Genome-Wide Characterization and Expression Profiling of the AP2/ERF Gene Family in Fragaria vesca L.
Abstract
:1. Introduction
2. Results
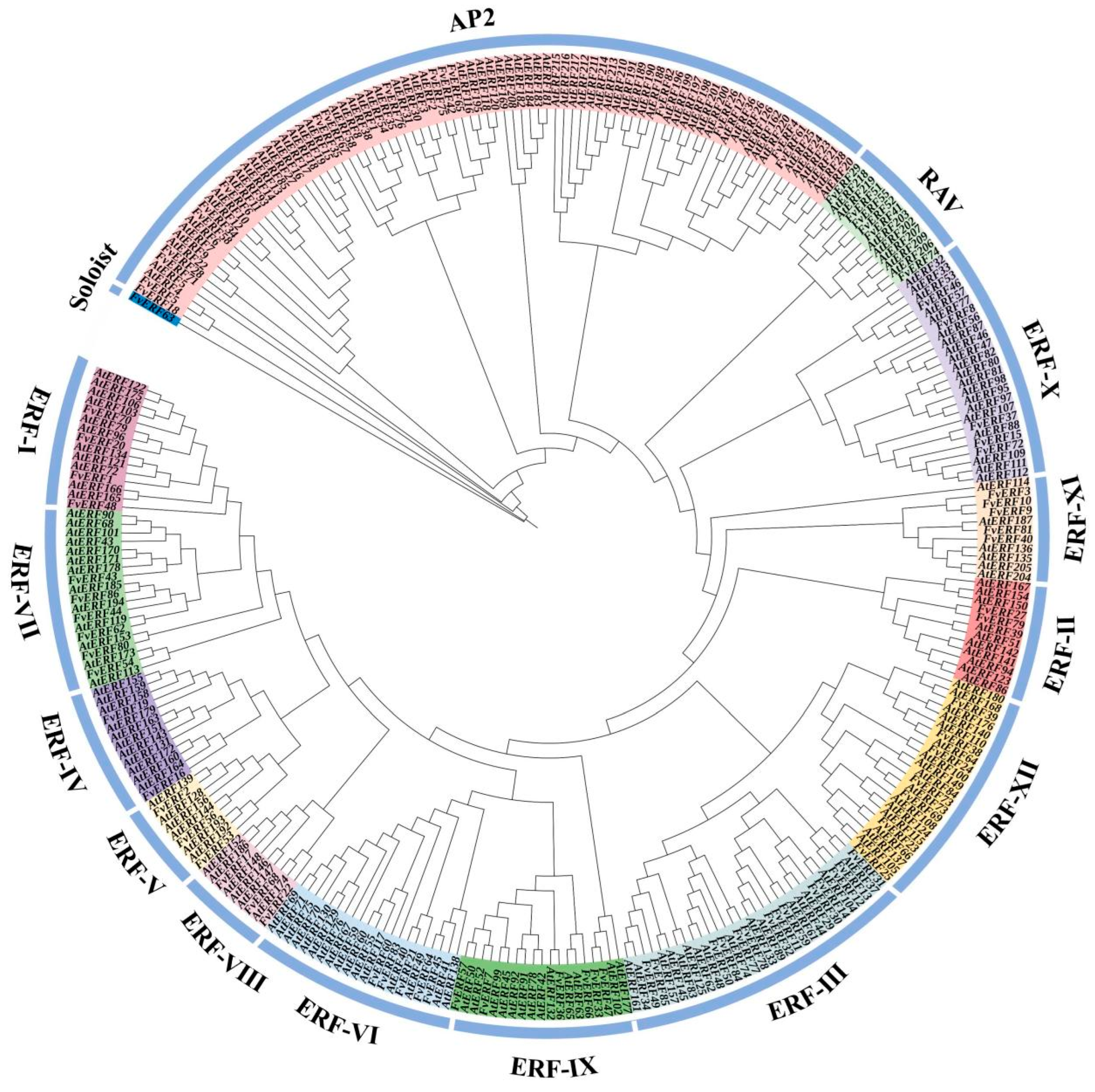
2.1. Wild Strawberry AP2/ERF Transcription Factor Family and Phylogenetic Analysis
2.2. Analysis of the Structural Composition and Physicochemical Properties of the FvERF Genes
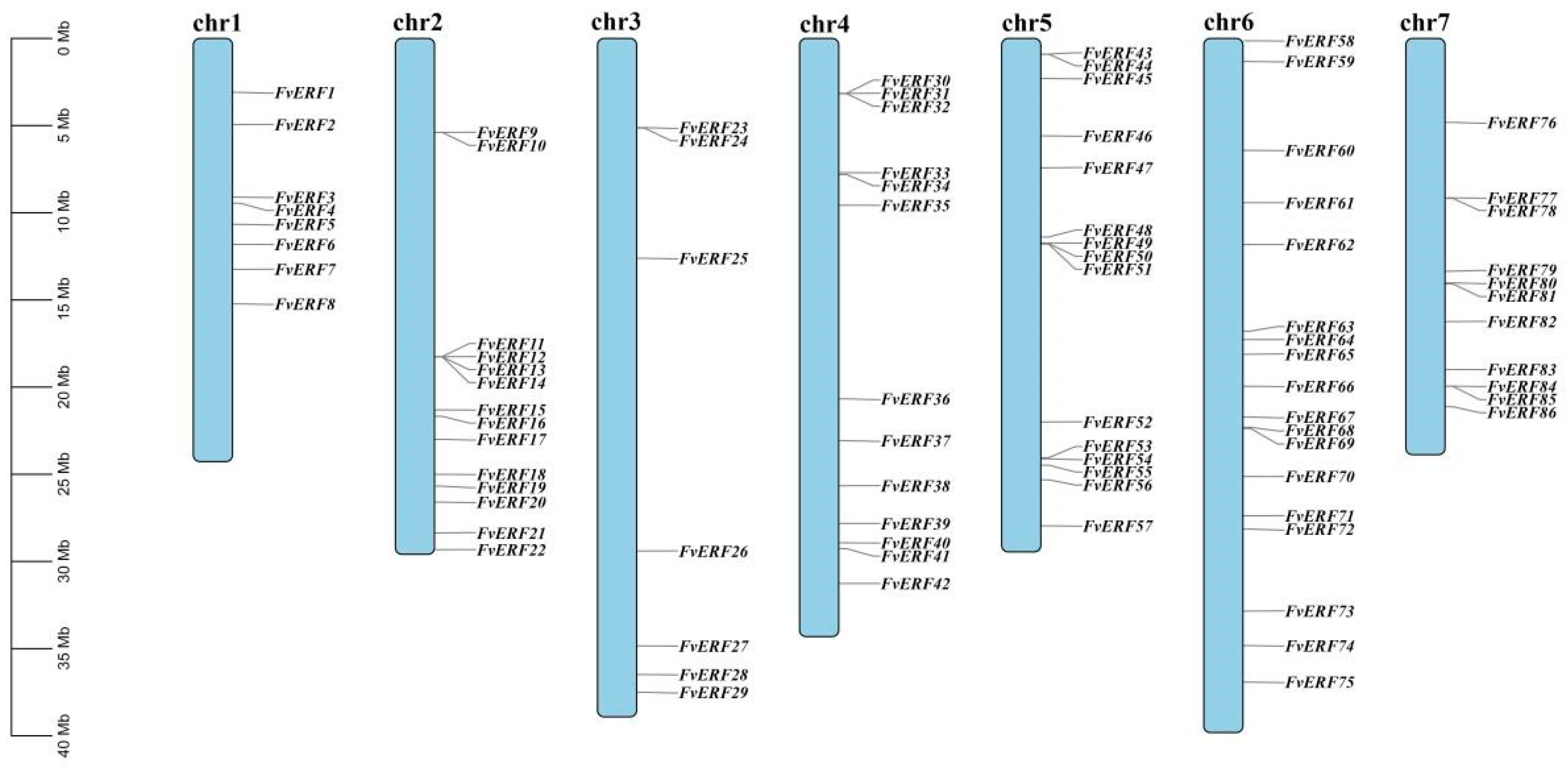
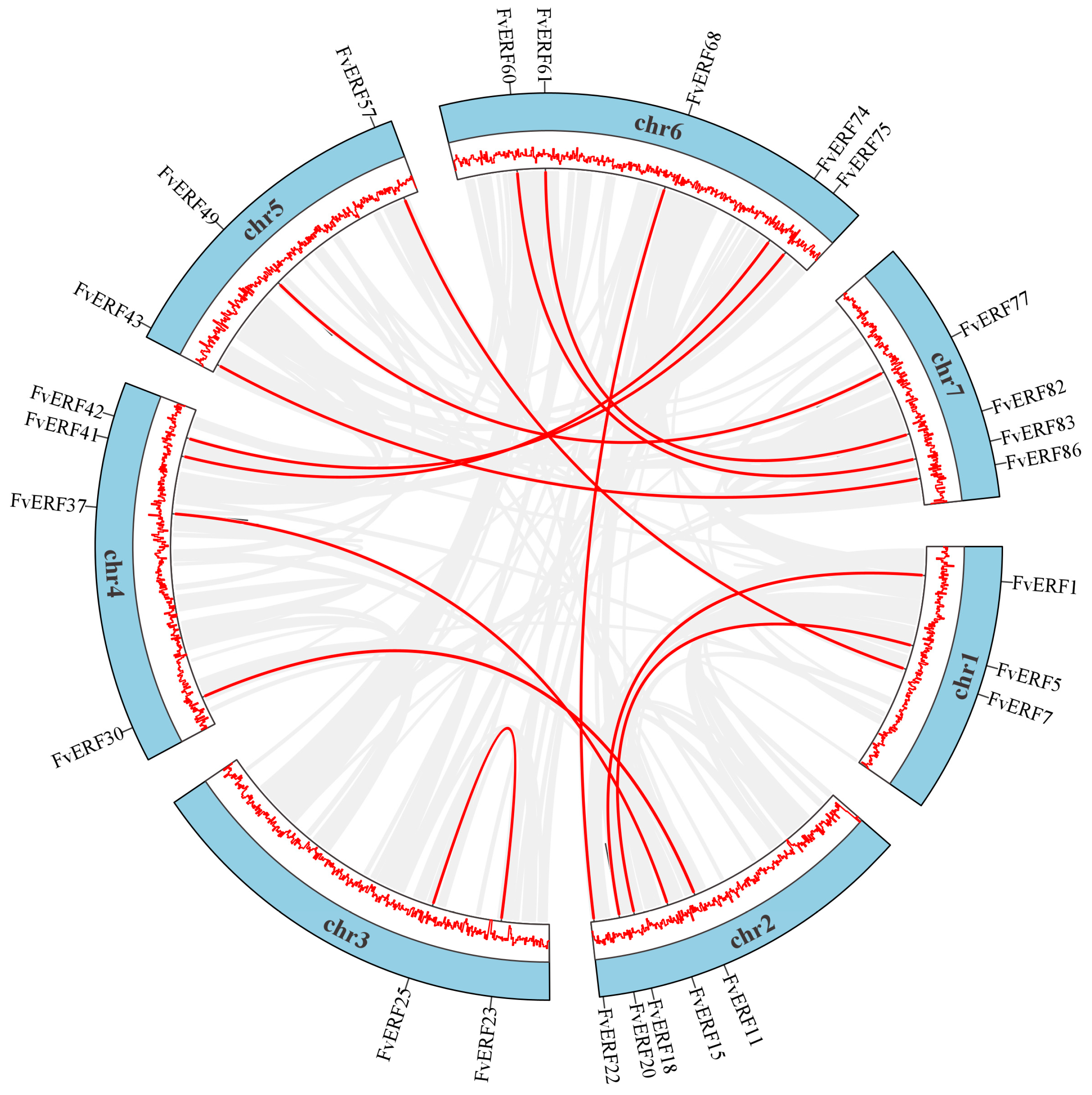
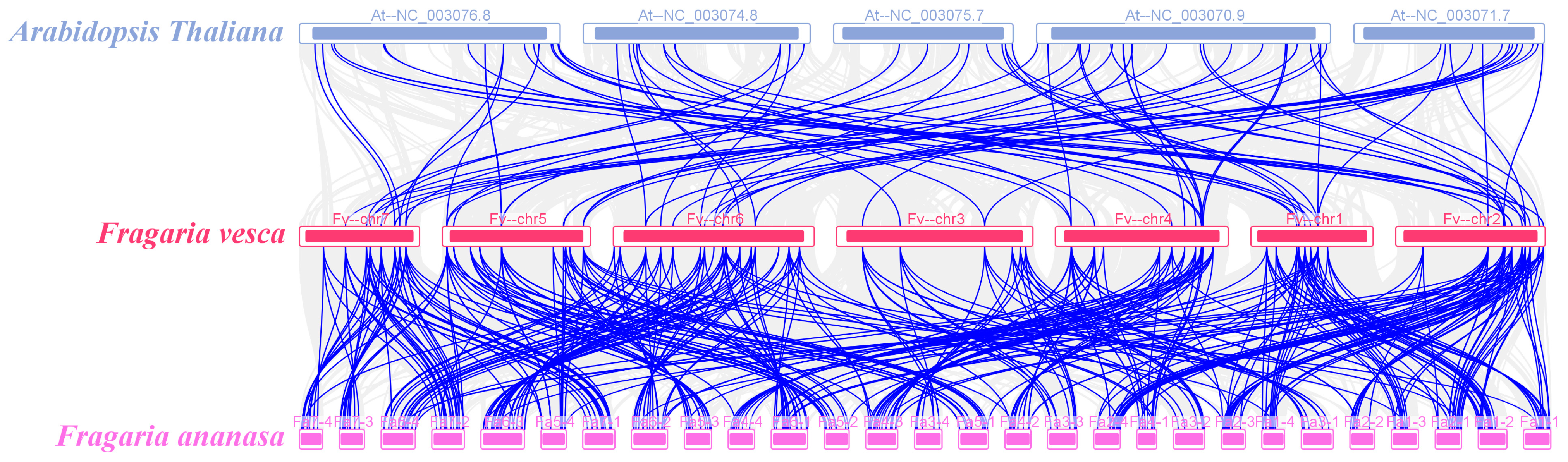
2.3. Chromosome Localization and Gene Family Evolutionary Analysis
2.4. AP2/ERF Promoter Region Cis-Acting Element Exploration
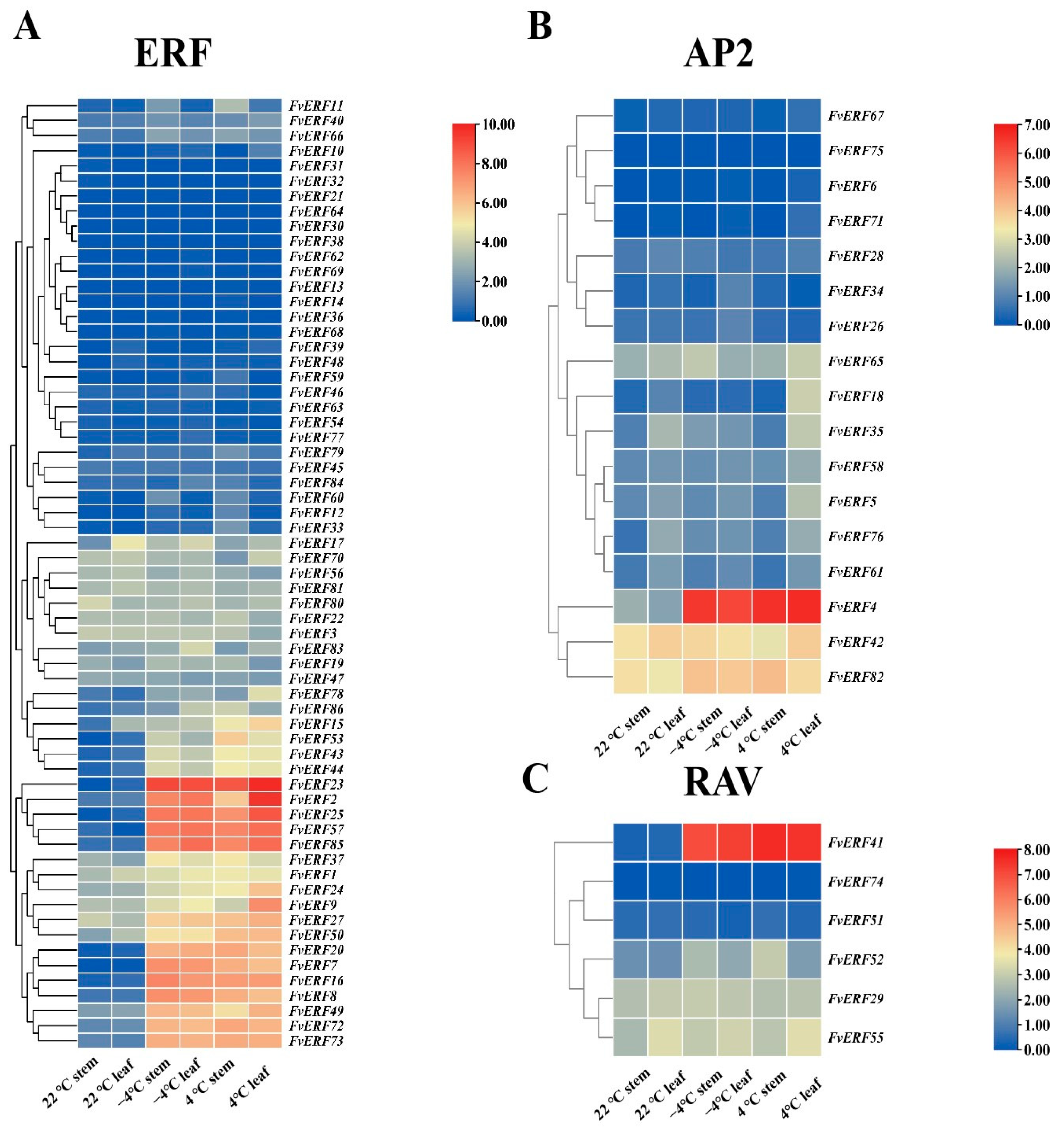
2.5. FvERF Genes Expression Profile in the Cryotranscriptome
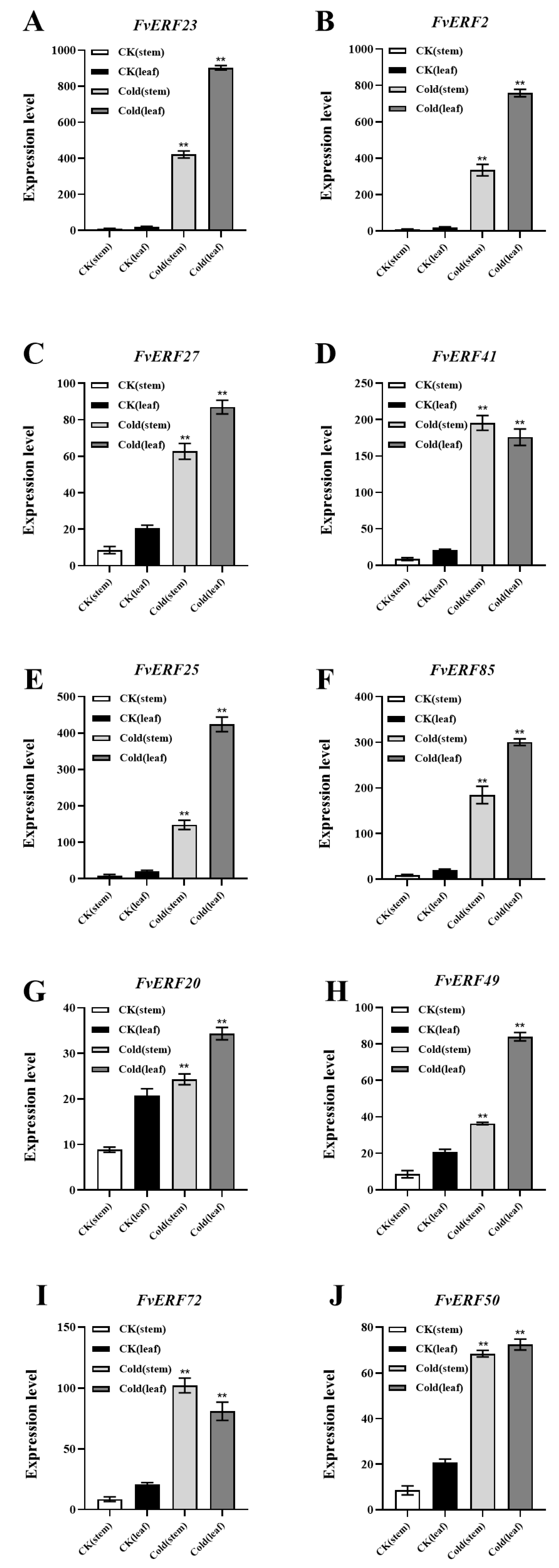
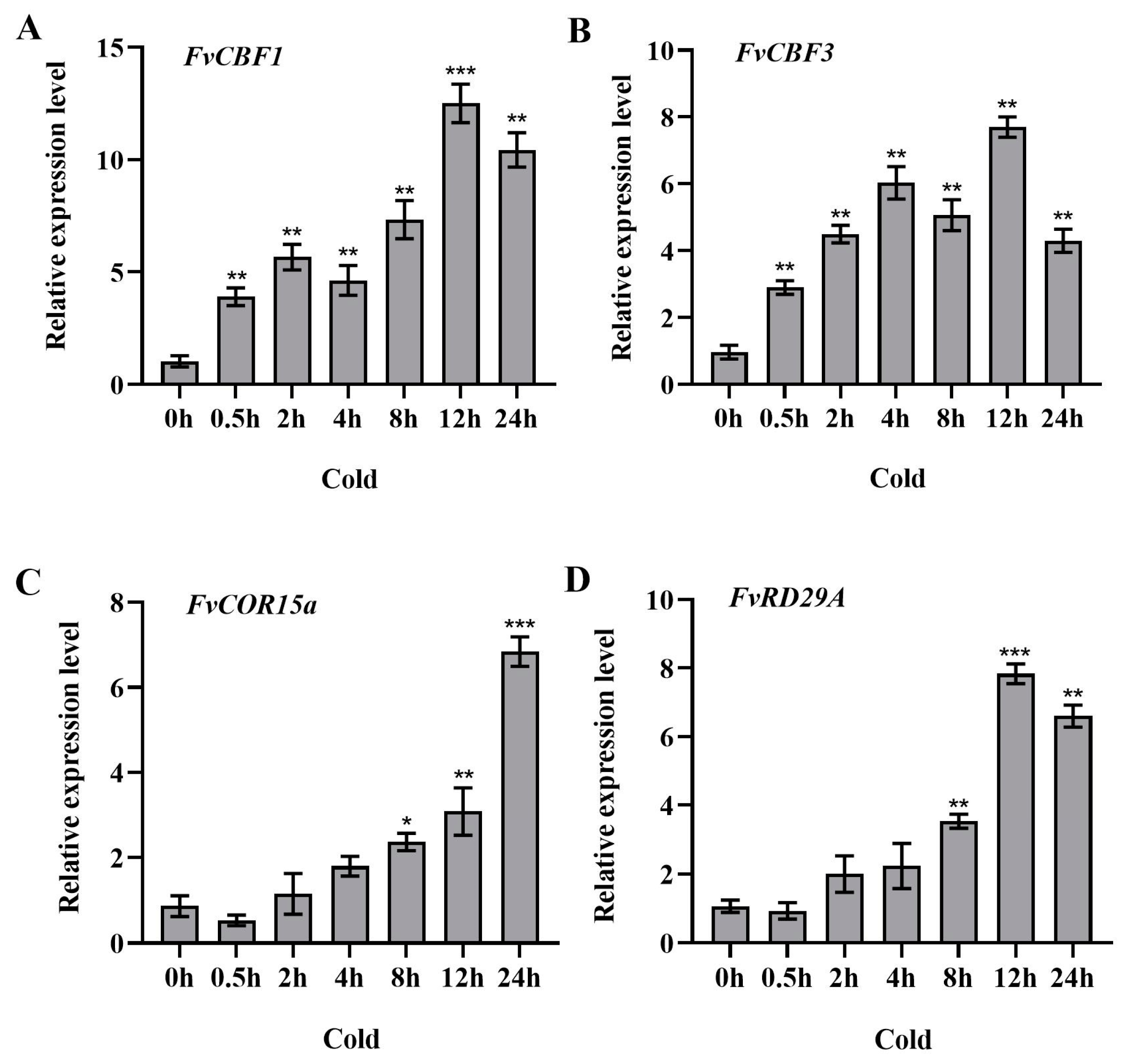
2.6. Expression Patterns of Cold-Stress-Responsive Genes in Strawberry
