Abstract
A series of quinoline derivatives was synthesized and biologically evaluated as Enhancer of Zeste Homologue 2 (EZH2) inhibitors. Structure-activity relationship (SAR) studies led to the discovery of 5-methoxy-2-(4-methyl-1,4-diazepan-1-yl)-N-(1-methylpiperidin-4-yl)quinolin-4-amine (5k), which displayed an IC50 value of 1.2 μM against EZH2, decreased global H3K27me3 level in cells and also showed good anti-viability activities against two tumor cell lines. Due to the low molecular weight and the fact that no quinoline derivative has been reported as an EZH2 inhibitor, this compound could serve as a lead compound for further optimization.
1. Introduction
Enhancer of Zeste Homologue 2 (EZH2) is a member of the histone-lysine N-methyltransferase (HKMT) family, which methylates K9 and K27 of histone H3, leading to transcriptional repression of the affected target genes. EZH2 has been found to be over-activated or over-expressed in many cancer types, such as lymphoma, colon, prostate, breast, and lung cancer [1,2]. The over-activation or over-expression of EZH2 has been linked to aberrant repression of some tumor suppressor genes, and is often implicated in tumor progression and correlates with poor prognosis [3,4]. Therefore, EZH2 has been highlighted as a promising intervention target for cancer therapy.
Due to the potential therapeutic value of EZH2 inhibitors in the treatment of cancer, many pharmaceutical companies and academic institutes have recently been involved in the development of small molecule inhibitors of EZH2. To date, a number of EZH2 inhibitors (see Figure 1) have been reported [5,6]. Unfortunately, the known EZH2 inhibitors have very limited scaffold structures. We thus decided to discover new classes of EZH2 inhibitors with scaffolds different from those of known EZH2 inhibitors. BIX-01294 (2-(hexahydro-4-methyl-1H-1,4-diazepin-1-yl)-6,7-dimethoxy-N-[1-(phenylmethyl)-4-piperidinyl]-4-quinazolinamine, Figure 2) is a well-known histone methyltransferase G9a/GLP inhibitor containing a 1-benzylpiperidin-4-ylamino group [7]. Simultaneously, quinoline derivatives are useful in diverse applications including pharmaceuticals and are available as drugs today, and have never been reported as EZH2 inhibitors. Thus we considered following the example of BIX-01294 to develop novel EZH2 inhibitors. We synthesized compound 1 (N-(1-benzylpiperidin-4-yl)-2-chloro-5-methoxyquinolin-4-amine, Figure 2) and then verified its activity against EZH2. This compound had a half maximal inhibitory concentration (IC50) value of 28 μM against EZH2. Obviously the potency of this compound was too low and needed further improvement. Therefore in this investigation we carried out structural optimization to compound 1. A series of quinoline derivatives were synthesized, and the structure–activity relationships (SAR) of these compounds will also be discussed.
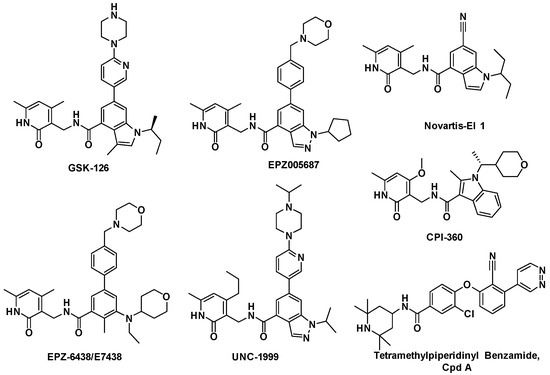
Figure 1.
Typical EZH2 inhibitors currently reported.
Figure 1.
Typical EZH2 inhibitors currently reported.
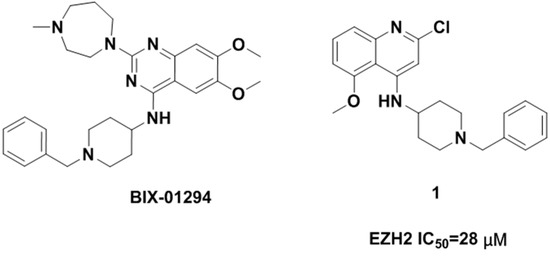
Figure 2.
Structure of BIX-01294 and hit compound 1.
Figure 2.
Structure of BIX-01294 and hit compound 1.
2. Results and Discussion
2.1. Chemistry
The general synthetic routes employed for the preparation of 5-methoxyquinoline derivatives 5a–v are outlined in Scheme 1. The intermediate 8-bromo-2,4-dichloro-5-methoxyquinoline (2) was prepared by cyclization of 2-bromo-5-methoxyaniline and malonic acid, using POCl3 as catalyst and solvent [8]. Then the bromine in the 8-positionof 2 was removed by n-BuLi in THF and CH3OH to give compound 3 [9], which reacted with a variety of primary or secondary amines at the 4-position of the quinoline moiety to produce compounds 4a–p by a nucleophilic substitution reaction [10]. Finally, the final products 5a–v were obtained by another nucleophilic substitution reaction of 4a–p with amines at the 2-position of the quinoline moiety in i-PrOH under microwave conditions, using TFA as an acid catalyst [11].
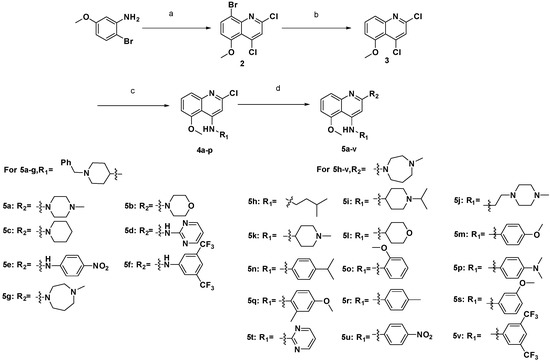
Scheme 1.
Synthetic routes of compounds 5a–v.
Scheme 1.
Synthetic routes of compounds 5a–v.
Reagents and conditions: (a) malonic acid, phosphorus oxychloride, reflux, 120 °C, 16 h; (b) n-Butyllithium, tetrahydrofuran, methanol, −78–0 °C, 2 h; (c) 1-Methyl-2-pyrrolidinone, ethyldiisopropylamine, 150 °C, R1NH2, 4 h; (d) trifluoroacetic acid, isopropanol, 160 °C, R2H, microwaves, 30 min.
The general routes for the preparation of quinoline derivatives 9a–i are outlined in Scheme 2. Phenylamines 6a–i with single or double substituted methoxy groups were cyclized with malonic acid, using POCl3 as catalyst and solvent, to give intermediates 7a–i, which reacted with 1-methylpiperidin-4-amine at the 4-position of the quinoline moiety to produce compounds 8a–i by a nucleophilic substitution reaction. Finally, the products 9a–i were obtained by another nucleophilic substitution reaction of 8a–i with 1-methyl-1,4-diazepane at the 2-position of the quinoline moiety in i-PrOH under microwave conditions, using TFA as an acid catalyst.
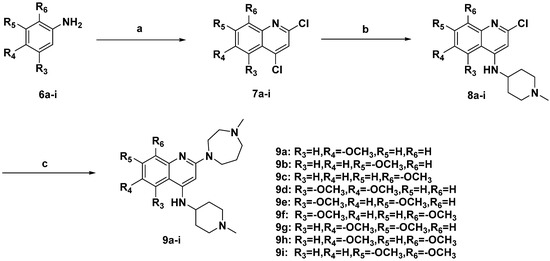
Scheme 2.
Synthetic routes of compounds 9a–i.
Scheme 2.
Synthetic routes of compounds 9a–i.
Reagents and conditions: (a) malonic acid, phosphorus oxychloride, reflux, 120 °C, 16 h; (b) 1-methyl-2-pyrrolidinone, ethyldiisopropylamine, 150 °C, 1-methylpiperidin-4-amine, 4 h; (c) trifluoroacetic acid, isopropanol, 160 °C, 1-methyl-1,4-diazepane, microwaves, 30 min.
2.2. SAR Analyses
The SAR analyses of the quinoline derivatives were based on both enzymatic and cellular assays, in which the enzymatic activities were determined by the AlphaLISA method, and the cell growth inhibition potencies were evaluated on the HCT15 and MD-MB-231 cell lines by the MTT method.
The SAR analysis started by varying substituents of the 2-position of quinoline moiety with the 4-position being substituted by 1-benzylpiperidin-4-amine. We first synthesized eight compounds (1, 5a–g) with each one bearing different substituents or no substituent at the 2-position of the quinoline. Bioactivities of these compounds are summarized in Table 1. Among these compounds, 5g, which contains a 1-methyl-1,4-diazepane group at the 2-position, was the most active compound at both the enzymatic and cellular levels. Though compound 5c, which bears a piperidine group at the 2-position, had a good activity at the cellular level, it did not show notable activity at the enzymatic level (IC50 > 10 μM). Compounds 5d–f, which contain a primary amine at the 2-position of quinoline moiety, only exhibited relatively weak potency.

Table 1.
