Crystal Structure of the 23S rRNA Fragment Specific to r-Protein L1 and Designed Model of the Ribosomal L1 Stalk from Haloarcula marismortui
Abstract
:1. Introduction
2. Results and Discussion
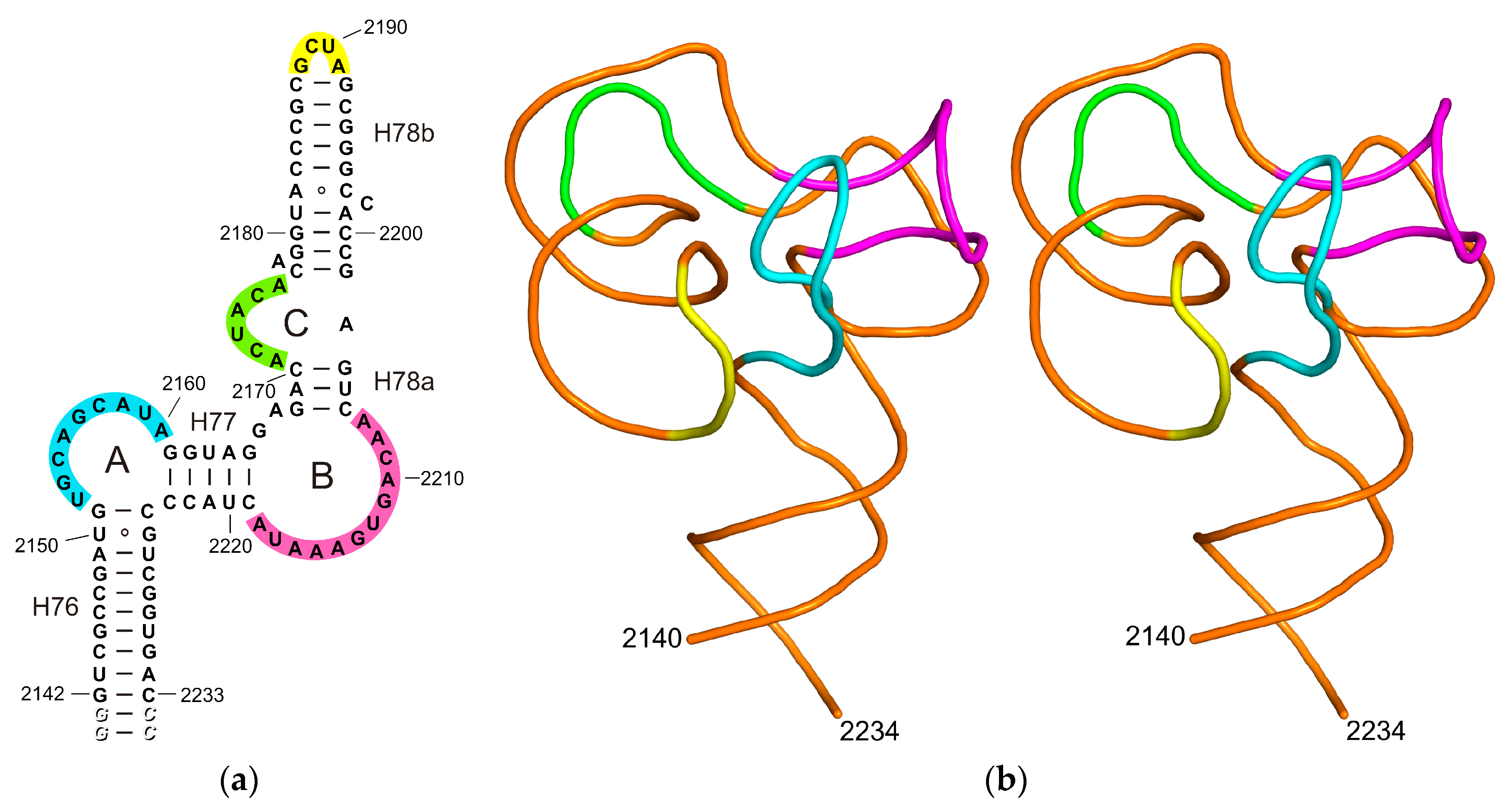
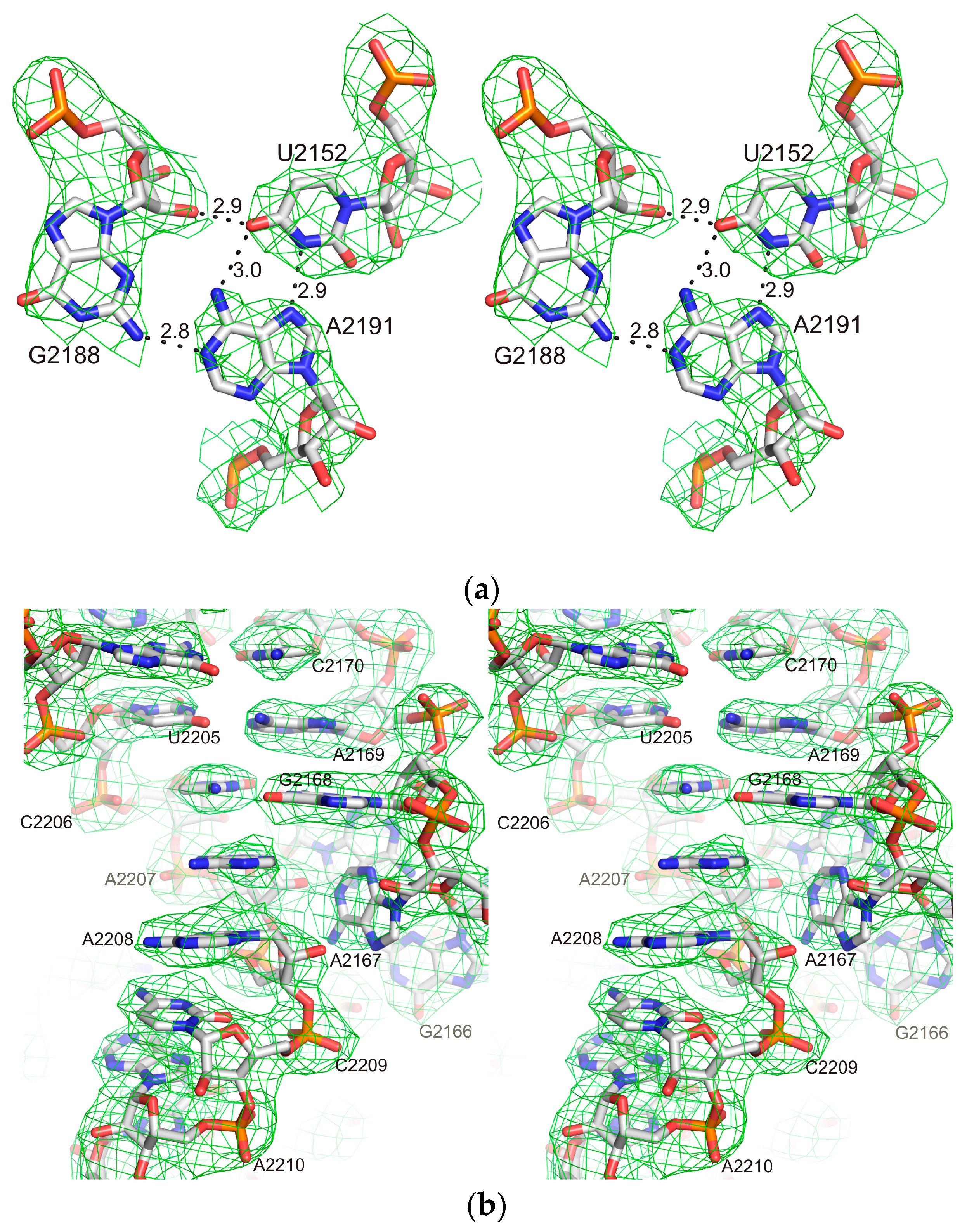
2.1. Structure of the L1-Specific rRNA Fragment from H. marismortui
2.2. The Designed Model of the HmaL1 Stalk
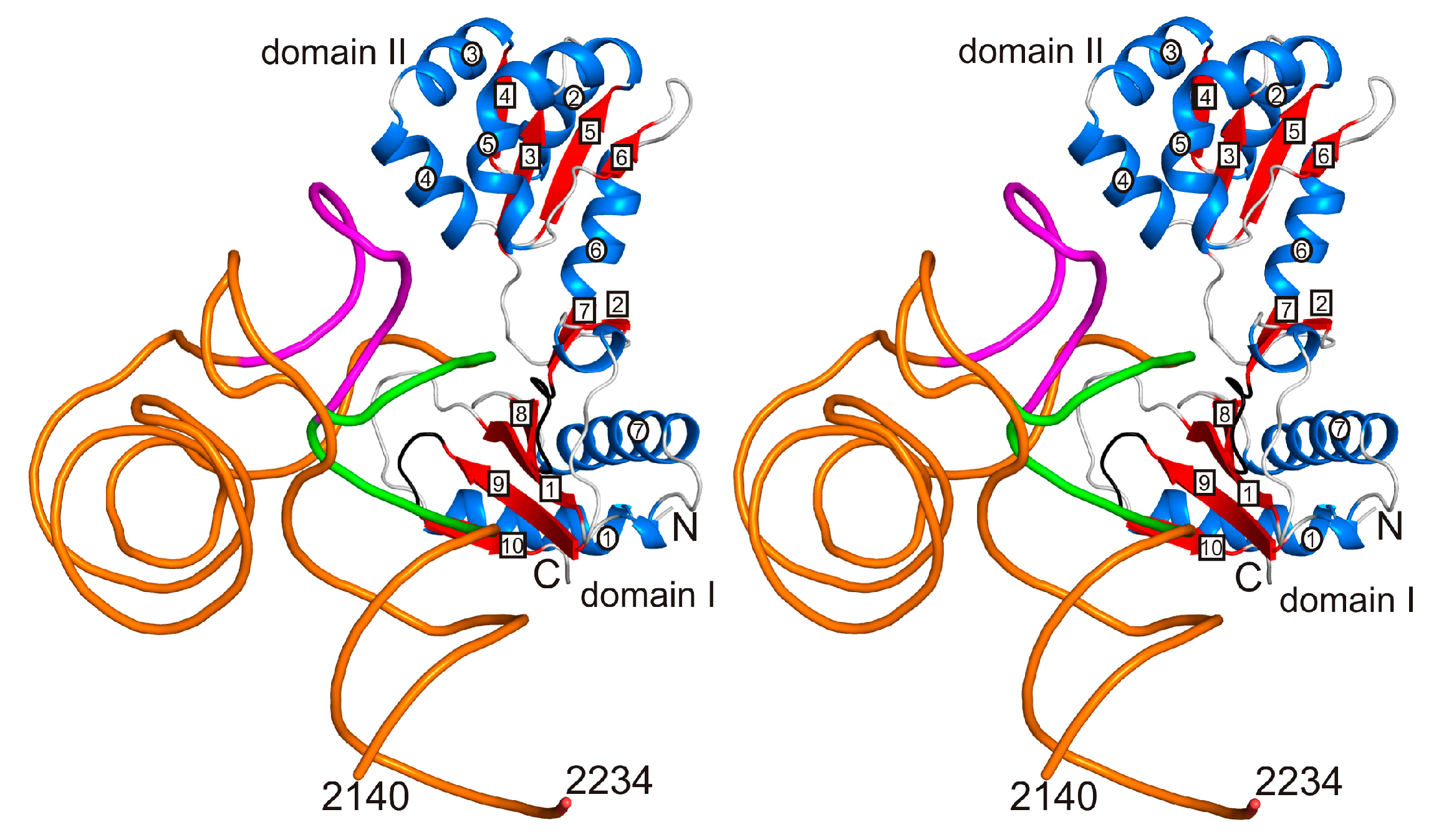
2.3. Incorporation of the L1-Specific 23S rRNA Fragment into 50S Subunit of the H. marismortui Ribosome
3. Materials and Methods
3.1. RNA Fragment Preparation
3.2. Crystallization
3.3. Data Collection and Structure Determination
4. Conclusions
Supplementary Materials
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Evers, U.; Franceschi, F.; Boddeker, N.; Yonath, A. Crystallography of halophilic ribosome: The isolation of an internal ribonucleoprotein complex. Biol. Chem. 1994, 50, 3–16. [Google Scholar] [CrossRef]
- Ban, N.; Nissen, P.; Hansen, J.; Moore, P.B.; Steitz, T.A. The complete atomic structure of the large ribosomal subunit at 2.4 Å resolution. Science 2000, 289, 905–920. [Google Scholar] [CrossRef] [PubMed]
- Gabdulkhakov, A.; Nikonov, S.; Garber, M. Revisiting the Haloarcula marismortui 50S ribosomal subunit model. Acta Crystallogr. Sect. D Biol. Crystallogr. 2013, 69, 997–1004. [Google Scholar] [CrossRef] [PubMed]
