Transcriptome Analysis Reveals the Effect of Low NaCl Concentration on Osmotic Stress and Type III Secretion System in Vibrio parahaemolyticus
Abstract
:1. Introduction
2. Results
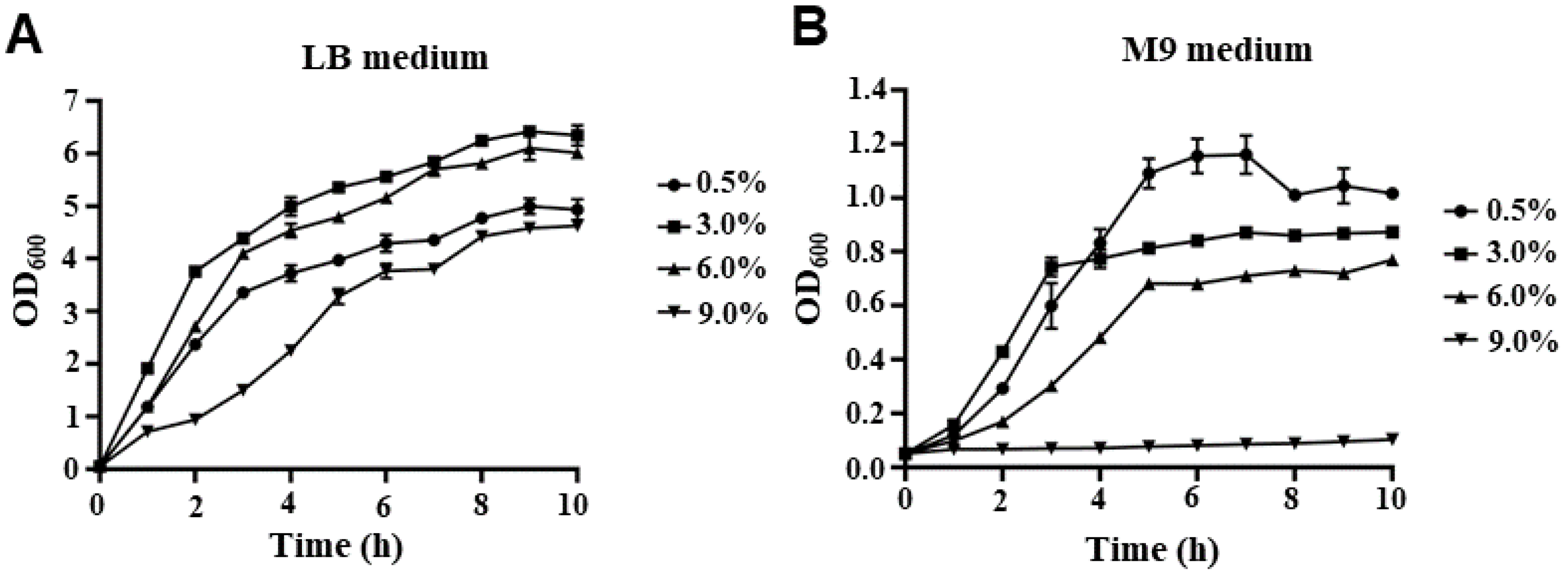
2.1. The Growth Curve of V. parahaemolyticus under Different NaCl Concentrations
2.2. Transcriptome Analysis of V. parahaemolyticus in M9 Medium with 0.5% or 3% NaCl
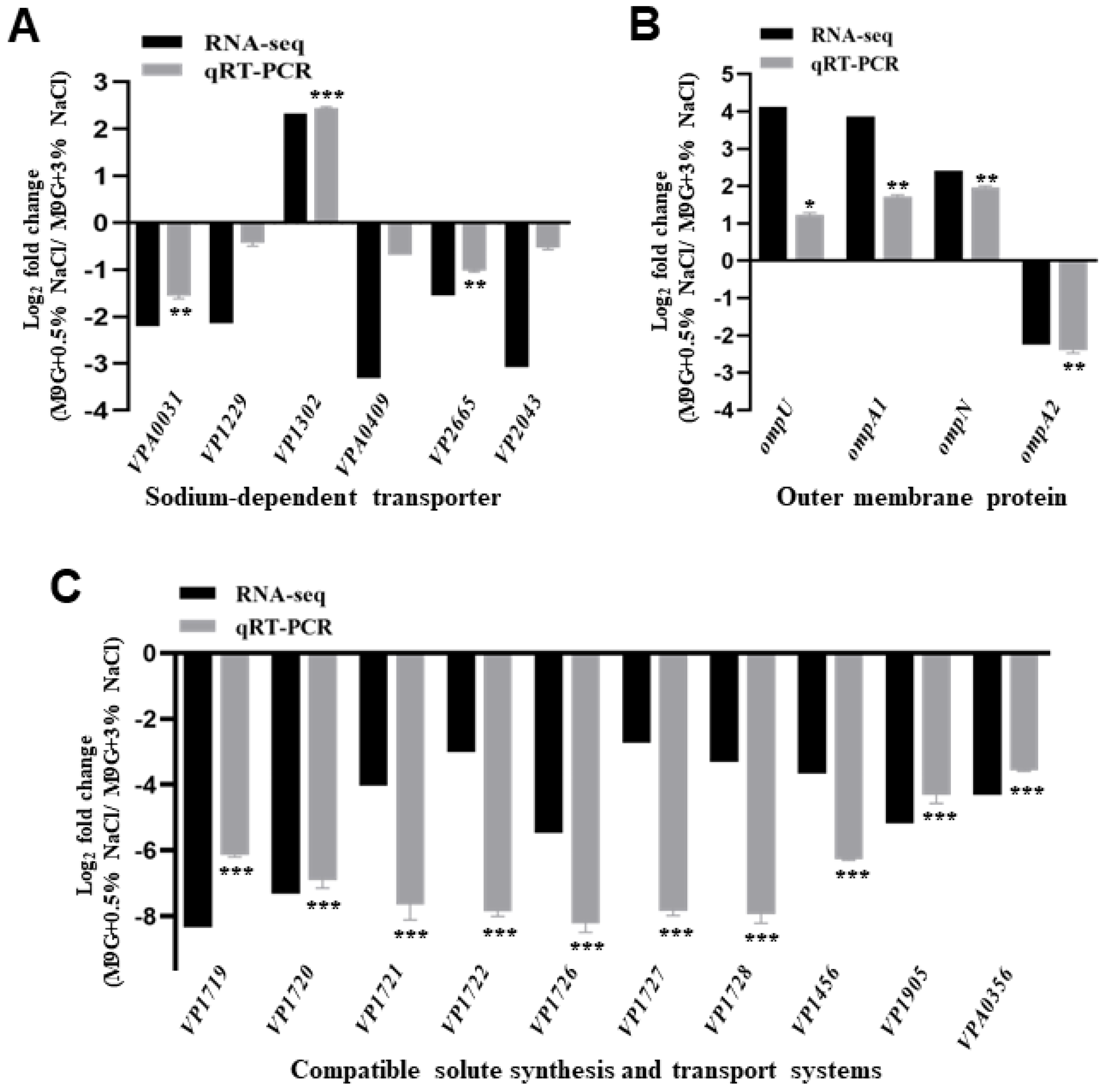
2.3. The Expression Levels of Osmotic Response Genes under Low NaCl Concentration
2.4. Low Concentration of NaCl Induces High Expression of T3SS1 and Contributes to the Cytotoxicity of V. parahaemolyticus
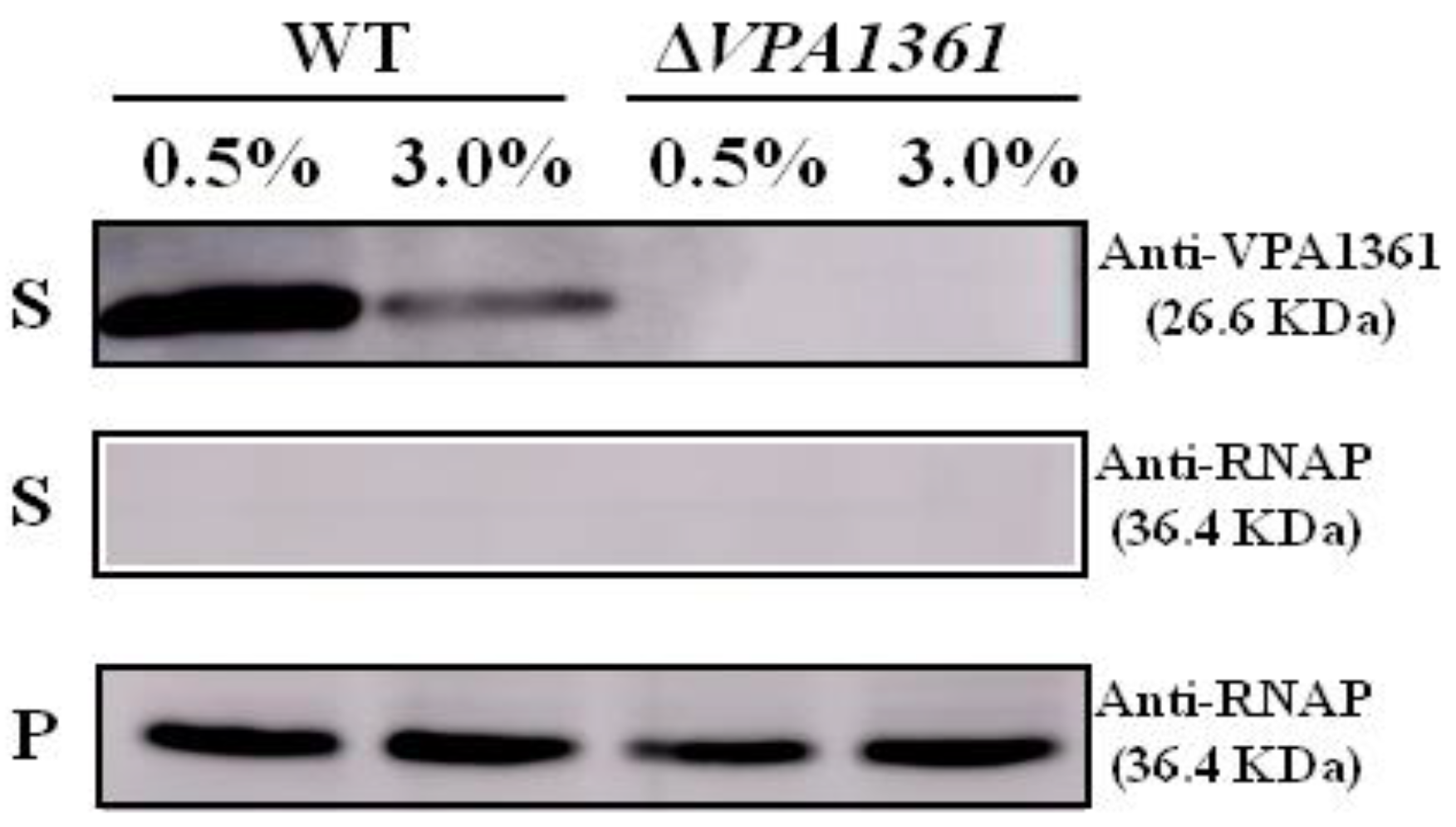
2.5. A Low NaCl Concentration Induces the Secretion of the T3SS2 Translocon Protein VPA1361
3. Discussion
4. Materials and Methods
4.1. Bacterial Strain and Culture Conditions
4.2. Growth Curve
4.3. RNA-Seq Analysis
4.4. qRT-PCR
4.5. Cytotoxicity Analysis
4.6. Western Blotting
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Yang, C.; Li, Y.; Jiang, M.; Wang, L.; Jiang, Y.; Hu, L.; Shi, X.; He, L.; Cai, R.; Wu, S.; et al. Outbreak dynamics of foodborne pathogen Vibrio parahaemolyticus over a seventeen year period implies hidden reservoirs. Nat. Microbiol. 2022, 7, 1221–1229. [Google Scholar] [CrossRef]
- Li, H.; Tang, R.; Lou, Y.; Cui, Z.; Chen, W.; Hong, Q.; Zhang, Z.; Malakar, P.K.; Pan, Y.; Zhao, Y. A Comprehensive Epidemiological Research for Clinical Vibrio parahaemolyticus in Shanghai. Front. Microbiol. 2017, 8, 1043. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Velazquez-Roman, J.; Leon-Sicairos, N.; de Jesus Hernandez-Diaz, L.; Canizalez-Roman, A. Pandemic Vibrio parahaemolyticus O3:K6 on the American continent. Front. Cell. Infect. Microbiol. 2014, 3, 110. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- McLaughlin, J.B.; DePaola, A.; Bopp, C.A.; Martinek, K.A.; Napolilli, N.P.; Allison, C.G.; Murray, S.L.; Thompson, E.C.; Bird, M.M.; Middaugh, J.P. Outbreak of Vibrio parahaemolyticus gastroenteritis associated with Alaskan oysters. N. Engl. J. Med. 2005, 353, 1463–1470. [Google Scholar] [CrossRef] [Green Version]