Bioactivities of compounds 1, 5a–g.
| Compound | IC50 a (μM) | ||
|---|---|---|---|
| EZH2(AlphaLISA) | HCT15 | MDA-MB-231 | |
| GSK-126 | 0.001 | N.T. | N.T. |
| 1 | 28.0 ± 2.23 | 26.0 ± 2.68 | >20.0 |
| 5a | 16.4 ± 1.24 | 12.3 ± 0.95 | 11.9 ± 1.83 |
| 5b | 23.0 ± 2.06 | 15.6 ± 1.27 | 14.2 ± 1.97 |
| 5c | 32.0 ± 2.78 | 7.80 ± 0.46 | 5.60 ± 0.14 |
| 5d | 17.5 ± 0.96 | >20 | >20 |
| 5e | 29.4 ± 1.01 | 17.8 ± 1.27 | 14.9 ± 2.20 |
| 5f | 21.6 ± 2.35 | 10.2 ± 1.24 | >20 |
| 5g | 6.6 ± 0.49 | 3.78 ± 0.07 | 4.56 ± 0.24 |
a Values are means of at least three measurements. N.T.: Not Tested.
We then fixed the 2-position of the quinoline nucleus with a 1-methyl-1,4-diazepane unit as the optimal group, and varied the 4-position of the quinoline moiety. A total of 15 compounds were synthesized. Bioactivities of these compounds are listed in Table 2. From Table 2, we can see that 5k, which contains a 1-methylpiperidin-4-amine group at the 4-position, was the most active compound in the enzymatic assay, with an IC50 value of 1.2 μM. It also showed a good cellular potency against HCT15 and MDA-MB-231, with IC50 values of 5.6 μM and 2.45 μM, respectively. Obviously, the bioactivities of 5k at both the enzymatic and cellular levels were increased by more than 20-fold compared with those of 1.

Table 2.
Bioactivities of compounds 5h–v.
| Compound | IC50 a (μM) | ||
|---|---|---|---|
| EZH2(AlphaLISA) | HCT15 | MDA-MB-231 | |
| 5h | 11.5 ± 1.21 | 5.46 ± 0.85 | 4.31 ± 0.57 |
| 5i | 17.4 ± 1.07 | >20 | 15.7 ± 1.27 |
| 5j | 4.20 ± 0.20 | 18.6 ± 2.20 | 12.4 ± 0.95 |
| 5k | 1.20 ± 0.11 | 7.60 ± 0.46 | 2.45 ± 0.24 |
| 5l | 2.80 ± 0.21 | 15.5 ± 0.20 | 2.40 ± 0.49 |
| 5m | 19.7 ± 2.56 | 18.9 ± 1.97 | >20 |
| 5n | 26.4 ± 2.04 | 14.6 ± 1.27 | 13.2 ± 2.53 |
| 5o | 17.5 ± 1.13 | >20 | >20 |
| 5p | 27.6 ± 0.97 | 17.4 ± 1.54 | 16.2 ± 1.67 |
| 5q | 34.8 ± 1.54 | 16.9 ± 0.36 | 14.5 ± 1.23 |
| 5r | 39.7 ± 1.26 | 12.9 ± 0.89 | >20 |
| 5s | 16.8 ± 0.94 | 19.6 ± 0.56 | 16.9 ± 1.06 |
| 5t | 35.3 ± 1.61 | >20 | >20 |
| 5u | 26.2 ± 2.54 | 17.8 ± 1.27 | 14.9 ± 2.20 |
| 5v | 34.1 ± 2.22 | 10.2 ± 1.24 | >20 |
a Values are means of at least three measurements.
We further explored the possible effect of a methoxy group at different substitution positions of the quinoline moiety. Single or double methoxy groups were introduced to the 5, 6, 7 or 8-positions of the quinoline moiety, which led to compounds 9a–i. As shown in Table 3, compound 9a (6-methoxy substituted), 9d (5,6-dimethoxy substituted), 9e (5,7-dimethoxy substituted), and 9f (5,8-dimethoxy substituted) displayed a slightly decreased potency in both enzymatic and cellular assays compared with 5k, while the other compounds exhibited a greatly decreased activity. This indicated that methoxy groups at the 5-position might be more important for the potency of the compounds, and single or double substitutions at other positions would not contribute to the potency.

Table 3.
Bioactivities of compounds 9a–i.
| Compound | IC50 a (μM) | ||
|---|---|---|---|
| EZH2(AlphaLISA) | HCT15 | MDA-MB-231 | |
| 9a | 8.40 ± 0.34 | 10.9 ± 0.35 | 14.6 ± 2.24 |
| 9b | 18.4 ± 1.65 | >20 | >20 |
| 9c | 20.1 ± 0.85 | >20 | >20 |
| 9d | 5.60 ± 0.45 | 8.60 ± 2.20 | 12.4 ± 0.95 |
| 9e | 6.70 ± 0.36 | 10.6 ± 0.83 | 12.5 ± 0.62 |
| 9f | 9.20 ± 0.51 | 15.5 ± 0.53 | 12.4 ± 0.49 |
| 9g | 17.8 ± 2.07 | 15.9 ± 1.85 | >20 |
| 9h | 14.9 ± 1.71 | 19.8 ± 1.27 | 13.2 ± 1.64 |
| 9i | 13.7 ± 1.32 | >20 | >20 |
a Values are means of at least three measurements.
2.3. Selectivity of 5k
To examine the selectivity of 5k, we measured the activities of 5k against several selected protein methyltransferases, including SMYD3, G9a, SUV39H1, SETDB1, PRMT1 and SETD8. As shown in Table 4, compound 5k showed very weak activity against the selected protein methyltransferases, indicating a good selectivity for EZH2.

Table 4.
Inhibition activity of 5k against EZH2 and several other selected histone methyltransferases.
| Targets | EZH2 | SMYD3 | G9a | SUV39H1 | SETDB1 | PRMT1 | SETD8 |
|---|---|---|---|---|---|---|---|
| IC50 a (μM) | 1.20 ± 0.11 | 35.4 ± 1.97 | 82.6 ± 2.23 | 21.2 ± 0.28 | 27.0 ± 1.36 | 28.8 ± 1.25 | 40.0 ± 2.71 |
a Values are means of at least three measurements.
2.4. Analysis of H3K27 Methylation in MDA-MB-231 Cell Line Treated with 5k
The main catalytic activity of EZH2 is methylating histone 3 lysine 27 (H3K27), so evaluation of H3K27me3 was conducted through a western blotting assay. As shown in Figure 3, the H3K27me3 level of MDA-MB-231cell treated with compound 5k was obviously down-regulated. These results indicated that compound 5k inhibited the methyltransferase activity of EZH2.
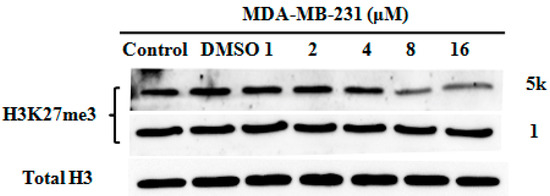
Figure 3.
Analysis of H3K27 methylation in MDA-MB-231 cell line treated with 1 and 5k. Evaluation of H3K27me3 following treatment for 72 h. Total H3 is shown as a loading control.
Figure 3.
Analysis of H3K27 methylation in MDA-MB-231 cell line treated with 1 and 5k. Evaluation of H3K27me3 following treatment for 72 h. Total H3 is shown as a loading control.
2.5. The Possible Binding Mode of 5k with EZH2
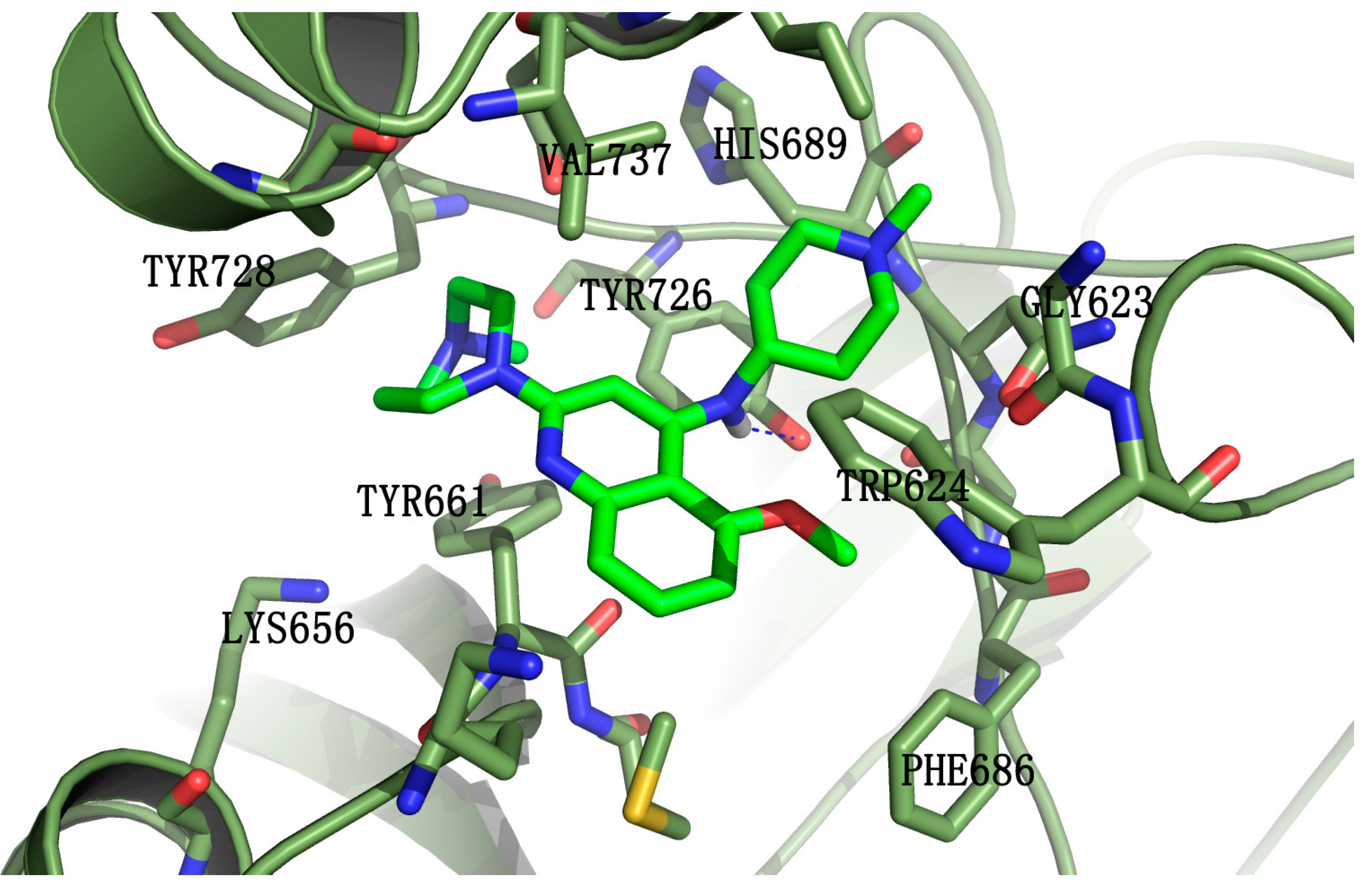
Molecular docking was employed to understand the binding mode of the most potent compound, 5k, with EZH2. Some of the amino acid residues in the active binding pocket of EZH2 are deleted in the crystal structure (PDB ID: 4MI5). Hence this crystal structure is not an ideal receptor structure, and it could not be used in molecular docking studies. Therefore a complete structure containing all the amino acid residues in the active binding pocket should be constructed by the homology modeling method. The homology model of the EZH2 active pocket was built with the SETD8 X-ray crystal structure (PDB ID: 1ZKK) because of their homology (Figure 4). Figure 5 shows a predicted binding mode of 5k with the SET domain of EZH2. Compound 5k occupies the substrate binding groove, and the hydrogen of the secondary amino group at the 4-position of the quinoline ring indeed forms a hydrogen bond with TYR726.