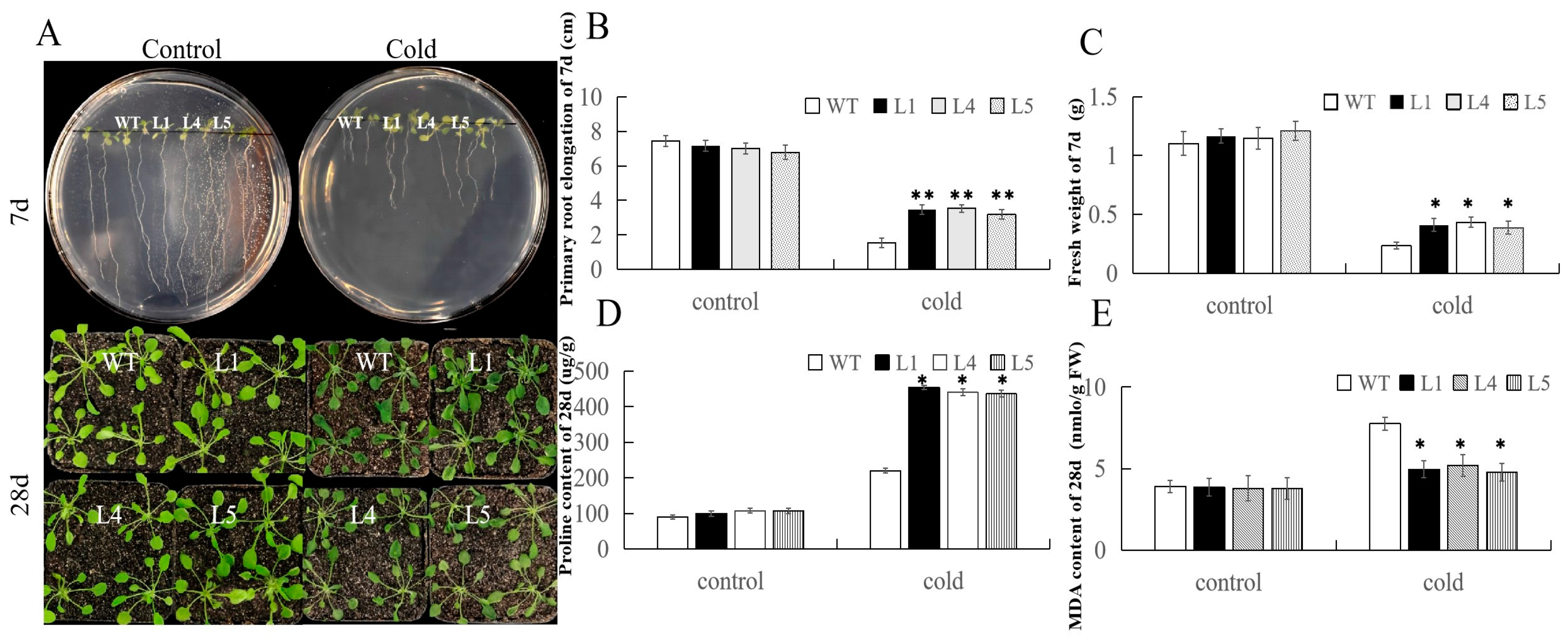
2.7. Overexpression of FvERF23 in Arabidopsis thaliana
3. Discussion
4. Materials and Methods
4.1. Determination of the Wild Strawberry AP2/ERF Gene Family
4.2. Conserved Motifs, Domain, Gene Structure, and Physicochemical Properties
4.3. Chromosomal Localization and Collinearity Analysis of Genes
4.4. Study of Cis-Acting Elements
4.5. Transcriptome Data Processing and Expression Profiling
4.6. Plant Material and Real-Time Fluorescence Quantitative PCR Validation
4.7. Plant Growth Stress Experiment
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Sanghera, G.S.; Wani, S.H.; Hussain, W.; Singh, N.B. Engineering Cold Stress Tolerance in Crop Plants. Curr. Genom. 2011, 12, 30–43. [Google Scholar] [CrossRef] [PubMed]