- Ali, S.; Hossain, M.; Azad, A.B.; Siddique, A.B.; Moniruzzaman, M.; Ahmed, M.A.; Amin, M.B.; Islam, M.S.; Rahman, M.M.; Mondal, D.; et al. Diversity of Vibrio parahaemolyticus in marine fishes of Bangladesh. J. Appl. Microbiol. 2021, 131, 2539–2551. [Google Scholar] [CrossRef] [PubMed]
- Yu, W.T.; Jong, K.J.; Lin, Y.R.; Tsai, S.E.; Tey, Y.H.; Wong, H.C. Prevalence of Vibrio parahaemolyticus in oyster and clam culturing environments in Taiwan. Int. J. Food Microbiol. 2013, 160, 185–192. [Google Scholar] [CrossRef]
- Dong, X.; Li, Z.; Wang, X.; Zhou, M.; Lin, L.; Zhou, Y.; Li, J. Characteristics of Vibrio parahaemolyticus isolates obtained from crayfish (Procambarus clarkii) in freshwater. Int. J. Food Microbiol. 2016, 238, 132–138. [Google Scholar] [CrossRef]
- Sadat, A.; El-Sherbiny, H.; Zakaria, A.; Ramadan, H.; Awad, A. Prevalence, antibiogram and virulence characterization of Vibrio isolates from fish and shellfish in Egypt: A possible zoonotic hazard to humans. J. Appl. Microbiol. 2021, 131, 485–498. [Google Scholar] [CrossRef]
- Jiang, H.; Yu, T.; Yang, Y.; Yu, S.; Wu, J.; Lin, R.; Li, Y.; Fang, J.; Zhu, C. Co-occurrence of antibiotic and heavy metal resistance and sequence type diversity of Vibrio parahaemolyticus isolated from Penaeus vannamei at freshwater farms, seawater farms, and markets in Zhejiang Province, China. Front. Microbiol. 2020, 11, 1294. [Google Scholar] [CrossRef]
- Lee, L.H.; Ab Mutalib, N.S.; Law, J.W.; Wong, S.H.; Letchumanan, V. Discovery on antibiotic resistance patterns of Vibrio parahaemolyticus in Selangor reveals carbapenemase producing Vibrio parahaemolyticus in marine and freshwater fish. Front. Microbiol. 2018, 9, 2513. [Google Scholar] [CrossRef]
- Meletis, G. Carbapenem resistance: Overview of the problem and future perspectives. Ther. Adv. Infect. Dis. 2016, 3, 15–21. [Google Scholar] [CrossRef] [Green Version]
- Chen, H.; Dong, S.; Yan, Y.; Zhan, L.; Zhang, J.; Chen, J.; Zhang, Z.; Zhang, Y.; Mei, L. Prevalence and population analysis of Vibrio parahaemolyticus isolated from freshwater fish in Zhejiang Province, China. Foodborne Pathog. Dis. 2021, 18, 139–146. [Google Scholar] [CrossRef] [PubMed]
- Wang, B.; Jun, L.V. Pathogen identification of food poison caused by small lobster contaminated by Vibrio parahaemolyticus. Chin. J. Health Lab. 2011, 21, 1969–1970. [Google Scholar] [CrossRef]