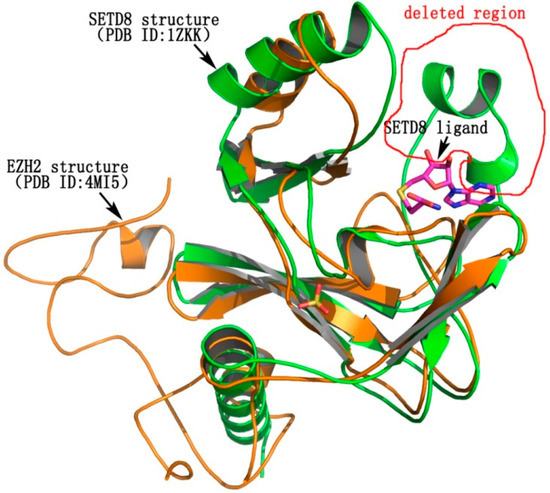
Figure 4.
The homology of EZH2 (PDB ID: 4MI5) and SETD8 (PDB ID: 1ZKK). EZH2 is shown in yellow, while SETD8 is shown in green. The deleted region in the active binding pocket of EZH2 is circled in red.
Figure 4.
The homology of EZH2 (PDB ID: 4MI5) and SETD8 (PDB ID: 1ZKK). EZH2 is shown in yellow, while SETD8 is shown in green. The deleted region in the active binding pocket of EZH2 is circled in red.
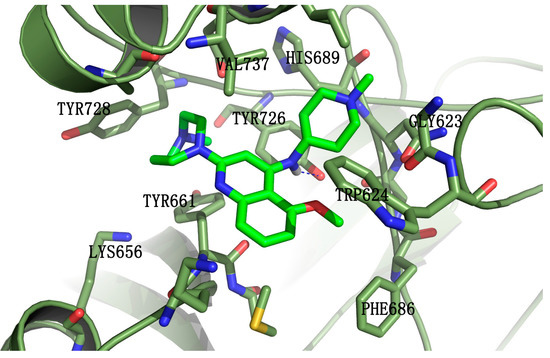
Figure 5.
Predicted binding mode of compound 5k with the SET-domain of EZH2. Compound 5k is shown in green. EZH2 is represented by ribbons with the interacting residues represented as sticks. Hydrogen-bonding is drawn as dashed blue.
Figure 5.
Predicted binding mode of compound 5k with the SET-domain of EZH2. Compound 5k is shown in green. EZH2 is represented by ribbons with the interacting residues represented as sticks. Hydrogen-bonding is drawn as dashed blue.
3. Experimental Section
3.1. General
The human cancer cell lines HCT15 and MDA-MB-231 were purchased from the American Type Culture Collection (ATCC, Rockville, MD, USA). Dulbecco’s modified Eagle medium (DMEM) and RPMI 1640 were purchased from Gibco (Grand Island, NY, USA). Fetal bovine serum (FBS) was obtained from Hyclone (Logan, UT, USA). The purity of compound screened in biological assays was determined to be >98% by HPLC analysis. A Symmetry C18 (75 mm × 4.6 mm, i.d. 3.5 μm) (Waters, Milford, MA, USA) was used with acetonitrile and HPLC-grade water as mobile phase. 1H-NMR were recorded at 400 MHz, mass spectra (MS) were measured by a Q-TOF Premier (Micromass, Manchester, UK) mass spectrometer utilizing electrospray ionization (ESI).
3.2. AlphaLISA Technology Assay for EZH2
The AlphaLISA assay and the enzyme selectivity assay were performed by Shanghai ChemPartner Co., Ltd. (Shanghai, China). Briefly, compounds were transferred to the assay plate (OptiPlate-384, PerkinElmer, Waltham, MA, USA) with 3-fold dilution by Echo (compound total volume 100 nL), and 5 μL enzyme solution (final concentration was 2 nM) or pH 9.0 TRIS-based assay buffer (for Min well) to the assay plate were added, and centrifuged at 1000 rpm for 1 min. The assay plate was incubated for 15 min at RT prior to initiation of the reaction by addition of 5 μL biotinylated peptide/SAM mix (final concentrations were 100 nM and 3000 nM, respectively). The plate was covered with TopSeal-A film and incubated for 1 h at RT after centrifuging at 1000 rpm for 1 min (DMSO final concentration 1%). Acceptor beads (15 μL) and donor beads mixed were added to stop the enzymatic reaction. After incubating for 1 h again in subdued light, the assay plate was read in Alpha mode on an EnVision instrument (PerkinElmer, Waltham, MA, USA). GSK-126 was used as the positive control.
3.3. MTT Assay
Briefly, cells were seeded in 96-well plates and cultured for 24 h, followed by treatment with the compounds for 96 h. Ten microliters of 10 mg/mL MTT was added per well and incubated for another 2.5 h at 37 °C. Then the supernatant fluid was removed and 150 μL/well DMSO was added for 15–20 min. The absorbance (OD) of each well was measured at 570 nm, using a SpectraMAX M5 microplate spectrophotometer (Molecular Devices, CA, USA).
3.4. Western Blotting Assay of H3K27me3 Status
The western blotting assay of H3K27 status was performed as previously described [12].
3.5. Homology Modeling, Molecular Docking and Molecular Dynamic (MD) Simulation
The homology model of the SET domain of EZH2 was built using the Build Homology Model module of Discovery Studio (Accelrys, San Diego, CA, USA). For this step, the MODELER program was applied. The SET domain of N-lysine methyltransferase SETD8 (PDB accession code1ZKK) was used as template, because of its homology to EZH2, and similar structure in the SET domain.
All the molecular docking simulation is performed using the GOLD program in Discovery Studio. GOLD is a genetic algorithm-based molecular docking method that uses the CHARMM force field. All the molecules were prepared before docking using the prepare ligands protocol in Discovery Studio. The docking parameters are the default setting.
All the molecular dynamics (MD) simulations of the protein-ligand complex obtained by docking was carried out using the Standard Dynamics Cascade protocol in the Discovery Studio environment. For the Standard Dynamics Cascade, the CHARMM force field was employed for the protein and the GAFF force field for small molecules. During the simulation, five stages was experienced, which according to the order, are the minimization 1 using the robust steepest descent algorithm, minimization 2 using the conjugate gradient method, 10 ps heating stage (the temperature was changed from 50 to 300 K), 10 ps Equilibration stage to equilibrate the system at the target temperature 300 K, and finally, and 10 ps Production stage using a leap-frog Verlet integration algorithm, respectively.
3.6. General Procedure for the Synthesis of 2
A mixture of 2-bromo-5-methoxyaniline (10 mmol, 2.02 g), malonic acid (15 mmol) and POCl3 (25 mL) was refluxed for 16 h. The reaction mixture was slowly poured into water and extracted with DCM. The organic layers was dried over Na2SO4, filtered, and concentrated under reduced pressure. The crude material was purified on silica gel, eluted with DCM, to afford the final product as a light yellow solid in 90% yield [8].1H-NMR (400 MHz, CDCl3): δ 7.96 (t, J = 8 Hz, 1H), 7.47 (s, 1H), 6.82 (t, J = 8 Hz, 1H), 3.96 (s, 3H). 13C-NMR (100 MHz, CDCl3): δ 156.2, 151.4, 144.2, 143.5, 138.0, 118.8, 122.0, 114.6, 105.0, 56.2. ESI-MS (m/z, %): 303.8989 [M−H]−. Mp: 204–206 °C.
