Catalytic Activity Prediction of α-Diimino Nickel Precatalysts toward Ethylene Polymerization by Machine Learning
Abstract
:1. Introduction
2. Results and Discussions
Interpretation of XGBoost Model
3. Computational Methods
3.1. Geometry Optimization
3.2. Descriptor Calculation and Selection
3.3. Machine Learning Modelling
3.4. Model Interpretation
3.4.1. Permutation Feature Importance (PFI)
3.4.2. SHapley Additive exPlanations (SHAP)
3.4.3. Individual Conditional Expectations Plots (ICE)
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Ronca, S. Polyethylene. In Brydson’s Plastics Materials, 8th ed.; Gilbert, M., Ed.; Butterworth-Heinemann: Oxford, UK, 2017; pp. 247–278. [Google Scholar]
- Johnson, L.K.; Killian, M.C.; Brookhart, M. New Pd (II)-and Ni (II)-based catalysts for polymerization of ethylene and. alpha.-olefins. J. Am. Chem. Soc. 1995, 117, 6414–6415. [Google Scholar] [CrossRef]
- Wang, Z.; Liu, Q.; Solan, G.A.; Sun, W.-H. Recent advances in Ni-mediated ethylene chain growth: Nimine-donor ligand effects on catalytic activity, thermal stability and oligo-/polymer structure. Coord. Chem. Rev. 2017, 350, 68–83. [Google Scholar] [CrossRef]
- Soshnikov, I.E.; Bryliakov, K.P.; Antonov, A.A.; Sun, W.-H.; Talsi, E.P. Ethylene polymerization of nickel catalysts with α-diimine ligands: Factors controlling the structure of active species and polymer properties. Dalton Trans. 2019, 48, 7974–7984. [Google Scholar] [CrossRef] [PubMed]
- Qasim, M.; Bashir, M.S.; Iqbal, S.; Mahmood, Q. Recent advancements in α-diimine-nickel and-palladium catalysts for ethylene polymerization. Eur. Polym. J. 2021, 160, 110783. [Google Scholar] [CrossRef]
- Wu, R.; Wang, Y.; Guo, L.; Guo, C.Y.; Liang, T.; Sun, W.-H. Highly branched and high-molecular-weight polyethylenes produced by 1-[2, 6-bis (bis (4-fluorophenyl) methyl)-4-MeOC6H2N]-2-aryliminoacenaphthylnickel (II) halides. J. Polym. Sci. A Polym. Chem. 2019, 57, 130–145. [Google Scholar] [CrossRef]
- Zada, M.; Guo, L.; Zhang, R.; Zhang, W.; Ma, Y.; Solan, G.A.; Sun, Y.; Sun, W.-H. Moderately branched ultra-high molecular weight polyethylene by using N,N′-nickel catalysts adorned with sterically hindered dibenzocycloheptyl groups. Appl. Organomet. Chem. 2019, 33, e4749. [Google Scholar] [CrossRef]
