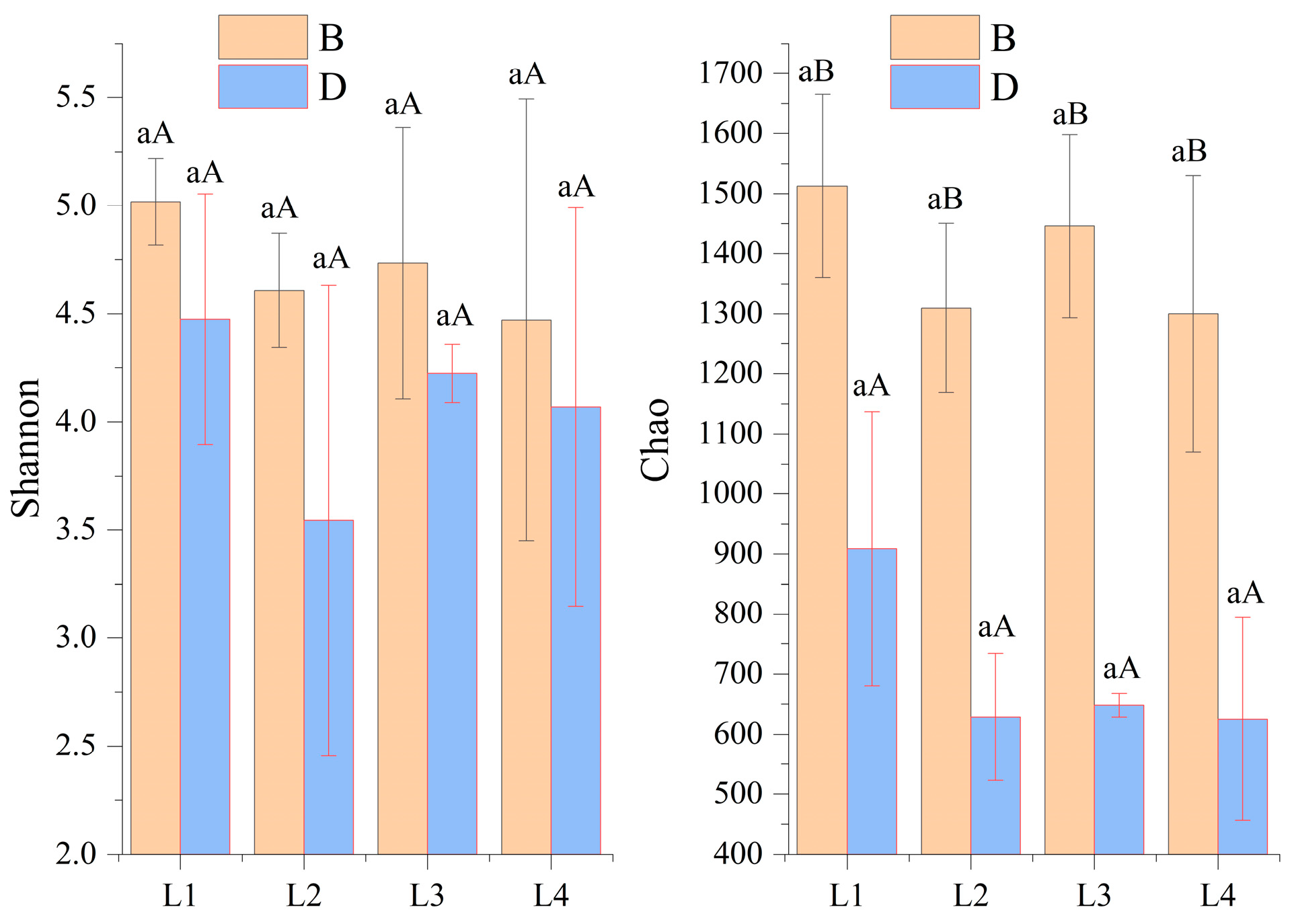
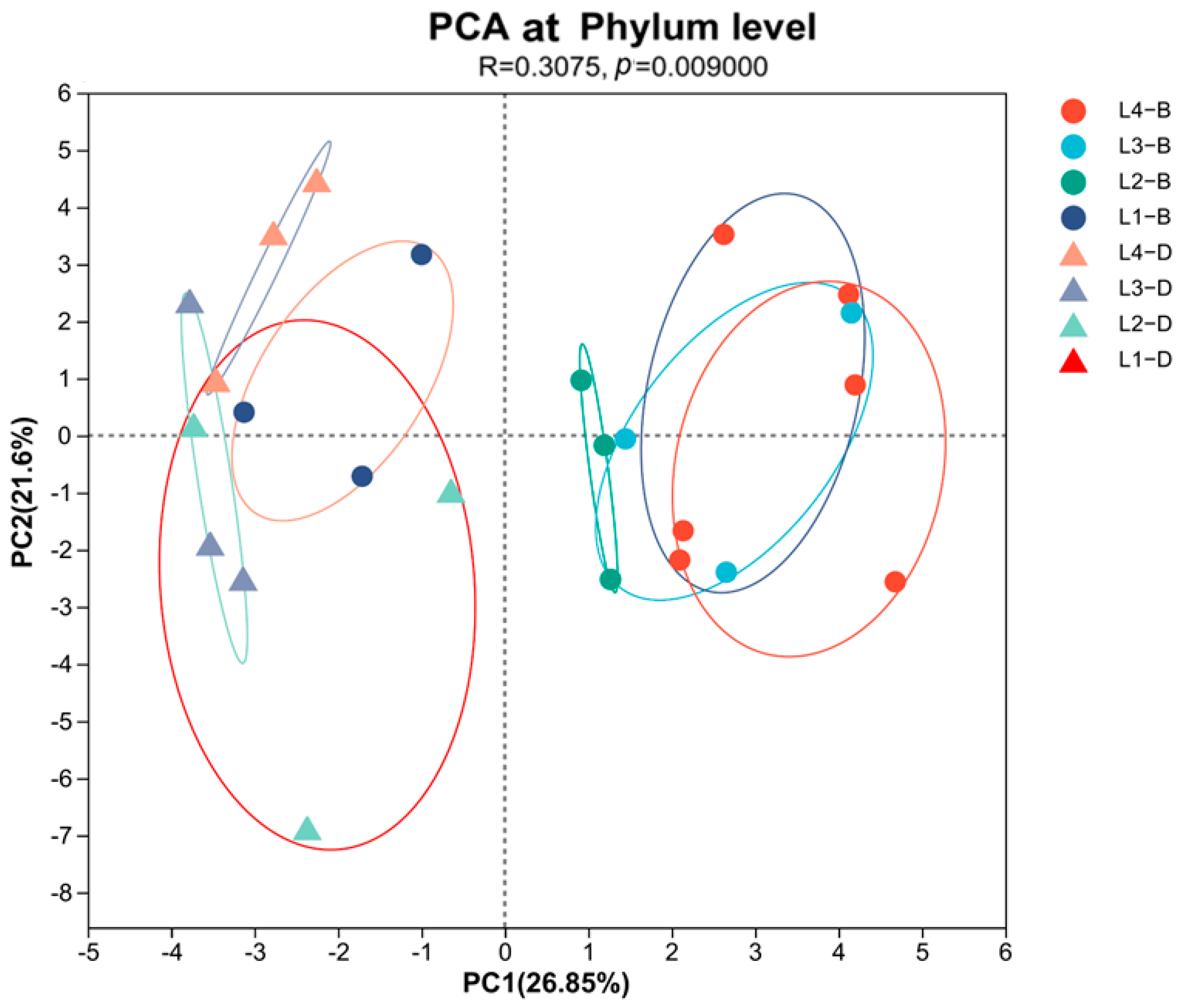
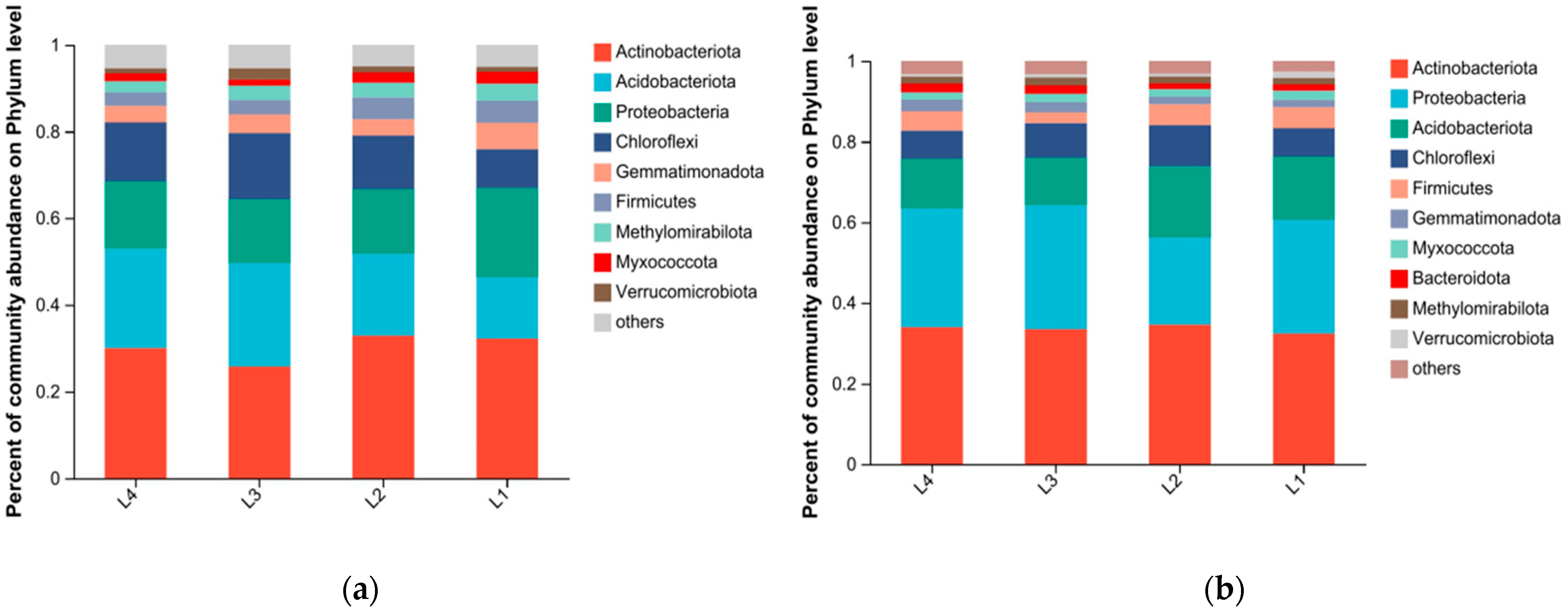


| Treatment | SOC (g/kg) | NO3−-N (mg/kg) | NH4+-N (mg/kg) | AK (mg/kg) | AP (mg/kg) | TN (g/kg) | TP (g/kg) | TK (g/kg) |
|---|---|---|---|---|---|---|---|---|
| L1-B | 30.12 ± 4.6 aB | 12.80 ± 3.6 aA | 11.42 ± 2.55 aA | 188.00 ± 51.88 aB | 17.48 ± 1.14 aA | 2.54 ± 0.4 aB | 0.55 ± 0.13 aA | 20.07 ± 1.1 bA |
| L2-B | 37.17 ± 0.6 abA | 18.96 ± 0.44 bB | 11.89 ± 2.80 aA | 221.33 ± 90.16 aA | 17.57 ± 1.58 aA | 3.18 ± 0.15 abA | 0.69 ± 0.17 aA | 19.93 ± 1.86 abA |
| L3-B | 32.47 ± 7.91 abB | 13.11 ± 2.33 aA | 13.49 ± 3.06 aA | 134.67 ± 36.50 aA | 17.92 ± 4.09 aA | 2.96 ± 0.58 abB | 0.52 ± 0.03 aA | 18.87 ± 0.12 abA |