3.7. General Procedure for the Synthesis of 3
To 8-bromo-2,4-dichloro-5-methoxyquinoline (2, 10 mmol, 3.06 g) in 20 mL of THF, n-BuLi (11 mmol, 2.5 M, 4.5 mL) was added dropwise at −78 °C. The resulting mixture was stirred at −78 °C for 1 h, and CH3OH (2 mL) was added dropwise at −78 °C. The mixture was stirred at 0 °C for 1 h, and 20 mL of water and 20 mL of dichloromethane were added. The two layers were separated, and the aqueous layer was extracted with dichloromethane (3 × 30 mL). The combined organic extracts were washed with brine, dried over anhydrous sodium sulfate, filtered, and concentrated to afford the final product in 92% yield [9].1H-NMR (400 MHz, CDCl3): δ 7.64–7.47 (m, 2H), 7.34 (s, 1H), 6.88 (dd, J = 7.3, 1.3 Hz, 1H), 3.91 (s, 3H). 13C-NMR (100 MHz, CDCl3): δ 156.7, 150.4, 149.2, 143.6, 135.3, 121.6, 120.1, 118.2, 107.7, 56.2. ESI-MS (m/z, %): 225.9875 [M−H]−. Mp: 198–200 °C.
3.8. Synthesis of 4a–p
The synthesis of compounds 4a–p was performed according to the literature method [10].
3.9. General Procedure for the Synthesis of 5a–v
To a solution of 4a–p (0.2 mmol) in isopropanol (3 mL) was added various primary amines or secondary amines (0.4 mmol) and TFA (0.6 mmol, 68 mg). The resulting solution was stirred in a sealed vessel inside a microwave apparatus at 160 °C for 20 min. After cooling, TLC indicated the completion of the reaction. After removal of the solvent by rotary evaporation, the residue was redissolved in DCM, and washed with saturated NaHCO3 solution. The organic layer was dried, concentrated and purified by column chromatography (1% DCM-methanol) to give the desired compound [11].
N-(1-Benzylpiperidin-4-yl)-5-methoxy-2-(4-methylpiperazin-1-yl)quinolin-4-amine (5a): Yellow solid, Mp: 268–269 °C. 80% yield from 4a. 1H-NMR (400 MHz, DMSO-d6): δ 7.79 (d, J = 7.6 Hz, 1H), 7.49 (t, J = 8.2 Hz, 1H), 7.38–7.29 (m, 4H), 7.21 (t, J = 6.9 Hz, 2H), 6.88 (d, J = 7.9 Hz, 1H), 6.39 (s, 1H), 3.94 (s, 3H), 3.79 (s, 3H), 3.32 (s, 4H), 3.07 (dd, J = 12.5Hz, 8.0 Hz, 6H), 2.79(s, 3H), 2.52 (s, 3H), 2.08 (s, 2H), 1.73 (d, J = 12.5 Hz, 1H). 13C-NMR (100 MHz, DMSO-d6): δ 174.2, 150.5, 149.8, 147.6, 138.6, 131.5, 128.8 (2C), 128.4 (2C), 127.2, 120.2, 105.7, 102.0, 94.6, 64.7, 57.2 (2C), 56.8, 56.2, 51.9 (2C), 47.6 (2C), 46.6, 30.3 (2C). ESI-MS: m/z 446.2875 [M+H]+.
N-(1-Benzylpiperidin-4-yl)-5-methoxy-2-morpholinoquinolin-4-amine (5b): Yellow solid, Mp: 248–249 °C. 82% yield from 4a. 1H-NMR (400 MHz, DMSO-d6): δ 7.78 (d, J = 7.6 Hz, 1H), 7.52 (t, J = 8.2 Hz, 1H), 7.34–7.29 (m, 4H), 7.25 (t, J = 6.9 Hz, 2H), 6.91 (d, J = 7.9 Hz, 1H), 6.39 (s, 1H), 3.95 (s, 3H), 3.71(d, J = 4.1 Hz, 4H), 3.61(s, 4H), 3.06–2.88 (m, 2H), 2.01 (s, 2H), 1.64 (s, 2H), 1.20 (q, J = 8.0 Hz, 5H). 13C-NMR (100 MHz, DMSO-d6): δ 174.2, 150.5, 149.8, 147.6, 138.6, 131.5, 128.8 (2C), 128.4 (2C), 127.2, 120.2, 105.7, 102.0, 94.6, 66.3 (2C), 64.7, 56.8, 56.2, 51.9 (2C), 48.7 (2C), 30.3 (2C). ESI-MS: m/z 433.2559 [M+H]+.
N-(1-Benzylpiperidin-4-yl)-5-methoxy-2-(piperidin-1-yl)quinolin-4-amine (5c): Yellow solid, Mp: 244–246 °C. 85% yield from 4a. 1H-NMR (400 MHz, DMSO-d6): δ 7.79 (d, J = 7.6 Hz, 1H), 7.45 (t, J = 8.2 Hz, 1H), 7.39–7.30 (m, 4H), 7.25 (t, J = 6.9 Hz, 2H), 6.88 (d, J = 7.9 Hz, 1H), 6.39 (s, 1H), 3.94 (s, 3H), 3.79–3.43 (m, 6H), 2.70 (s, 2H), 2.33 (s, 2H), 1.98 (s, 2H), 1.68 (s, 7H), 1.25(d, 2H). 13C-NMR (100 MHz, DMSO-d6): δ 174.2, 150.5, 149.8, 147.6, 138.6, 131.5, 128.8 (2C), 128.4 (2C), 127.2, 120.2, 105.7, 102.0, 94.6, 64.7, 56.8, 56.2, 51.9 (2C), 47.2 (2C), 30.3 (2C), 25.5 (2C), 24.5. ESI-MS: m/z 431.2766 [M+H]+.
N4-(1-Benzylpiperidin-4-yl)-5-methoxy-N2-(pyrimidin-2-yl)quinoline-2,4-diamine (5d): Yellow solid, Mp: 224–226 °C. 84% yield from 4a. 1H-NMR (400 MHz, DMSO-d6): δ 7.58–7.29 (m, 10H), 6.86 (d, J = 8.0 Hz, 1H), 6.70 (d, J = 8.0 Hz, 1H), 5.29 (s, 1H), 4.44 (br s, 1H), 3.90 (s, 3H), 3.61 (s, 2H), 3.12–3.05 (m, 3H), 2.26 (d, J = 8.0 Hz, 2H), 2.09 (s, 2H), 1.73 (d, J = 8.0 Hz, 2H). 13C-NMR (100 MHz, DMSO-d6): δ 169.3, 157.9 (2C), 155.5, 148.4, 148.0, 146.0, 138.6, 129.9, 128.8 (2C), 128.4 (2C), 127.2, 117.8, 115.3, 104.6, 100.6, 94.5, 64.7, 56.8, 56.2, 51.9 (2C), 30.3 (2C). ESI-MS: m/z 441.2358 [M+H]+.
N4-(1-Benzylpiperidin-4-yl)-5-methoxy-N2-(4-nitrophenyl)quinoline-2,4-diamine (5e): Yellow solid, Mp: 226–228 °C. 88% yield from 4a. 1H-NMR (400 MHz, DMSO-d6): δ 8.36 (d, J = 8.1 Hz, 2H), 8.19 (d, J = 8.6 Hz, 2H), 7.94 (d, J = 8.0 Hz, 1H), 7.81 (d, J = 8.0 Hz, 1H), 7.64 (d, J = 4.2 Hz, 1H), 7.35 (s, 2H), 6.73 (d, J = 8.8 Hz, 2H), 6.60 (d, J = 8.0 Hz, 1H), 6.13 (s, 1H), 3.96 (s, 3H), 3.54 (s, 2H), 2.25 (br s, 4H), 2.05 (br s, 4H). 13C-NMR (100 MHz, DMSO-d6): δ 155.5, 148.4, 148.0, 147.0, 146.0, 138.6, 137.9, 129.9, 128.8 (2C), 128.4 (2C), 127.2, 124.7 (2C), 119.2 (2C), 117.8, 104.6, 100.6, 94.5, 64.7, 56.8, 56.2, 51.9 (2C), 30.3 (2C). ESI-MS: m/z 484.2304 [M+H]+.
N4-(1-Benzylpiperidin-4-yl)-N2-(3,5-bis(trifluoromethyl)phenyl)-5-methoxyquinoline-2,4-diamine (5f): Yellow solid, Mp: 221–223 °C. 88% yield from 4a. 1H-NMR (400 MHz, DMSO-d6): δ 10.79 (s, 2H), 7.54–7.29 (m, 6H), 7.40–7.20 (m, 2H), 6.86 (d, J = 8.1 Hz, 1H), 6.71 (d, J = 8.0 Hz, 1H), 5.34 (s, 1H) , 4.33 (s, 1H), 3.91 (s, 3H), 3.59 (s, 2H), 3.21–2.94 (m, 3H), 2.27 (d, J = 12.2 Hz, 2H), 2.08 (s, 2H), 1.68 (d, J = 12.2 Hz, 2H). 13C-NMR (100 MHz, DMSO-d6): δ 155.5, 148.4, 148.0, 146.0, 140.7, 138.6, 132.1 (2C), 129.9, 128.8 (2C), 128.4 (2C), 127.2, 124.2 (2C), 119.3 (2C), 117.8, 113.1, 104.6, 100.6, 94.5, 64.7, 56.8, 56.2, 51.9 (2C), 30.3 (2C). ESI-MS: m/z 575.2201 [M+H]+.