- Yuan, S.; Duan, T.; Zhang, R.; Solan, G.A.; Ma, Y.; Liang, T.; Sun, W.-H. Alkylaluminum activator effects on polyethylene branching using a N,N′-nickel precatalyst appended with bulky 4,4′-dimethoxybenzhydryl groups. Appl. Organomet. Chem. 2019, 33, e4785. [Google Scholar] [CrossRef]
- Zhang, R.; Wang, Z.; Ma, Y.; Solan, G.A.; Sun, Y.; Sun, W.-H. Plastomeric-like polyethylenes achievable using thermally robust N,N′-nickel catalysts appended with electron withdrawing difluorobenzhydryl and nitro groups. Dalton Trans. 2019, 48, 1878–1891. [Google Scholar] [CrossRef]
- Zhang, Q.; Zhang, R.; Ma, Y.; Solan, G.A.; Liang, T.; Sun, W.-H. Branched polyethylenes attainable using thermally enhanced bis (imino) acenaphthene-nickel catalysts: Exploring the effects of temperature and pressure. Appl. Catal. A-Gen. 2019, 573, 73–86. [Google Scholar] [CrossRef]
- Guan, B.; Jiang, H.; Wei, Y.; Liu, Z.; Wu, X.; Lin, H.; Huang, Z. Density functional theory researches for atomic structure, properties prediction, and rational design of selective catalytic reduction catalysts: Current progresses and future perspectives. Mol. Catal. 2021, 510, 111704. [Google Scholar] [CrossRef]
- Kitchin, J.R. Machine learning in catalysis. Nat. Catal. 2018, 1, 230–232. [Google Scholar] [CrossRef]
- Yang, W.; Fidelis, T.T.; Sun, W.-H. Machine learning in catalysis, from proposal to practicing. ACS Omega 2019, 5, 83–88. [Google Scholar] [CrossRef] [PubMed]
- Yang, W.; Sun, W.-H. Application and prospect of machine learning in polyolefin catalysts. Chin. Sci. Bull. 2022, 67, 1870–1880. [Google Scholar] [CrossRef]
- Hayatzadeh, A.; Fattahi, M.; Rezaveisi, A. Machine learning algorithms for operating parameters predictions in proton exchange membrane water electrolyzers: Anode side catalyst. Int. J. Hydrogen Energy 2024, 56, 302–314. [Google Scholar] [CrossRef]
- Yang, W.; Ma, Z.; Sun, W.-H. Modeling study on the catalytic activities of 2-imino-1, 10-phenanthrolinylmetal (Fe, Co, and Ni) precatalysts in ethylene oligomerization. RSC Adv. 2016, 6, 79335–79342. [Google Scholar] [CrossRef]
- Malik, A.A.; Yang, W.; Ma, Z.; Sun, W.-H. The Catalytic Activities of Carbocyclic Fused Pyridineimine Nickel Complexes Analogues in Ethylene Polymerization by Modeling Study. Catalysts 2019, 9, 520. [Google Scholar] [CrossRef]
- Yang, W.; Meraz, M.; Fidelis, T.T.; Sun, W.-H. The Quantitative Influence of Coordinated Halogen Atoms on the Catalytic Performance of Bisiminoacenaphthylnickel Complexes in Ethylene Polymerization. ChemPhysChem 2021, 22, 585–592. [Google Scholar] [CrossRef] [PubMed]