- Jan, N.; Hussain, M.U.; Andrabi, K.I. Cold resistance in plants: A mystery unresolved. Electron. J. Biotechnol. 2009, 12, 14–15. [Google Scholar] [CrossRef]
- Akula, R.; Ravishankar, G.A. Influence of abiotic stress signals on secondary metabolites in plants. Plant Signal. Behav. 2011, 6, 1720–1731. [Google Scholar] [CrossRef] [PubMed]
- Licausi, F.; Ohme-Takagi, M.; Perata, P. APETALA2/Ethylene Responsive Factor (AP2/ERF) transcription factors: Mediators of stress responses and developmental programs. New Phytol. 2013, 199, 639–649. [Google Scholar] [CrossRef] [PubMed]
- Feng, K.; Hou, X.; Xing, G.; Liu, J.; Duan, A.; Xu, Z.; Li, M.; Jing, Z.; Xiong, A. Advances in AP2/ERF super-family transcription factors in plant. Crit. Rev. Biotechnol. 2020, 40, 750–776. [Google Scholar] [CrossRef] [PubMed]
- Nakano, T.; Suzuki, K.; Fujimura, T.; Shinshi, H. Genome-Wide Analysis of the ERF Gene Family in Arabidopsis and Rice. Plant Physiol. 2006, 140, 411–432. [Google Scholar] [CrossRef] [PubMed]
- Thomashow, M.F. Plant Cold Acclimation: Freezing Tolerance Genes and Regulatory Mechanisms. Annu. Rev. Plant Physiol. Plant Mol. Biol. 1999, 50, 571–599. [Google Scholar] [CrossRef] [PubMed]
- Riechmann, J.L.; Meyerowitz, E.M. The AP2/EREBP family of plant transcription factors. Biol. Chem. 1998, 379, 633–646. [Google Scholar] [PubMed]
- Licausi, F.; Giorgi, F.M.; Zenoni, S.; Osti, F.; Pezzotti, M.; Perata, P. Genomic and transcriptomic analysis of the AP2/ERF superfamily in Vitis vinifera. BMC Genom. 2010, 11, 719. [Google Scholar] [CrossRef] [PubMed]
- Irish, V.F.; Sussex, I.M. Function of the apetala-1 gene during Arabidopsis floral development. Plant Cell. 1990, 2, 741–753. [Google Scholar]