N-(1-Benzylpiperidin-4-yl)-5-methoxy-2-(4-methyl-1,4-diazepan-1-yl)quinolin-4-amine (5g): Yellow solid, Mp: 235–237 °C. 95% yield from 4a. 1H-NMR (400 MHz, DMSO-d6): δ 7.34–7.04 (m, 9H), 6.42 (d, J = 7.4 Hz, 1H), 5.51 (s, 1H), 3.89 (s, 2H), 3.83 (s, 3H), 3.76–3.60 (m, 3H), 3.43 (br s, 1H), 2.70 (s, 4H), 2.53 (t, J = 4.0 Hz, 2H), 2.24 (t, J = 4.0 Hz , 2H), 2.09–1.89 (m, 5H), 1.82–1.46 (m, 4H). 13C-NMR (100 MHz, DMSO-d6): δ 174.2, 150.5, 149.8, 147.6, 138.6, 131.5, 128.8 (2C), 128.4 (2C), 127.2, 120.2, 105.7, 102.0, 94.6, 64.7, 61.8, 59.0, 57.6, 56.8, 56.2, 51.9 (3C), 46.9, 30.3 (2C), 26.4. ESI-MS: m/z 460.3032 [M+H]+.
N-Isopentyl-5-methoxy-2-(4-methyl-1,4-diazepan-1-yl)quinolin-4-amine (5h): Yellow solid, Mp: 215–217 °C. 80% yield from 4b. 1H-NMR (400 MHz, DMSO-d6): δ 7.51 (d, J = 8.2 Hz, 2H), 7.27 (s, 1H), 6.74 (dd, J = 8.1, 2.2 Hz, 1H), 5.51 (s, 1H), 4.00 (s, 2H), 3.90 (s, 3H), 3.84 (t, J = 4.0 Hz, 2H), 3.74 (s, 1H), 3.29 (q, J = 8.0 Hz, 2H), 2.82 (t, J = 4.1 Hz, 2H), 2.62 (t, J = 4.0 Hz, 2H), 2.41(s, 3H), 2.10 (t, J = 8.1 Hz, 2H), 1.80–1.63 (m, 1H), 1.66 (q, J = 8.2 Hz, 2H), 1.22 (s, 3H), 1.00 (d, J = 8.1 Hz, 2H). 13C-NMR (100 MHz, DMSO-d6): δ 174.2 150.5, 149.8, 147.6, 131.5, 120.2, 105.7, 102.0, 94.6, 61.8, 59.0, 57.6, 56.2, 51.9, 46.9, 42.6, 26.4, 25.5, 22.9 (2C). ESI-MS: m/z 357.2610 [M+H]+.
N-(1-Isopropylpiperidin-4-yl)-5-methoxy-2-(4-methyl-1,4-diazepan-1-yl)quinolin-4-amine (5i): Yellow solid, Mp: 213–215 °C. 87% yield from 4c. 1H-NMR (400 MHz, DMSO-d6): δ 7.37 (d, J = 4.0 Hz, 1H), 7.21 (t, J = 8.1 Hz, 1H), 6.86 (d, J = 8.3 Hz, 1H), 6.33 (d, J = 7.9 Hz, 1H), 5.63 (s, 1H), 3.91 (s, 3H), 3.79 (s, 1H), 3.64 (t, J = 7.9 Hz, 2H), 3.51–3.22 (m, 1H), 2.61 (s, 4H), 2.32–2.07 (m, 4H), 2.07–1.76 (m, 3H), 1.55 (d, J = 8.0 Hz, 2H), 1.35–1.13 (m, 4H), 1.07–0.89 (m, 6H), 0.84 (d, J = 7.4 Hz, 2H). 13C-NMR (100 MHz, DMSO-d6): δ 174.2, 150.5, 149.8, 147.6, 131.5, 120.2, 105.7, 102.0, 94.6, 61.8, 59.0, 58.3, 57.6, 56.8, 56.2, 51.9, 49.5 (2C), 46.9, 30.6 (2C), 26.4, 18.1. ESI-MS: m/z 412.3032 [M+H]+.
5-Methoxy-2-(4-methyl-1,4-diazepan-1-yl)-N-(2-(4-methylpiperazin-1-yl)ethyl)quinolin-4-amine (5j): Yellow solid, Mp: 210–212 °C. 86% yield from 4d. 1H-NMR (400 MHz, DMSO-d6): δ 7.39 (d, J = 4.2 Hz, 1H), 7.23 (t, J = 7.9 Hz, 1H), 6.96 (d, J = 8.0 Hz, 1H), 6.53 (d, J = 8.1 Hz, 1H), 3.86 (s, 3H), 3.79 (s, 1H), 3.64 (t, J = 8.1 Hz, 2H), 3.51 (s, 3H), 3.45–3.21 (m, 2H), 2.61 (s, 4H), 2.53–2.07 (m, 6H), 2.03–1.65 (m, 3H), 1.47 (d, J = 7.8 Hz, 2H), 1.35–0.98 (m, 4H), 0.78 (d, J = 7.4 Hz, 2H). 13C-NMR (100 MHz, DMSO-d6): δ 174.2, 150.5, 149.8, 147.6, 131.5, 120.2, 105.7, 102.0, 94.6, 61.8, 59.0, 57.6 (5C), 56.2, 55.5, 51.9, 47.6, 47.6, 46.9, 46.6, 26.4. ESI-MS: m/z 413.2984 [M+H]+.
5-Methoxy-2-(4-methyl-1,4-diazepan-1-yl)-N-(1-methylpiperidin-4-yl)quinolin-4-amine (5k): Yellow solid, Mp: 212–220 °C. 86% yield from 4e. 1H-NMR (400 MHz, DMSO-d6): δ 7.40 (d, J = 4.0 Hz, 1H), 7.22 (t, J = 8.0 Hz, 1H), 6.96 (d, J = 8.1 Hz, 1H), 6.54 (d, J = 8.3 Hz, 1H), 5.69 (s, 1H), 3.91 (s, 3H), 3.79 (s, 2H), 3.64 (t, J = 8.5 Hz, 2H), 3.54 (s, 1H), 3.44 (q, J = 7.0 Hz, 1H), 2.71–2.55 (m, 4H), 2.29 (t, J = 3.4 Hz, 5H), 2.25–2.18 (m, 3H), 1.98 (d, J = 10.2 Hz, 2H), 1.94–1.85 (m, 2H), 1.56 (dd, J = 18.3, 9.0 Hz, 2H), 1.06 (t, J = 7.0 Hz, 1H). 13C-NMR (100 MHz, DMSO-d6): δ 157.3, 157.2, 151.2,150.6, 128.5, 119.6, 105.6, 100.7, 83.1, 57.8, 56.5, 56.0(2C), 53.1, 45.9(4C), 45.3, 30.5, 27.0, 18. ESI-MS: m/z 384.2719 [M+H]+.
5-Methoxy-2-(4-methyl-1,4-diazepan-1-yl)-N-(tetrahydro-2H-pyran-4-yl)quinolin-4-amine (5l): Yellow solid, Mp: 213–215 °C. 86% yield from 4f. 1H-NMR (400 MHz, DMSO-d6): δ 8.41 (s, 1H), 7.81 (s, 1H), 7.66 (s, 1H), 7.04 (s, 1H), 5.77 (s, 1H), 4.32 (s, 1H), 4.03 (s, 4H), 3.94–3.65 (m, 3H), 3.56 (t, J = 10.4 Hz, 3H), 3.37 (d, J = 7.4 Hz, 1H), 3.37 (d, J = 7.4 Hz, 1H), 3.07 (s, 1H), 2.80 (s, 2H), 2.22 (s, 1H), 1.99 (d, J = 12.3 Hz, 2H), 1.64 (d, J = 9.1 Hz, 2H), 1.20 (dd, J = 16.0, 8.6 Hz, 3H), 0.85 (s, 1H). 13C-NMR (100 MHz, DMSO-d6): δ 174.2, 150.5, 149.8, 147.6, 131.5, 120.2, 105.7, 102.0, 94.6, 65.5 (2C), 61.8, 59.0, 57.6, 56.2, 55.5, 51.9, 46.9, 36.8 (2C), 26.4. ESI-MS: m/z 371.2402 [M+H]+.
5-Methoxy-N-(4-methoxyphenyl)-2-(4-methyl-1,4-diazepan-1-yl)quinolin-4-amine (5m): Yellow solid, Mp: 268–270 °C. 81% yield from 4g. 1H-NMR (400 MHz, DMSO-d6): δ 8.85 (s, 1H), 7.70 (s, 1H), 7.42 (t, J = 7.5 Hz, 1H), 7.18 (d, J = 8.2 Hz, 2H), 6.91 (d, J = 8.5 Hz, 2H), 6.75 (d, J = 7.7 Hz, 1H), 6.33 (s, 1H), 3.88 (s, 3H), 3.74 (s, 3H), 3.49 (br s, 4H), 3.16 (t, J = 8.5 Hz, 2H), 2.85 (s, 3H), 2.59 (t, J = 8.5 Hz, 2H), 1.23–1.08 (m, 2H). 13C-NMR (100 MHz, DMSO-d6): δ 155.8, 153.3, 153.1, 150.1, 149.5, 133.2, 130.6, 121.7 (2C), 118.5, 115.1 (2C), 110.9, 101.6, 99.8, 61.8, 59.0, 58.0, 56.2, 55.8, 52.3, 46.9, 26.4. ESI-MS: m/z 393.2246 [M+H]+.