- Yang, W.; Yi, J.; Ma, Z.; Sun, W.-H. 2D-QSAR modeling on the catalytic activities of 2-azacyclyl-6-aryliminopyridylmetal precatalysts in ethylene oligomerization. Catal. Commun. 2017, 101, 40–43. [Google Scholar] [CrossRef]
- Meraz, M.M.; Malik, A.A.; Yang, W.; Sun, W.-H. Catalytic Performance of Cycloalkyl-Fused Aryliminopyridyl Nickel Complexes toward Ethylene Polymerization by QSPR Modeling. Catalysts 2021, 11, 920. [Google Scholar] [CrossRef]
- Meraz, M.M.; Yang, W.; Yang, W.; Sun, W.-H. Predicting the catalytic activities of transition metal (Cr, Fe, Co, Ni) complexes towards ethylene polymerization by machine learning. J. Comput. Chem. 2023, 1. [Google Scholar] [CrossRef]
- Yang, W.; Fidelis, T.T.; Sun, W.-H. Prediction of catalytic activities of bis (imino) pyridine metal complexes by machine learning. J. Comput. Chem. 2020, 41, 1064–1067. [Google Scholar] [CrossRef] [PubMed]
- Wu, R.; Wang, Y.; Zhang, R.; Guo, C.Y.; Flisak, Z.; Sun, Y.; Sun, W.-H. Finely tuned nickel complexes as highly active catalysts affording branched polyethylene of high molecular weight: 1-(2,6-Dibenzhydryl-4-methoxyphenylimino)-2-(arylimino) acenaphthylenenickel halides. Polymer 2018, 153, 574–586. [Google Scholar] [CrossRef]
- Mahmood, Q.; Zeng, Y.; Wang, X.; Sun, Y.; Sun, W.-H. Advancing polyethylene properties by incorporating NO2 moiety in 1, 2-bis (arylimino) acenaphthylnickel precatalysts: Synthesis, characterization and ethylene polymerization. Dalton Trans. 2017, 46, 6934–6947. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.; Du, S.; Huang, C.; Solan, G.A.; Hao, X.; Sun, W.-H. Balancing high thermal stability with high activity in diaryliminoacenaphthene-nickel (II) catalysts for ethylene polymerization. J. Polym. Sci. A Polym. Chem. 2017, 55, 1971–1983. [Google Scholar] [CrossRef]
- Wang, X.; Fan, L.; Ma, Y.; Guo, C.Y.; Solan, G.A.; Sun, Y.; Sun, W.-H. Elastomeric polyethylenes accessible via ethylene homo-polymerization using an unsymmetrical α-diimino-nickel catalyst. Polym. Chem. 2017, 8, 2785–2795. [Google Scholar] [CrossRef]
- Wang, X.; Fan, L.; Yuan, Y.; Du, S.; Sun, Y.; Solan, G.A.; Guo, C.Y.; Sun, W.-H. Raising the N-aryl fluoride content in unsymmetrical diaryliminoacenaphthylenes as a route to highly active nickel (II) catalysts in ethylene polymerization. Dalton Trans. 2016, 45, 18313–18323. [Google Scholar] [CrossRef] [PubMed]