- Yusupov, M.M.; Yusupova, G.Z.; Baucom, A.; Lieberman, K.; Earnest, T.N.; Cate, J.H.D.; Noller, H.F. Crystal structure of the ribosome at 5.5 Å resolution. Science 2001, 292, 883–896. [Google Scholar] [CrossRef] [PubMed]
- Gao, Y.-G.; Selmer, M.; Dunham, C.M.; Weixlbaumer, A.; Kelley, A.C.; Ramakrishnan, V. The structure of the ribosome with elongation factor G trapped in the posttranslocation state. Science 2009, 326, 694–699. [Google Scholar] [CrossRef] [PubMed]
- Harms, J.; Schluenzen, F.; Zarivach, R.; Bashan, A.; Gat, S.; Agmon, I.; Bartels, H.; Franceschi, F.; Yonath, A. High resolution structure of the large ribosomal subunit from mesophilic eubacterium. Cell 2001, 107, 679–688. [Google Scholar] [CrossRef]
- Ben-Shem, A.; Jenner, L.; Yusupova, G.; Yusupov, M. Crystal structure of the eukaryotic ribosome. Science 2010, 320, 1203–1209. [Google Scholar] [CrossRef] [PubMed]
- Klein, D.J.; Moore, P.B.; Stetz, T.A. The role of ribosomal proteins in the structure assembly, and evolution of the large ribosomal subunit. J. Mol. Biol. 2004, 340, 141–177. [Google Scholar] [CrossRef] [PubMed]
- Córdova, D.I.C.; Ruíz, R.M.C.; Díaz, J.C.M.; López, J.A.C.; Rodríguez, J.A.G. Haloarcula marismortui, eighty-four years after its discovery in the Dead Sea. Int. J. Eng. Res. Technol. 2014, 3, 1257–1267. [Google Scholar]
- Nevskaya, N.; Tishchenko, S.; Fedorov, R.; Al-Karadaghi, S.; Liljas, A.; Kraft, A.; Piendl, W.; Garber, M.; Nikonov, S. Archaeal ribosomal protein L1: The structure provides new insights into RNA binding of the L1 protein family. Structure 2000, 8, 363–371. [Google Scholar] [CrossRef]