- Bowman, J.L.; Sakai, H.; Jack, T.P.; Weigel, D.; Mayer, U.; Meyerowitz, E.M. Superman, a regulator of floral homeotic genes in Arabidopsis. Development 1992, 114, 599–615. [Google Scholar] [CrossRef]
- Huala, E.; Sussex, I.M. LEAFY interacts with floral homeotic genes to regulate Arabidopsis floral development. Plant Cell. 1992, 4, 901–913. [Google Scholar] [CrossRef] [PubMed]
- Weigel, D. The APETALA2 domain is related to a novel type of DNA binding domain. Plant Cell. 1995, 7, 388–389. [Google Scholar] [PubMed]
- Alonso, J.M.; Stepanova, A.N.; Leisse, T.J.; Kim, C.J.; Chen, H.; Shinn, P.; Stevenson, D.K.; Zimmerman, J.; Barajas, P.; Cheuk, R.; et al. Genome-Wide Insertional Mutagenesis of Arabidopsis thaliana. Science 2003, 301, 653–657. [Google Scholar] [CrossRef] [PubMed]
- Hu, Y.; Wang, Y.; Liu, X.F.; Li, J. Arabidopsis RAV1 is down-regulated by brassinosteroid and may act as a negative regulator during plant development. Cell Res. 2004, 14, 8–15. [Google Scholar] [CrossRef] [PubMed]
- Yamasaki, K.; Kigawa, T.; Inoue, M.; Tateno, M.; Yamasaki, T.; Yabuki, T.; Aoki, M.; Seki, E.; Matsuda, T.; Tomo, Y.; et al. Solution Structure of the B3 DNA Binding Domain of the Arabidopsis Cold-Responsive Transcription Factor RAV1. Plant Cell. 2004, 16, 3448–3459. [Google Scholar] [CrossRef] [PubMed]
- Lee, D.; Yoon, S.I.; Kim, Y.; Kim, J. Rice OsERF71-mediated root modification affects shoot drought tolerance. Plant Signal. Behav. 2017, 12, e1268311. [Google Scholar] [CrossRef] [PubMed]
- Zhang, G.; Chen, M.; Li, L.; Xu, Z.; Chen, X.; Guo, J.; Ma, Y. Overexpression of the soybean GmERF3 gene, an AP2/ERF type transcription factor for increased tolerances to salt, drought, and diseases in transgenic tobacco. J. Exp. Bot. 2009, 60, 3781–3796. [Google Scholar] [CrossRef]