- Du, S.; Xing, Q.; Flisak, Z.; Yue, E.; Sun, Y.; Sun, W.-H. Ethylene polymerization by the thermally unique 1-[2-(bis (4-fluoro phenyl) methyl)-4, 6-dimethylphenylimino]-2-aryliminoacenaphthylnickel precursors. Dalton Trans. 2015, 44, 12282–12291. [Google Scholar] [CrossRef]
- Fan, L.; Du, S.; Guo, C.Y.; Hao, X.; Sun, W.-H. 1-(2, 6-dibenzhydryl-4-fluorophenylimino)-2-aryliminoacenaphthylylnickel halides highly polymerizing ethylene for the polyethylenes with high branches and molecular weights. J. Polym. Sci. A Polym. Chem. 2015, 53, 1369–1378. [Google Scholar] [CrossRef]
- Du, S.; Kong, S.; Shi, Q.; Mao, J.; Guo, C.; Yi, J.; Liang, T.; Sun, W.-H. Enhancing the activity and thermal stability of nickel complex precatalysts using 1-[2, 6-bis (bis (4-fluorophenyl) methyl)-4-methyl phenylimino]-2-aryliminoacenaphthylene derivatives. Organometallics 2015, 34, 582–590. [Google Scholar] [CrossRef]
- Fan, L.; Yue, E.; Du, S.; Guo, C.Y.; Hao, X.; Sun, W.-H. Enhancing thermo-stability to ethylene polymerization: Synthesis, characterization and the catalytic behavior of 1-(2,4-dibenzhydryl-6-chlorophenylimino)-2-aryliminoacenaphthylnickel halides. RSC Adv. 2015, 5, 93274–93282. [Google Scholar] [CrossRef]
- Yuan, S.F.; Fan, Z.; Zhang, Q.; Flisak, Z.; Ma, Y.; Sun, Y.; Sun, W.-H. Enhancing performance of α-diiminonickel precatalyst for ethylene polymerization by substitution with the 2,4-bis (4,4’-dimethoxybenzhydryl)-6-methylphenyl group. Appl. Organomet. Chem. 2020, 34, e5638. [Google Scholar] [CrossRef]
- Liu, H.; Zhao, W.; Yu, J.; Yang, W.; Hao, X.; Redshaw, C.; Chen, L.; Sun, W.-H. Synthesis, characterization and ethylene polymerization behavior of nickel dihalide complexes bearing bulky unsymmetrical α-diimine ligands. Catal. Sci. Technol. 2012, 2, 415–422. [Google Scholar] [CrossRef]
- Mahmood, Q.; Zeng, Y.; Yue, E.; Solan, G.A.; Liang, T.; Sun, W.-H. Ultra-high molecular weight elastomeric polyethylene using an electronically and sterically enhanced nickel catalyst. Polym. Chem. 2017, 8, 6416–6430. [Google Scholar] [CrossRef]
- Pedregosa, F.; Varoquaux, G.; Gramfort, A.; Michel, V.; Thirion, B.; Grisel, O.; Blondel, M.; Prettenhofer, P.; Weiss, R.; Dubourg, V.; et al. Scikit-learn: Machine learning in Python. J. Mach. Learn. Res. 2011, 12, 2825–2830. [Google Scholar]