- Tishchenko, S.; Gabdulkhakov, A.; Nevskaya, N.; Sarskikh, A.; Kostareva, O.; Nikonova, E.; Sycheva, A.; Moshkovskii, S.; Garber, M.; Nikonov, S. High-resolution crystal structure of the isolated ribosomal. Acta Crystallogr. Sect. D Biol. Crystallogr. 2012, 68, 1051–1057. [Google Scholar] [CrossRef] [PubMed]
- Feng, S.; Chen, Y.; Gao, Y.-G. Crystal structure of 70S ribosome with both cognate tRNAs in the E and P sites representing an authentic elongation complex. PLoS ONE 2013, 8, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Polikanov, Y.S.; Melnikov, S.V.; Söll, D.; Steitz, T.A. Structural insights into the role of rRNA modifications in protein synthesis and ribosome assembly. Nat. Struct. Mol. Biol. 2015, 22, 342–344. [Google Scholar] [CrossRef] [PubMed]
- Afonine, P.V.; Grosse-Kunstleve, R.W.; Echols, N.; Headd, J.J.; Moriarty, N.W.; Mustyakimov, M.; Terwilliger, T.C.; Urzhumtsev, A.; Zwart, P.H.; Adams, P.D. Towards automated crystallographic structure refinement with phenix.refine. Acta Crystallogr. Sect. D Biol. Crystallogr. 2012, 68, 352–367. [Google Scholar] [CrossRef] [PubMed]
- Nikulin, A.; Eliseikina, I.; Tischenko, S.; Nevskaya, N.; Davydova, N.; Platonova, O.; Piendl, W.; Selmer, M.; Liljas, A.; Drygin, D.; et al. Structure of the L1 protuberance in the ribosome. Nat. Struct. Biol. 2003, 10, 104–108. [Google Scholar] [CrossRef] [PubMed]