- Chakraborty, S.; Kenney, L.J. A new role of OmpR in acid and osmotic stress in Salmonella and E. coli. Front. Microbiol. 2018, 9, 2656. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhou, K.; George, S.M.; Métris, A.; Li, P.L.; Baranyi, J. Lag phase of Salmonella enterica under osmotic stress conditions. Appl. Environ. Microbiol. 2011, 77, 1758–1762. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gregory, G.J.; Morreale, D.P.; Boyd, E.F. CosR is a global regulator of the osmotic stress response with widespread distribution among bacteria. Appl. Environ. Microbiol. 2020, 86, e00120-20. [Google Scholar] [CrossRef] [PubMed]
- Allakhverdiev, S.I.; Murata, N. Salt stress inhibits photosystems II and I in cyanobacteria. Photosynth. Res. 2008, 98, 529–539. [Google Scholar] [CrossRef]
- Yang, L.; Zhan, L.; Han, H.; Gao, H.; Guo, Z.; Qin, C.; Yang, R.; Liu, X.; Zhou, D. The low-salt stimulon in Vibrio parahaemolyticus. Int. J. Food Microbiol. 2010, 137, 49–54. [Google Scholar] [CrossRef]
- da Costa, M.S.; Santos, H.; Galinski, E.A. An overview of the role and diversity of compatible solutes in Bacteria and Archaea. Adv. Biochem. Eng. Biotechnol. 1998, 61, 117–153. [Google Scholar] [CrossRef] [PubMed]
- Welsh, D.T. Ecological significance of compatible solute accumulation by micro-organisms: From single cells to global climate. FEMS Microbiol. Rev. 2000, 24, 263–290. [Google Scholar] [CrossRef]
- Sleator, R.D.; Hill, C. Bacterial osmoadaptation: The role of osmolytes in bacterial stress and virulence. FEMS Microbiol. Rev. 2002, 26, 49–71. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Begic, S.; Worobec, E.A. Regulation of Serratia marcescens ompF and ompC porin genes in response to osmotic stress, salicylate, temperature and pH. Microbiology 2006, 152, 485–491. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, L.C.; Morgan, L.K.; Godakumbura, P.; Kenney, L.J.; Anand, G.S. The inner membrane histidine kinase EnvZ senses osmolality via helix-coil transitions in the cytoplasm. EMBO J. 2012, 31, 2648–2659. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, L.; Meng, H.; Gu, D.; Li, Y.; Jia, M. Molecular mechanisms of Vibrio parahaemolyticus pathogenesis. Microbiol. Res. 2019, 222, 43–51. [Google Scholar] [CrossRef] [PubMed]