- Yap, C.W. PaDEL-descriptor: An open source software to calculate molecular descriptors and fingerprints. J. Comput. Chem. 2011, 32, 1466–1474. [Google Scholar] [CrossRef] [PubMed]
- Yang, W.; Ma, Z.; Yi, J.; Ahmed, S.; Sun, W.-H. Catalytic performance of bis (imino) pyridine Fe/Co complexes toward ethylene polymerization by 2D-/3D-QSPR modeling. J. Comput. Chem. 2019, 40, 1374–1386. [Google Scholar] [CrossRef] [PubMed]
- Mayo, S.L.; Olafson, B.D.; Goddard, W.A. DREIDING: A generic force field for molecular simulations. J. Phys. Chem. 1990, 94, 8897–8909. [Google Scholar] [CrossRef]
- Meunier, M.; Robertson, S. Materials Studio 20th anniversary. Mol. Simul. 2021, 47, 537–539. [Google Scholar] [CrossRef]
- Frisch, M.J.; Trucks, G.W.; Schlegel, H.B.; Scuseria, G.E.; Robb, M.A.; Cheeseman, J.R.; Scalmani, G.; Barone, V.; Mennucci, B.; Petersson, G.A.; et al. Gaussian 09; Revision C.01; Gaussian, Inc.: Wallingford, CT, USA, 2016. [Google Scholar]
- Katritzky, A.R.; Lobanov, V.S. Comprehensive Descriptors for Structural and Statistical Analysis (Codessa); Semichem, Inc.: Shawnee Mission, KS, USA, 2004. [Google Scholar]
- Chen, T.; Guestrin, C. XGBoost: A Scalable Tree Boosting System. In Proceedings of the 22nd ACM SIGKDD International Conference on Knowledge Discovery and Data Mining, San Francisco, CA, USA, 13–17 August 2016. [Google Scholar]
- Prokhorenkova, L.; Gusev, G.; Vorobev, A.; Dorogush, A.V.; Gulin, A. CatBoost: Unbiased boosting with categorical features. Adv. Neural. Inf. Process. Syst. 2018, 31. [Google Scholar] [CrossRef]
- Ke, G.; Meng, Q.; Finley, T.; Wang, T.; Chen, W.; Ma, W.; Ye, Q.; Liu, T.Y. Lightgbm: A highly efficient gradient boosting decision tree. Adv. Neural. Inf. Process. Syst. 2017, 30, 3149–3157. [Google Scholar]
- Geurts, P.; Ernst, D.; Wehenkel, L. Extremely randomized trees. Mach. Learn. 2006, 63, 3–42. [Google Scholar] [CrossRef]
- Breiman, L. Random forests. Mach. Learn. 2001, 45, 5–32. [Google Scholar] [CrossRef]
- Larose, D.T.; Larose, C.D. Discovering Knowledge in Data: An Introduction to Data Mining, 2nd ed.John Wiley & Sons: Hoboken, NJ, USA, 2014; pp. 149–164. [Google Scholar]
- Hasebrook, N.; Morsbach, F.; Kannengießer, N.; Franke, J.; Hutter, F.; Sunyaev, A. Practitioner Motives to Select Hyperparameter Optimization Methods. arXiv 2022, arXiv:2203.01717. [Google Scholar]