- Schuwirth, B.S.; Borovinskaya, M.A.; Hau, C.W.; Zhang, W.; Vila-Sanjurjo, A.; Holton, J.M.; Cate, J.H.D. Structures of the Bacterial Ribosome at 3.5 Å Resolution. Science 2005, 310, 827–834. [Google Scholar] [CrossRef] [PubMed]
- Kabsch, W. XDS. Acta Crystallogr. Sect. D Biol. Crystallogr. 2010, 66, 125–132. [Google Scholar] [CrossRef] [PubMed]
- McCoy, A.J.; Grosse-Kunstleve, R.W.; Adams, P.D.; Winn, M.D.; Storoni, L.C.; Read, R.J. Phaser crystallographic software. J. Appl. Crystallogr. 2007, 40, 658–674. [Google Scholar] [CrossRef] [PubMed]
- Murshudov, G.N.; Skubak, P.; Lebedev, A.A.; Pannu, N.S.; Steiner, R.A.; Nicholls, R.A.; Winn, M.D.; Long, F.; Vagin, A.A. REFMAC5 for the refinement of macromolecular crystal structures. Acta Crystallogr. Sect. D Biol. Crystallogr. 2011, 67, 355–367. [Google Scholar] [CrossRef] [PubMed]
- Emsley, P.; Lohkamp, B.; Scott, W.G.; Cowtan, K. Features and development of Coot. Acta Crystallogr. Sect. D Biol. Crystallogr. 2010, 66, 486–501. [Google Scholar] [CrossRef] [PubMed]
- DeLano, W.L. The PyMOL Molecular Graphics System. Available online: http://www.pymol.org/ (accessed on 11 January 2016).


| Data collection | |
|---|---|
| Wavelength (Å) | 0.8266 |
| Space group | P41212 (No. 92) |
| Unit-cell parameters (Å, °) | a = b = 136.84, c = 70.11, α = β = γ = 90 |
| Resolution limits (Å) | 30.0–3.3 (3.5–3.3) |
| Completeness (%) | 96.6 (93.0) |
| Measured reflections | 53948 (7363) |
| Unique reflections | 10134 (1551) |