| L4-B | 41.03 ± 5.21 bB | 16.96 ± 3.76 abA | 24.67 ± 7.12 bB | 162.67 ± 9.50 aA | 15.82 ± 1.97 aA | 3.39 ± 0.22 bB | 0.70 ± 0.04 aA | 17.87 ± 0.31 aA |
| L1-D | 13.11 ± 7.84 aA | 11.21 ± 3.77 aA | 9.44 ± 6.37 aA | 76.00 ± 28.48 aA | 13.72 ± 0.30 aB | 1.07 ± 0.72 aA | 0.34 ± 0.15 aA | 20.40 ± 1.22 bA |
| L2-D | 34.90 ± 5.68 cA | 29.93 ± 6.28 bA | 10.71 ± 1.87 aA | 126.00 ± 53.67 aA | 16.43 ± 1.39 bA | 3.16 ± 0.89 bA | 0.69 ± 0.21 bA | 19.87 ± 1.55 abA |
| L3-D | 17.10 ± 2.06 abA | 10.71 ± 2.17 aA | 9.68 ± 2.25 aA | 81.00 ± 13.53 aA | 14.25 ± 1.71 aA | 1.27 ± 0.36 aA | 0.37 ± 0.11 aA | 18.07 ± 0.92 aA |
| L4-D | 23.94 ± 0.87 bA | 19.32 ± 4.47 aA | 9.77 ± 4.49 aA | 88.00 ± 35.79 aA | 14.07 ± 0.26 aA | 1.94 ± 0.53 abA | 0.66 ± 0.03 bA | 18.13 ± 0.12 aA |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Chen, Z.; Li, S.; Sun, X.; He, L.; Zhou, W.; Zhao, G.; Yu, J.; Bai, X.; Zhang, J. Characteristics of Bacterial Communities under Different Tree Species and Their Response to Soil Physicochemical Properties. Forests 2024, 15, 740. https://doi.org/10.3390/f15050740
Chen Z, Li S, Sun X, He L, Zhou W, Zhao G, Yu J, Bai X, Zhang J. Characteristics of Bacterial Communities under Different Tree Species and Their Response to Soil Physicochemical Properties. Forests. 2024; 15(5):740. https://doi.org/10.3390/f15050740
Chicago/Turabian StyleChen, Zhe, Suyan Li, Xiangyang Sun, Libing He, Wenzhi Zhou, Guanyu Zhao, Jiantao Yu, Xueting Bai, and Jinshuo Zhang. 2024. "Characteristics of Bacterial Communities under Different Tree Species and Their Response to Soil Physicochemical Properties" Forests 15, no. 5: 740. https://doi.org/10.3390/f15050740