- Altmann, A.; Toloşi, L.; Sander, O.; Lengauer, T. Permutation importance: A corrected feature importance measure. Bioinformatics 2010, 26, 1340–1347. [Google Scholar] [CrossRef] [PubMed]
- Shapley, L.S. A Value for n-Person Games. In Contributions to the Theory of Games (AM-28), 1st ed.; Tucker, A.W., Kuhn, H.W., Eds.; Princeton University Press: Princeton, NJ, USA, 1953; pp. 307–318. [Google Scholar]
- Lundberg, S.M.; Lee, S.I. A Unified Approach to Interpreting Model Predictions. In Proceedings of the 31st Conference on Neural Information Processing Systems (NIPS 2017), Long Beach, CA, USA, 4–9 December 2017. [Google Scholar]
- Goldstein, A.; Kapelner, A.; Bleich, J.; Pitkin, E. Peeking inside the black box: Visualizing statistical learning with plots of individual conditional expectation. J. Comput. Graph. Stat. 2015, 24, 44–65. [Google Scholar] [CrossRef]





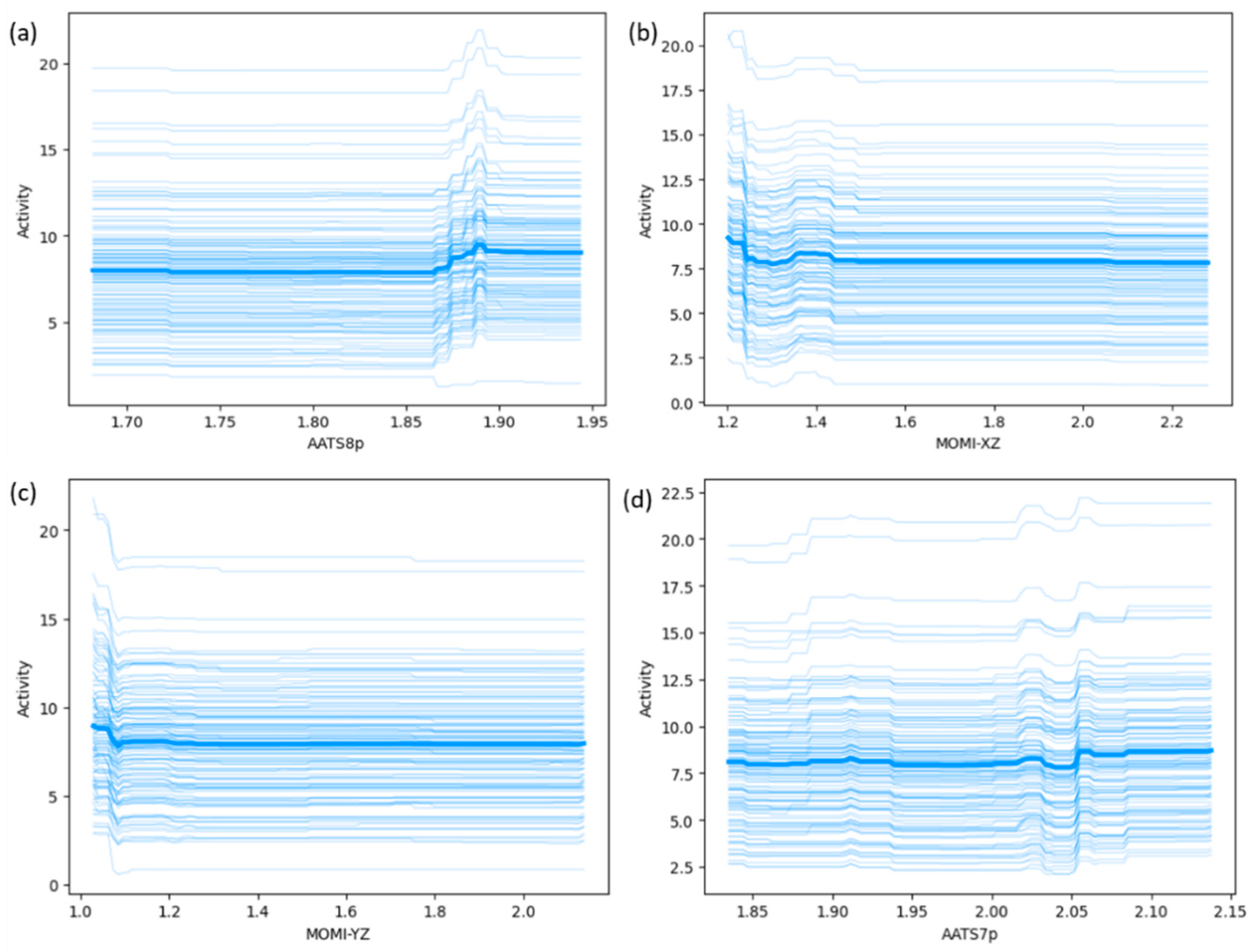
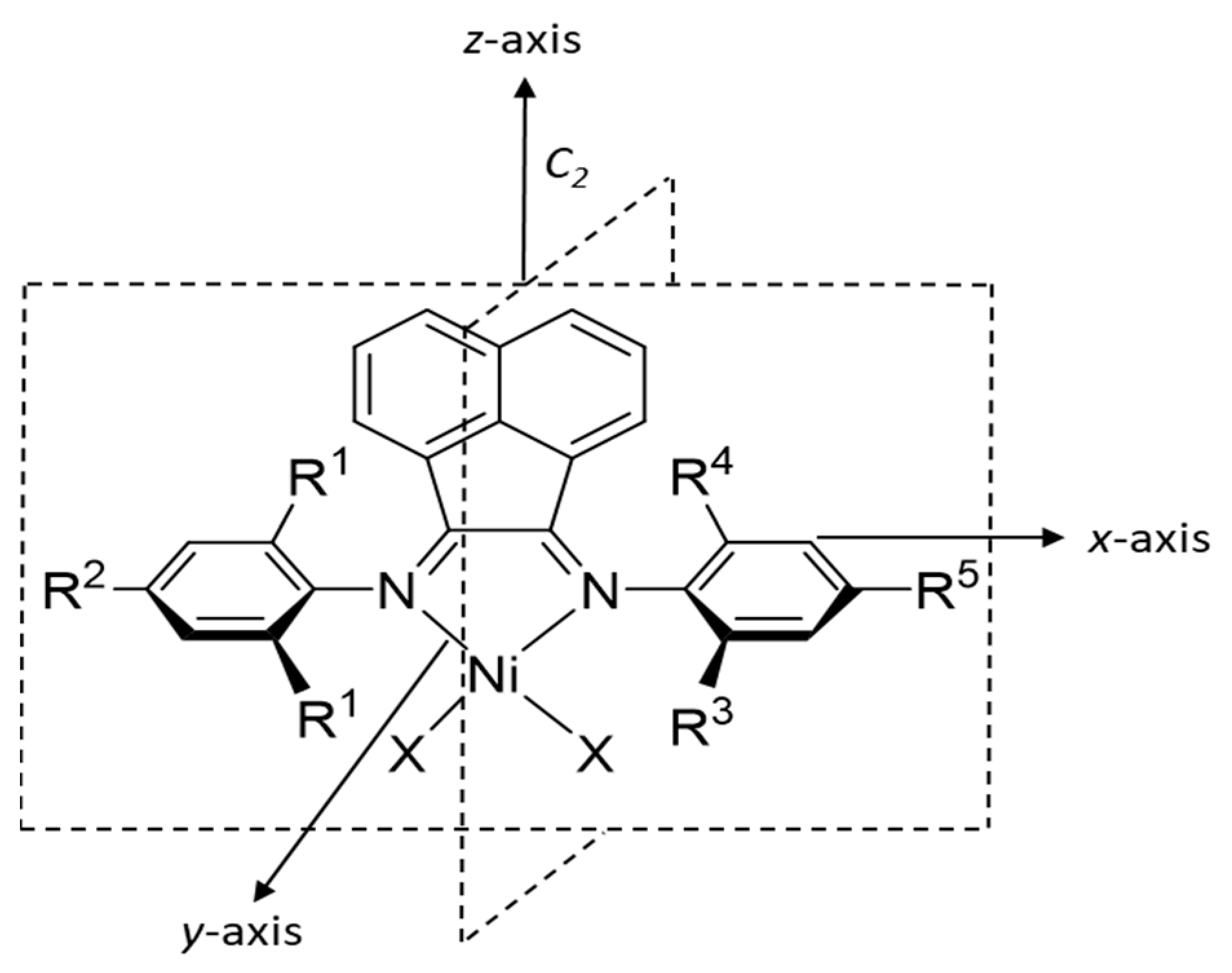
| Algorithm | Train R2 | Test Rt2 | Train MAE | Test MAEt | Train RMSE | Test RMSEt | Q2 (n_split = 4) |
|---|---|---|---|---|---|---|---|
| XGBoost | 0.999 | 0.921 | 0.094 | 0.697 | 0.120 | 0.859 | 0.561 |
| CatBoost | 0.991 | 0.896 | 0.285 | 0.714 | 0.346 | 0.978 | 0.537 |
| Extra Trees | 1.00 | 0.841 | 0.00 | 0.914 | 0.00 | 1.285 | 0.542 |
| Random Forest | 0.910 | 0.824 | 0.798 | 0.937 | 1.091 | 1.272 | 0.560 |
| k-Nearest Neighbors | 0.999 | 0.801 | 0.00 | 0.982 | 0.00 | 1.355 | 0.474 |
| LightGBM | 0.923 | 0.761 | 0.729 | 1.284 | 0.986 | 1.705 | 0.437 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Abbas, Z.; Meraz, M.M.; Yang, W.; Yang, W.; Sun, W.-H. Catalytic Activity Prediction of α-Diimino Nickel Precatalysts toward Ethylene Polymerization by Machine Learning. Catalysts 2024, 14, 195. https://doi.org/10.3390/catal14030195
Abbas Z, Meraz MM, Yang W, Yang W, Sun W-H. Catalytic Activity Prediction of α-Diimino Nickel Precatalysts toward Ethylene Polymerization by Machine Learning. Catalysts. 2024; 14(3):195. https://doi.org/10.3390/catal14030195
Chicago/Turabian StyleAbbas, Zaheer, Md Mostakim Meraz, Wenhong Yang, Weisheng Yang, and Wen-Hua Sun. 2024. "Catalytic Activity Prediction of α-Diimino Nickel Precatalysts toward Ethylene Polymerization by Machine Learning" Catalysts 14, no. 3: 195. https://doi.org/10.3390/catal14030195