N-(4-Isopropylphenyl)-5-methoxy-2-(4-methyl-1,4-diazepan-1-yl)quinolin-4-amine (5n): Yellow solid, Mp: 278–280 °C. 81% yield from 4h. 1H-NMR (400 MHz, DMSO-d6): δ 8.82 (s, 1H),7.62 (d, J = 9.0 Hz, 2H), 7.35 (t, J = 8.1 Hz, 1H), 7.11 (d, J = 8.2 Hz, 1H), 6.73 (d, J = 9.0 Hz, 2H), 6.67 (d, J = 7.9 Hz, 1H), 6.32 (s, 1H), 3.85 (s, 3H), 3.49 (br s, 4H), 3.16 (t, J = 8.5 Hz, 2H), 2.88 (s, 1H), 2.85 (s, 3H), 2.59 (t, J = 8.5 Hz, 2H), 1.20 (d, J = 8.5 Hz, 6H), 1.08–0.97 (m, 2H). 13C-NMR (100 MHz, DMSO-d6): δ 155.8, 153.1, 150.1, 149.5, 138.4, 138.1, 130.6, 126.9 (2C), 120.1 (2C), 118.5, 110.9, 101.6, 99.8, 61.8, 59.0, 58.0, 56.2, 52.3, 46.9, 33.2, 26.4, 23.3 (2C). ESI-MS: m/z 405.2610 [M+H]+.
5-Methoxy-N-(2-methoxyphenyl)-2-(4-methyl-1,4-diazepan-1-yl)quinolin-4-amine (5o): Yellow solid, Mp: 265–267 °C. 81% yield from 4i. 1H-NMR (400 MHz, DMSO-d6): δ 8.89 (s, 1H), 8.67 (s, 1H), 8.23 (s, 1H), 7.38 (t, J = 7.5 Hz, 1H), 7.18 (d, J = 7.8 Hz, 1H), 7.01 (s, 1H), 6.93 (s, 2H), 6.73 (s, 1H), 3.87 (s, 6H), 3.49 (br s, 4H), 3.16 (t, J = 8.5 Hz, 2H), 2.85 (s, 3H), 2.59 (t, J = 8.5 Hz, 2H), 1.23–1.08 (m, 2H). 13C-NMR (100 MHz, DMSO-d6): δ 155.8, 153.1, 150.1, 149.5, 147.4, 132.6, 130.6, 122.6, 121.8, 118.5, 113.4, 112.0, 110.9, 101.6, 99.8, 61.8, 59.0, 58.0, 56.2, 55.8, 52.3, 46.9, 26.4. ESI-MS: m/z 393.2246 [M+H]+.
N1-(5-Methoxy-2-(4-methyl-1,4-diazepan-1-yl)quinolin-4-yl)-N4,N4-dimethylbenzene-1,4-diamine (5p): Yellow solid, Mp: 268–270 °C. 81% yield from 4l. 1H-NMR (400 MHz, DMSO-d6): δ 8.82 (s, 1H),7.62 (d, J = 9.0 Hz, 2H), 7.35 (t, J = 8.1 Hz, 1H), 7.11 (d, J = 8.2 Hz, 1H), 6.73 (d, J = 9.0 Hz, 2H), 6.67 (d, J = 7.9 Hz, 1H), 6.32 (s, 1H), 3.85 (s, 3H), 3.49 (br s, 4H), 3.16 (t, J = 8.5 Hz, 2H), 3.02 (s, 6H), 2.85 (s, 3H), 2.59 (t, J = 8.5 Hz, 2H), 1.08–0.97 (m, 2H). 13C-NMR (100 MHz, DMSO-d6): δ 155.8, 153.1, 150.1, 149.5, 138.4, 138.1, 130.6, 126.9 (2C), 120.1 (2C), 118.5, 110.9, 101.6, 99.8, 61.8, 59.0, 58.0, 56.2, 52.3, 46.9, 26.4, 23.3 (2C). ESI-MS: m/z 406.2562 [M+H]+.
5-Methoxy-N-(4-methoxy-2-methylphenyl)-2-(4-methyl-1,4-diazepan-1-yl)quinolin-4-amine (5q): Yellow solid, Mp: 258–260 °C. 88% yield from 4j. 1H-NMR (400 MHz, DMSO-d6): δ 8.85 (s, 1H), 7.70 (s, 2H), 7.42 (t, J = 7.5 Hz, 1H), 7.18 (d, J = 8.2 Hz, 1H), 6.91 (d, J = 8.5 Hz, 1H), 6.75 (d, J = 7.7 Hz, 1H), 6.33 (s, 1H), 3.88 (s, 3H), 3.74 (s, 3H), 3.49 (br s, 4H), 3.16 (t, J = 8.5 Hz, 2H), 2.85 (s, 3H), 2.59 (t, J = 8.5 Hz, 2H), 2.12 (s, 3H), 1.23–1.08 (m, 2H). 13C-NMR (100 MHz, DMSO-d6): δ 155.8, 153.1, 151.1, 150.1, 149.5, 134.3, 130.6, 130.0, 121.4, 118.5, 117.2, 112.1, 110.9, 101.6, 99.8, 66.3 (2C), 56.2, 55.8, 53.7 (2C), 17.9. ESI-MS: m/z 407.2402 [M+H]+.
5-Methoxy-2-(4-methyl-1,4-diazepan-1-yl)-N-(p-tolyl)quinolin-4-amine (5r): Yellow solid, Mp: 246–248 °C. 88% yield from 4m. 1H-NMR (400 MHz, DMSO-d6): δ 7.23 (d, J = 9.0 Hz, 4H), 7.35 (t, J = 8.1 Hz, 1H), 7.11 (d, J = 8.2 Hz, 1H), 6.73 (d, J = 9.0 Hz, 1H), 6.67 (d, J = 7.9 Hz, 1H), 6.32 (s, 1H), 3.85 (s, 3H), 3.51 (s, 2H), 3.45 (br s, 2H), 3.03–2.88 (m, 2H), 2.84 (s, 6H), 2.58 (s, 2H), 1.23–1.08 (m, 2H). 13C-NMR (100 MHz, DMSO-d6): δ 155.8, 153.3, 153.1, 150.1, 149.5, 133.2, 131.2, 129.8 (2C), 118.5, 116.3 (2C), 110.9, 101.6, 99.8, 61.8, 59.0, 58.0, 56.2, 52.3, 46.9, 26.4, 21.3. ESI-MS: m/z 377.2297 [M+H]+.
5-Methoxy-N-(3-methoxyphenyl)-2-(4-methyl-1,4-diazepan-1-yl)quinolin-4-amine (5s): Yellow solid, Mp: 244–246 °C. 86% yield from 4k. 1H-NMR (400 MHz, DMSO-d6): δ7.73–7.45 (m, 2H), 7.25 (dd, J = 14.5 HZ, 8.0 HZ, 3H), 6.85 (d, J =7.5 HZ, 1H), 6.62 (s, 1H), 6.58 (s, 1H), 6.47 (s, 1H), 3.90 (s, 3H), 3.51 (s, 2H), 3.45 (br s, 2H), 3.03–2.88 (m, 2H), 2.84 (s, 6H), 2.58 (s, 2H), 1.23–1.08 (m, 2H). 13C-NMR (100 MHz, DMSO-d6): δ 155.8, 153.1, 150.1, 149.5, 147.4, 132.6, 130.6, 122.6, 121.8, 118.5, 113.4, 112.0, 110.9, 101.6, 99.8, 61.8, 59.0, 58.0, 56.2, 55.8, 52.3, 46.9, 26.4. ESI-MS: m/z 393.2246 [M+H]+.
5-Methoxy-2-(4-methyl-1,4-diazepan-1-yl)-N-(pyrimidin-5-yl)quinolin-4-amine (5t): Yellow solid, Mp: 254–256 °C. 88% yield from 4n. 1H-NMR (400 MHz, DMSO-d6): δ 8.90 (s, 1H), 8.47 (d, J = 8.5 Hz, 2H), 7.19–7.17 (m, 2H), 6.91 (s, 1H), 6.87 (t, J = 8.5 Hz, 1H), 6.19 (d, J = 8.5 Hz, 1H), 3.90 (s, 3H), 3.49 (br s, 4H), 3.16 (t, J = 8.5 Hz, 2H), 2.85 (s, 3H), 2.59 (t, J = 8.5 Hz, 2H), 1.23–1.08 (m, 2H). 13C-NMR (100 MHz, DMSO-d6): δ 174.2, 150.5, 147.6, 147.4, 144.8, 143.7 (2C), 142.9, 131.5, 120.2, 105.7, 102.0, 94.6, 61.8, 59.0, 57.6, 56.2, 51.9, 46.9, 26.4. ESI-MS: m/z 365.2045 [M+H]+.
5-Methoxy-2-(4-methyl-1,4-diazepan-1-yl)-N-(4-nitrophenyl)quinolin-4-amine (5u): Yellow solid, Mp: 283–285 °C. 88% yield from 4o. 1H-NMR (400 MHz, DMSO-d6): δ 8.85 (s, 1H), 8.03 (d, J = 8.5 Hz, 2H), 7.70 (s, 1H), 7.42 (t, J = 7.5 Hz, 1H), 7.18 (d, J = 8.2 Hz, 2H), 6.75 (d, J = 7.7 Hz, 1H), 6.33 (s, 1H), 3.90 (s, 3H), 3.49 (br s, 4H), 3.16 (t, J = 8.5 Hz, 2H), 2.85 (s, 3H), 2.59 (t, J = 8.5 Hz, 2H), 1.23–1.08 (m, 2H). 13C-NMR (100 MHz, DMSO-d6): δ 174.2, 152.0, 150.5, 147.6, 144.8, 137.9, 131.5, 124.7 (2C), 120.2, 119.2 (2C), 105.7, 102.0, 94.6, 61.8, 59.0, 57.6, 56.2, 51.9, 46.9, 26.4. ESI-MS: m/z 408.1991 [M+H]+.