- Salomon, D.; Gonzalez, H.; Updegraff, B.L.; Orth, K. Vibrio parahaemolyticus type VI secretion system 1 is activated in marine conditions to target bacteria, and is differentially regulated from system 2. PLoS ONE 2013, 8, e61086. [Google Scholar] [CrossRef] [PubMed]
- Gu, D.; Zhang, Y.; Wang, Q.; Zhou, X. S-nitrosylation-mediated activation of a histidine kinase represses the type 3 secretion system and promotes virulence of an enteric pathogen. Nat. Commun. 2020, 11, 5777. [Google Scholar] [CrossRef]
- Yu, Y.; Yang, H.; Li, J.; Zhang, P.; Wu, B.; Zhu, B.; Zhang, Y.; Fang, W. Putative type VI secretion systems of Vibrio parahaemolyticus contribute to adhesion to cultured cell monolayers. Arch. Microbiol. 2012, 194, 827–835. [Google Scholar] [CrossRef]
- Nydam, S.D.; Shah, D.H.; Call, D.R. Transcriptome analysis of Vibrio parahaemolyticus in type III secretion system 1 inducing conditions. Front. Cell. Infect. Microbiol. 2014, 4, 1. [Google Scholar] [CrossRef]
- Hubbard, T.P.; Chao, M.C.; Abel, S.; Blondel, C.J.; Abel Zur Wiesch, P.; Zhou, X.; Davis, B.M.; Waldor, M.K. Genetic analysis of Vibrio parahaemolyticus intestinal colonization. Proc. Natl. Acad. Sci. USA 2016, 113, 6283–6288. [Google Scholar] [CrossRef] [Green Version]
- Hiyoshi, H.; Kodama, T.; Saito, K.; Gotoh, K.; Matsuda, S.; Akeda, Y.; Honda, T.; Iida, T. VopV, an F-actin-binding type III secretion effector, is required for Vibrio parahaemolyticus-induced enterotoxicity. Cell Host Microbe 2011, 10, 401–409. [Google Scholar] [CrossRef]
- Gu, D.; Meng, H.; Li, Y.; Ge, H.; Jiao, X. A GntR family transcription factor (VPA1701) for swarming motility and colonization of Vibrio parahaemolyticus. Pathogens 2019, 8, 235. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, X.; Sun, J.; Zhang, M.; Xue, X.; Wu, Q.; Yang, W.; Yin, Z.; Zhou, D.; Lu, R.; Zhang, Y. The effect of salinity on biofilm formation and c-di-GMP production in Vibrio parahaemolyticus. Curr. Microbiol. 2021, 79, 25. [Google Scholar] [CrossRef] [PubMed]
- Kimbrough, J.H.; Cribbs, J.T.; McCarter, L.L. Homologous c-di-GMP-binding Scr transcription factors orchestrate biofilm development in Vibrio parahaemolyticus. J. Bacterial. 2020, 202, e00723-19. [Google Scholar] [CrossRef] [PubMed]