| Rmerge (%) | 15.0 (45.2) |
| Mean I/σ(I) | 6.68 (3.2) |
| Redundancy | 5.32 (4.74) |
| Refinement | |
| Resolution (Å) | 30.0–3.30 (3.39–3.30) |
| No. of reflections | 9 674 (665) |
| R factor (%) | 22.6 (35.4) |
| Free R factor (%) | 29.14 (50.6) |
| Average overall B factor (Å2) | 65.3 |
| R.m.s. deviations | |
| Bond lengths (Å) | 0.008 |
| Bond angles (°) | 1.72 |
| PDB code | 5ML7 |
© 2017 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license ( http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Gabdulkhakov, A.; Tishchenko, S.; Mikhaylina, A.; Garber, M.; Nevskaya, N.; Nikonov, S. Crystal Structure of the 23S rRNA Fragment Specific to r-Protein L1 and Designed Model of the Ribosomal L1 Stalk from Haloarcula marismortui. Crystals 2017, 7, 37. https://doi.org/10.3390/cryst7020037
Gabdulkhakov A, Tishchenko S, Mikhaylina A, Garber M, Nevskaya N, Nikonov S. Crystal Structure of the 23S rRNA Fragment Specific to r-Protein L1 and Designed Model of the Ribosomal L1 Stalk from Haloarcula marismortui. Crystals. 2017; 7(2):37. https://doi.org/10.3390/cryst7020037
Chicago/Turabian StyleGabdulkhakov, Azat, Svetlana Tishchenko, Alisa Mikhaylina, Maria Garber, Natalia Nevskaya, and Stanislav Nikonov. 2017. "Crystal Structure of the 23S rRNA Fragment Specific to r-Protein L1 and Designed Model of the Ribosomal L1 Stalk from Haloarcula marismortui" Crystals 7, no. 2: 37. https://doi.org/10.3390/cryst7020037