N-(3,5-Bis(trifluoromethyl)phenyl)-5-methoxy-2-(4-methyl-1,4-diazepan-1-yl)quinolin-4-amine (5v): Yellow solid, Mp: 297–299 °C. 78% yield from 4p. 1H-NMR (400 MHz, DMSO-d6): δ 8.85 (s, 1H), 8.03 (d, J = 8.5 Hz, 2H), 7.56 (s, 2H), 7.49 (s, 1H), 7.42 (t, J = 7.5 Hz, 1H), 6.33 (s, 1H), 3.90 (s, 3H), 3.49 (br s, 4H), 3.16 (t, J = 8.5 Hz, 2H), 2.85 (s, 3H), 2.59 (t, J = 8.5 Hz, 2H), 1.23–1.08 (m, 2H). 13C-NMR (100 MHz, DMSO-d6): δ 174.2, 150.5, 147.6, 144.8, 143.0, 132.1 (2C), 131.5, 124.4 (2C), 120.2, 119.3 (2C), 113.1, 105.7, 102.0, 94.6, 61.8, 59.0, 57.6, 56.2, 51.9, 46.9, 26.4. ESI-MS: m/z 499.1888 [M+H]+.
3.10. Synthesis of 7a–i
Compounds 7a‒i were synthesized according to the literature method [8].
3.11. Synthesis of 8a–i
Compounds 8a‒i were synthesized according to the literature method [10].
3.12. General Procedure for the Synthesis of 9a–i
The general procedure for the synthesis of 9a–i was the same as that described for 5a–v.
6-Methoxy-2-(4-methyl-1,4-diazepan-1-yl)-N-(1-methylpiperidin-4-yl)quinolin-4-amine (9a): Yellow solid, Mp: 266–268 °C. 75% yield from 8a. 1H-NMR (400 MHz, DMSO-d6): δ 7.55 (d, J = 8 Hz, 1H), 7.26 (s, 1H), 6.97 (d, J = 8 Hz, 1H), 6.36 (s, 1H), 5.65 (s, 1H), 3.81 (s, 3H), 3.79 (s, 1H), 3.66 (t, J = 8 Hz, 2H), 3.48 (s, 3H), 2.61 (s, 4H), 2.25–2.05 (m, 4H), 2.05–1.76 (m, 3H), 1.58 (d, J = 8 Hz, 2H), 1.35–0.98 (m, 4H), 0.84 (d, J = 7.4 Hz, 2H). 13C-NMR (100 MHz, DMSO-d6): δ 171.7, 154.7, 152.5, 145.4, 129.6 122.7, 116.7, 101.6, 96.0, 61.8, 59.0, 57.6, 56.8, 56.2, 54.5 (2C), 51.9, 46.9, 30.3 (2C), 26.4. ESI-MS: m/z 384.2719 [M+H]+.
7-Methoxy-2-(4-methyl-1,4-diazepan-1-yl)-N-(1-methylpiperidin-4-yl)quinolin-4-amine (9b): Yellow solid, Mp: 261–263 °C. 65% yield from 8b. 1H-NMR (400 MHz, DMSO-d6): δ 7.42 (d, J = 8 Hz, 1H), 7.09 (s, 1H), 6.85 (d, J = 8 Hz, 1H), 6.40 (s, 1H), 5.65 (s, 1H), 3.87 (s, 3H), 3.79 (s, 1H), 3.62 (t, J = 8 Hz, 2H), 3.48 (s, 3H), 2.65 (s, 4H), 2.15–2.05 (m, 4H), 2.05–1.66 (m, 3H), 1.45 (d, J = 8 Hz, 2H), 1.35–0.88 (m, 4H), 0.84 (d, J = 7.4 Hz, 2H). 13C-NMR (100 MHz, DMSO-d6): δ 174.4, 155.6, 150.7, 147.8, 122.5, 112.9, 109.2, 106.4, 93.3, 61.8, 59.0, 57.6, 56.8, 56.2, 54.5 (2C), 51.9, 46.9, 30.3 (2C), 26.4. ESI-MS: m/z 384.2719 [M+H]+.
8-Methoxy-2-(4-methyl-1,4-diazepan-1-yl)-N-(1-methylpiperidin-4-yl)quinolin-4-amine (9c): Yellow solid, Mp: 251–253 °C. 69% yield from 8c. 1H-NMR (400 MHz, DMSO-d6): δ 8.01 (d, J = 8 Hz, 1H), 7.33 (s, 1H), 7.17 (d, J = 8 Hz, 1H), 6.38 (s, 1H), 5.65 (s, 1H), 4.07 (s, 3H), 3.79 (s, 1H), 3.68 (t, J = 8 Hz, 2H), 3.56 (s, 3H), 2.45 (s, 4H), 2.18–2.05 (m, 4H), 2.05–1.66 (m, 3H), 1.45 (d, J = 8 Hz, 2H), 1.35–0.88 (m, 4H), 0.84 (d, J = 7.4 Hz, 2H). 13C-NMR (100 MHz, DMSO-d6): δ 173.0, 155.8, 155.2, 141.6, 117.4, 116.5, 116.3, 107.5, 95.8, 61.8, 59.0, 57.6, 56.8, 56.2, 54.5 (2C), 51.9, 46.9, 30.3 (2C), 26.4. ESI-MS: m/z 384.2717 [M+H]+.
5,6-Dimethoxy-2-(4-methyl-1,4-diazepan-1-yl)-N-(1-methylpiperidin-4-yl)quinolin-4-amine (9d): Yellow solid, Mp: 272–274 °C. 73% yield from 8d. 1H-NMR (400 MHz, DMSO-d6): δ 7.00 (d, J = 8 Hz, 1H), 6.81 (s, 1H), 6.23 (d, J = 8 Hz, 1H), 5.65 (s, 1H), 3.83 (s, 6H), 3.79 (s, 1H), 3.68 (t, J = 8 Hz, 2H), 3.56 (s, 3H), 2.45 (s, 4H), 2.19–2.09 (m, 4H), 2.09–1.66 (m, 3H), 1.43 (d, J = 8 Hz, 2H), 1.35–0.88 (m, 4H), 0.84 (d, J = 7.4 Hz, 2H). 13C-NMR (100 MHz, DMSO-d6): δ 172.0, 151.1, 148.6, 146.2, 133.8, 122.5, 121.7, 106.7, 95.1, 61.8, 59.0, 57.6, 56.8, 56.2, 55.8, 54.5 (2C), 51.9, 46.9, 30.3 (2C), 26.4. ESI-MS: m/z 414.2824 [M+H]+.
5,7-Dimethoxy-2-(4-methyl-1,4-diazepan-1-yl)-N-(1-methylpiperidin-4-yl)quinolin-4-amine (9e): Yellow solid, Mp: 272–274 °C. 73% yield from 8e. 1H-NMR (400 MHz, DMSO-d6): δ 6.54 (s, 1H), 6.27 (s, 1H), 6.22 (s, 1H), 5.65 (s, 1H), 3.81 (s, 6H), 3.77 (s, 1H), 3.69 (t, J = 8 Hz, 2H), 3.57 (s, 3H), 2.45 (s, 4H), 2.19–2.09 (m, 4H), 2.09–1.66 (m, 3H), 1.43 (d, J = 8 Hz, 2H), 1.35–0.88 (m, 4H), 0.84 (d, J = 7.4 Hz, 2H). 13C-NMR (100 MHz, DMSO-d6): δ 174.4, 155.8, 154.7, 151.5, 149.5, 101.4, 98.5, 94.6, 92.4, 61.8, 59.0, 57.6, 56.8, 56.2, 55.8, 54.5 (2C), 51.9, 46.9, 30.3 (2C), 26.4. ESI-MS: m/z 414.2822 [M+H]+.
5,8-Dimethoxy-2-(4-methyl-1,4-diazepan-1-yl)-N-(1-methylpiperidin-4-yl)quinolin-4-amine (9f): Yellow solid, Mp: 276–278 °C. 69% yield from 8f. 1H-NMR (400 MHz, DMSO-d6): δ 6.25 (s, 1H), 5.97 (d, J = 8 Hz, 1H), 5.88 (d, J = 8 Hz, 1H), 5.65 (s, 1H), 3.80 (s, 6H), 3.77 (s, 1H), 3.69 (t, J = 8 Hz, 2H), 3.57 (s, 3H), 2.45 (s, 4H), 2.19–2.09 (m, 4H), 2.09–1.66 (m, 3H), 1.43 (d, J = 8 Hz, 2H), 1.35–0.88 (m, 4H), 0.84 (d, J = 7.4 Hz, 2H). 13C-NMR (100 MHz, DMSO-d6): δ 173.3, 149.1, 147.9, 142.4, 139.7, 107.3, 106.5, 99.3, 94.9, 61.8, 59.0, 57.6, 56.8, 56.2, 55.8, 54.5 (2C), 51.9, 46.9, 30.3 (2C), 26.4. ESI-MS: m/z 414.2824 [M+H]+.