- Hiyoshi, H.; Kodama, T.; Iida, T.; Honda, T. Contribution of Vibrio parahaemolyticus virulence factors to cytotoxicity, enterotoxicity, and lethality in mice. Infect. Immun. 2010, 78, 1772–1780. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gotoh, K.; Kodama, T.; Hiyoshi, H.; Izutsu, K.; Park, K.S.; Dryselius, R.; Akeda, Y.; Honda, T.; Iida, T. Bile acid-induced virulence gene expression of Vibrio parahaemolyticus reveals a novel therapeutic potential for bile acid sequestrants. PLoS ONE 2010, 5, e13365. [Google Scholar] [CrossRef] [Green Version]
- Kodama, T.; Hiyoshi, H.; Gotoh, K.; Akeda, Y.; Matsuda, S.; Park, K.S.; Cantarelli, V.V.; Iida, T.; Honda, T. Identification of two translocon proteins of Vibrio parahaemolyticus type III secretion system 2. Infect. Immun. 2008, 76, 4282–4289. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kelting, D.L.; Laxson, C.L.; Yerger, E.C. Regional analysis of the effect of paved roads on sodium and chloride in lakes. Water Res. 2012, 46, 2749–2758. [Google Scholar] [CrossRef] [PubMed]
- Whitaker, W.B.; Parent, M.A.; Naughton, L.M.; Richards, G.P.; Blumerman, S.L.; Boyd, E.F. Modulation of responses of Vibrio parahaemolyticus O3:K6 to pH and temperature stresses by growth at different salt concentrations. Appl. Environ. Microbiol. 2010, 76, 4720–4729. [Google Scholar] [CrossRef] [Green Version]
- Hu, S.; Li, Y.; Wang, B.; Yin, L.; Jia, X. Effects of NaCl Concentration on the Behavior of Vibrio brasiliensis and Transcriptome Analysis. Foods 2022, 11, 840. [Google Scholar] [CrossRef]
- Kuroda, T.; Mizushima, T.; Tsuchiya, T. Physiological roles of three Na+/H+ antiporters in the halophilic bacterium Vibrio parahaemolyticus. Microbiol. Immunol. 2005, 49, 711–719. [Google Scholar] [CrossRef]
- Paz, A.; Claxton, D.P.; Kumar, J.P.; Kazmier, K.; Bisignano, P.; Sharma, S.; Nolte, S.A.; Liwag, T.M.; Nayak, V.; Wright, E.M.; et al. Conformational transitions of the sodium-dependent sugar transporter, vSGLT. Proc. Natl. Acad. Sci. USA 2018, 115, E2742–E2751. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Xiong, X.P.; Zhang, B.W.; Yang, M.J.; Ye, M.Z.; Peng, X.X.; Li, H. Identification of vaccine candidates from differentially expressed outer membrane proteins of Vibrio alginolyticus in response to NaCl and iron limitation. Fish Shellfish Immunol. 2010, 29, 810–816. [Google Scholar] [CrossRef]
- Xu, C.; Ren, H.; Wang, S.; Peng, X. Proteomic analysis of salt-sensitive outer membrane proteins of Vibrio parahaemolyticus. Res. Microbiol. 2004, 155, 835–842. [Google Scholar] [CrossRef] [PubMed]
- Nie, D.; Hu, Y.; Chen, Z.; Li, M.; Hou, Z.; Luo, X.; Mao, X.; Xue, X. Outer membrane protein A (OmpA) as a potential therapeutic target for Acinetobacter baumannii infection. J. Biomed. Sci. 2020, 27, 26. [Google Scholar] [CrossRef] [Green Version]
- Xu, C.; Soyfoo, D.M.; Wu, Y.; Xu, S. Virulence of Helicobacter pylori outer membrane proteins: An updated review. Eur. J. Clin. Microbiol. Infect. Dis. 2020, 39, 1821–1830. [Google Scholar] [CrossRef]
- Rueter, C.; Bielaszewska, M. Secretion and Delivery of Intestinal Pathogenic Escherichia coli Virulence Factors via Outer Membrane Vesicles. Front. Cell. Infect. Microbiol. 2020, 10, 91. [Google Scholar] [CrossRef] [PubMed]
- Pennetzdorfer, N.; Höfler, T.; Wölflingseder, M.; Tutz, S.; Schild, S.; Reidl, J. σ(E) controlled regulation of porin OmpU in Vibrio cholerae. Mol. Microbiol. 2021, 115, 1244–1261. [Google Scholar] [CrossRef] [PubMed]
- Pazhani, G.P.; Chowdhury, G.; Ramamurthy, T. Adaptations of Vibrio parahaemolyticus to stress during environmental survival, host colonization, and infection. Front. Microbiol. 2021, 12, 737299. [Google Scholar] [CrossRef]
- Kodama, T.; Gotoh, K.; Hiyoshi, H.; Morita, M.; Izutsu, K.; Akeda, Y.; Park, K.S.; Cantarelli, V.V.; Dryselius, R.; Iida, T.; et al. Two regulators of Vibrio parahaemolyticus play important roles in enterotoxicity by controlling the expression of genes in the Vp-PAI region. PLoS ONE 2010, 5, e8678. [Google Scholar] [CrossRef]
- Tang, Y.; Li, F.; Gu, D.; Wang, W.; Huang, J.; Jiao, X. Antimicrobial effect and the mechanism of diallyl trisulfide against Campylobacter jejuni. Antibiotics 2021, 10, 246. [Google Scholar] [CrossRef]
- Bechlars, S.; Jäckel, C.; Diescher, S.; Wüstenhagen, D.A.; Kubick, S.; Dieckmann, R.; Strauch, E. Characterization of trh2 harbouring Vibrio parahaemolyticus strains isolated in Germany. PLoS ONE 2015, 10, e0118559. [Google Scholar] [CrossRef] [PubMed]