6,7-Dimethoxy-2-(4-methyl-1,4-diazepan-1-yl)-N-(1-methylpiperidin-4-yl)quinolin-4-amine (9g): Yellow solid, Mp: 253–255 °C. 69% yield from 8g. 1H-NMR (400 MHz, DMSO-d6): δ 6.73 (s, 1H), 6.54 (s, 1H), 6.26 (s, 1H), 5.65 (s, 1H), 3.80 (s, 6H), 3.77 (s, 1H), 3.69 (t, J = 8 Hz, 2H), 3.57 (s, 3H), 2.45 (s, 4H), 2.19–2.09 (m, 4H), 2.09–1.66 (m, 3H), 1.43 (d, J = 8 Hz, 2H), 1.35–0.88 (m, 4H), 0.84 (d, J = 7.4 Hz, 2H). 13C-NMR (100 MHz, DMSO-d6): δ 172.2, 154.4, 151.2, 146.4, 145.6, 112.4, 105.8, 100.0, 93.8, 61.8, 59.0, 57.6, 56.8, 56.1 (2C), 54.5 (2C), 51.9, 46.9, 30.3 (2C), 26.4. ESI-MS: m/z 414.2820 [M+H]+.
6,8-Dimethoxy-2-(4-methyl-1,4-diazepan-1-yl)-N-(1-methylpiperidin-4-yl)quinolin-4-amine (9h): Yellow solid, Mp: 258–260 °C. 73% yield from 8h. 1H-NMR (400 MHz, DMSO-d6): δ 6.71 (s, 1H), 6.33 (s, 1H), 6.24 (s, 1H), 5.65 (s, 1H), 3.93 (s, 6H), 3.77 (s, 1H), 3.69 (t, J = 8 Hz, 2H), 3.57 (s, 3H), 2.45 (s, 4H), 2.19–2.09 (m, 4H), 2.09–1.66 (m, 3H), 1.43 (d, J = 8 Hz, 2H), 1.35–0.88 (m, 4H), 0.84 (d, J = 7.4 Hz, 2H). 13C-NMR (100 MHz, DMSO-d6): δ 170.8, 156.1, 154.8, 154.0, 137.3, 117.5, 96.3, 93.7, 93.0, 61.8, 59.0, 57.6, 56.8, 56.1 (2C), 55.8 (2C), 51.9, 46.9, 30.3 (2C), 26.4. ESI-MS: m/z 414.2824 [M+H]+.
7,8-Dimethoxy-2-(4-methyl-1,4-diazepan-1-yl)-N-(1-methylpiperidin-4-yl)quinolin-4-amine (9i): Yellow solid, Mp: 258–260 °C. 73% yield from 8i. 1H-NMR (400 MHz, DMSO-d6): δ 6.87 (d, J = 8 Hz, 1H), 6.70 (d, J = 8 Hz, 1H), 6.28 (s, 1H), 5.65 (s, 1H), 3.83 (s, 6H), 3.77 (s, 1H), 3.69 (t, J = 8 Hz, 2H), 3.57 (s, 3H), 2.45 (s, 4H), 2.19–2.09 (m, 4H), 2.09–1.66 (m, 3H), 1.43 (d, J = 8 Hz, 2H), 1.35–0.88 (m, 4H), 0.84 (d, J = 7.4 Hz, 2H). 13C-NMR (100 MHz, DMSO-d6): δ 173.5, 154.9, 142.6, 141.8 (2C), 114.6, 112.2, 108.4, 93.6, 93.0, 61.8, 60.9, 59.0, 57.6, 56.1, 55.8 (2C), 51.9, 46.9, 30.3 (2C), 26.4. ESI-MS: m/z 414.2824 [M+H]+.
4. Conclusions
In summary, a series of quinoline derivatives were synthesized, and SAR analysis led to the discovery of a number of compounds that showed good activities in both enzymatic and cellular assays. The most active compound, 5k, exhibited an IC50 value of 1.2 μM against EZH2, decreased global H3K27me3 level in cells and also showed good selectivity for EZH2 against several selected protein methyltransferases. Compound 5k could be a lead compound deserving further structural optimization due to the fact that it represents a new scaffold and possesses a low molecular weight.
Acknowledgments
This work was supported by National Science and Technology Major Projects (2011ZX09102-001-13).
Author Contributions
Ying-Lan Zhao, Sheng-Yong Yang, Pu Xiang and Bo Yang, designed research; Hui Jie, Hui-Juan Wang and Jing Hu performed biological data; Pu Xiang, Yang Zhou and Jian Hu performed research and analyzed the data; Pu Xiang wrote the paper. All authors read and approved the final manuscript.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Varambally, S.; Dhanasekaran, S.M.; Zhou, M.; Barrette, T.R.; Kumar-Sinha, C.; Sanda, M.G.; Ghosh, D.; Pienta, K.J.; Sewalt, R.G.A.B.; Otte, A.P.; et al. The polycomb group protein EZH2 is involved in progression of prostate cancer. Nature 2002, 419, 624–629. [Google Scholar] [PubMed]
- Kleer, C.G.; Cao, Q.; Varambally, S.; Shen, R.; Ota, I.; Tomlins, S.A.; Ghosh, D.; Sewalt, R.G.A.B.; Otte, A.P.; Hayes, D.F.; et al. EZH2 is a marker of aggressive breast cancer and promotes neoplastic transformation of breast epithelial cells. Proc. Natl. Acad. Sci. USA 2003, 100, 11606–11611. [Google Scholar] [CrossRef] [PubMed]
- Wagener, N.; Macher-Goeppinger, S.; Pritsch, M.; Hüsing, J.; Hoppe-Seyler, K.; Schirmacher, P.; Pfitzenmaier, J.; Haferkamp, A.; Hoppe-Seyler, F.; Hohenfellner, M. Enhancer of zeste homolog 2 (EZH2) expression is an independent prognostic factor in renal cell carcinoma. BMC Cancer 2010, 10, 524. [Google Scholar] [CrossRef] [PubMed]
- Takawa, M.; Masuda, K.; Kunizaki, M.; Daigo, Y.; Takagi, K.; Iwai, Y.; Cho, H.S.; Toyokawa, G.; Yamane, Y.; Maejima, K.; et al. Validation of the histone methyltransferase EZH2 as a therapeutic target for various types of human cancer and as a prognostic marker. Cancer Sci. 2011, 102, 1298–1305. [Google Scholar] [CrossRef] [PubMed]
- Nasveschuk, C.G.; Gagnon, A.; Garapaty-Rao, S.; Balasubramanian, S.; Campbell, R.; Lee, C.; Zhao, F.; Bergeron, L.; CμMmings, R.; Trojer, P.; et al. Discovery and Optimization of Tetramethylpiperidinyl Benzamides as Inhibitors of EZH2. ACS. Med. Chem. Lett. 2014, 5, 378–383. [Google Scholar] [CrossRef] [PubMed]
- Bradley, W.D.; Arora, S.; Busby, J.; Balasubramanian, S.; Gehling, V.S.; Nasveschuk, C.G.; Vaswani, R.G.; Yuan, C.C.; Hatton, C.; Zhao, F.; et al. EZH2 inhibitor efficacy in non-Hodgkin's lymphoma does not require suppression of H3K27 monomethylation. Chem. Biol. 2014, 21, 1463–1475. [Google Scholar] [CrossRef] [PubMed]
- Kubicek, S.; O’Sullivan, R.J.; August, E.M.; Hickey, E.R.; Zhang, Q.; Teodoro, M.L.; Rea, S.; Mechtler, K.; Kowalski, J.A.; Homon, C.A.; et al. Reversal of H3K9me2 by a small-molecule inhibitor for the G9a histone methyltransferase. Mol. Cell 2007, 25, 473–481. [Google Scholar] [CrossRef] [PubMed]
- Parsy, C.C.; Alexandre, F.; Leroy, F.; Convard, T.; Surleraux, D.; Derock, M.; Meillon, J. Macrocyclic Serine Protease Inhibitors Useful against Viral Infections, Particularly hcv. WO2011017389A1, 10 February 2011. [Google Scholar]
- Li, H.J.; Guillot, R.; Gandon, V. A Gallium-Catalyzed Cycloisomerization/Friedel−Crafts Tandem. J. Org. Chem. 2010, 75, 8435–8449. [Google Scholar] [CrossRef] [PubMed]
- Wynne, G.M.; de Moor, O.; Johnson, P.D.; Vickers, R.J. Use of Compounds for Preparing Anti-Tuberculosis Agents. WO2009001060A2, 31 December 2008. [Google Scholar]
- Liu, F.; Chen, X.; Allali-Hassani, A.; Quinn, A.M.; Wasney, G.A.; Dong, A.; Barsyte, D.; Kozieradzki, I.; Senisterra, G.; Chau, I.; et al. Discovery of a 2,4-Diamino-7-aminoalkoxyquinazoline as a Potent and Selective Inhibitor of Histone Lysine Methyltransferase G9a. J. Med. Chem. 2009, 52, 7950–7953. [Google Scholar] [CrossRef] [PubMed]
- Knutson, S.K.; Wigle, T.J.; Warholic, N.M.; Sneeringer, C.J.; Allain, C.J.; Klaus, C.R.; Sacks, J.D.; Raimondi, A.; Majer, C.R.; Song, J.; et al. A selective inhibitor of EZH2 blocks H3K27 methylation and kills mutant lymphoma cells. Nat. Chem. Biol. 2012, 8, 890–896. [Google Scholar]
- Sample Availability: Samples of the compounds 5a–v, 9a–i are available from the authors.
© 2015 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution license ( http://creativecommons.org/licenses/by/4.0/).