- Wang, Q.; Millet, Y.A.; Chao, M.C.; Sasabe, J.; Davis, B.M.; Waldor, M.K. A genome-wide screen reveals that the Vibrio cholerae phosphoenolpyruvate phosphotransferase system modulates virulence gene expression. Infect. Immun. 2015, 83, 3381–3395. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liang, W.; Wang, S.; Yu, F.; Zhang, L.; Qi, G.; Liu, Y.; Gao, S.; Kan, B. Construction and evaluation of a safe, live, oral Vibrio cholerae vaccine candidate, IEM108. Infect. Immun. 2003, 71, 5498–5504. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Makino, K.; Oshima, K.; Kurokawa, K.; Yokoyama, K.; Uda, T.; Tagomori, K.; Iijima, Y.; Najima, M.; Nakano, M.; Yamashita, A.; et al. Genome sequence of Vibrio parahaemolyticus: A pathogenic mechanism distinct from that of V cholerae. Lancet 2003, 361, 743–749. [Google Scholar] [CrossRef]
- Milton, D.L.; O’Toole, R.; Horstedt, P.; Wolf-Watz, H. Flagellin A is essential for the virulence of Vibrio anguillarum. J. Bacterial. 1996, 178, 1310–1319. [Google Scholar] [CrossRef] [PubMed]


Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhang, Y.; Tan, X.; Li, M.; Liu, P.; Jiao, X.; Gu, D. Transcriptome Analysis Reveals the Effect of Low NaCl Concentration on Osmotic Stress and Type III Secretion System in Vibrio parahaemolyticus. Int. J. Mol. Sci. 2023, 24, 2621. https://doi.org/10.3390/ijms24032621
Zhang Y, Tan X, Li M, Liu P, Jiao X, Gu D. Transcriptome Analysis Reveals the Effect of Low NaCl Concentration on Osmotic Stress and Type III Secretion System in Vibrio parahaemolyticus. International Journal of Molecular Sciences. 2023; 24(3):2621. https://doi.org/10.3390/ijms24032621
Chicago/Turabian StyleZhang, Youkun, Xiaotong Tan, Mingzhu Li, Peng Liu, Xinan Jiao, and Dan Gu. 2023. "Transcriptome Analysis Reveals the Effect of Low NaCl Concentration on Osmotic Stress and Type III Secretion System in Vibrio parahaemolyticus" International Journal of Molecular Sciences 24, no. 3: 2621. https://doi.org/10.3390/ijms24032621
APA StyleZhang, Y., Tan, X., Li, M., Liu, P., Jiao, X., & Gu, D. (2023). Transcriptome Analysis Reveals the Effect of Low NaCl Concentration on Osmotic Stress and Type III Secretion System in Vibrio parahaemolyticus. International Journal of Molecular Sciences, 24(3), 2621. https://doi.org/10.3390/ijms24032621