- Chen, Y.; Cai, X.; Tang, B.; Xie, Q.; Chen, G.; Chen, X.; Hu, Z. SlERF.J2 Reduces Chlorophyll Accumulation and Inhibits Chloroplast Biogenesis and Development in Tomato Leaves. Plant Sci. 2023, 328, 111578. [Google Scholar] [CrossRef]
- Li, D.; He, Y.; Li, S.; Shi, S.Q.; Li, L.; Liu, Y.; Chen, H. Genome-wide characterization and expression analysis of AP2/ERF genes in eggplant (Solanum melongena L.). Plant Physiol. Biochem. 2021, 167, 492–503. [Google Scholar] [CrossRef]
- Jin, J.; Wang, M.; Zhang, H.; Khan, A.; Wei, A.; Luo, D.; Gong, Z. Genome-wide identification of the AP2/ERF transcription factor family in pepper (Capsicum annuum L.). Genome 2018, 61, 663–674. [Google Scholar] [CrossRef]
- Charfeddine, M.; Saïdi, M.N.; Charfeddine, S.; Hammami, A.; Gargouri Bouzid, R. Genome-Wide Analysis and Expression Profiling of the ERF Transcription Factor Family in Potato (Solanum tuberosum L.). Mol. Biotechnol. 2015, 57, 348–358. [Google Scholar] [CrossRef] [PubMed]
- Liang, X.; Luo, G.; Li, W.; Yao, A.; Liu, W.; Xie, L.; Han, M.; Li, X.; Han, D. Overexpression of a Malus baccata CBF transcription factor gene, MbCBF1, increases cold and salinity tolerance in Arabidopsis thaliana. Plant Physiol. Biochem. 2022, 192, 230–242. [Google Scholar] [CrossRef] [PubMed]
- Han, J.; Li, X.; Li, W.; Yang, Q.; Li, Z.; Cheng, Z.; Lv, L.; Zhang, L.; Han, D. Isolation and preliminary functional analysis of FvICE1, involved in cold and drought tolerance in Fragaria vesca through overexpression and CRISPR/Cas9 technologies. Plant Physiol. Biochem. 2023, 196, 270–280. [Google Scholar] [CrossRef] [PubMed]
- Edger, P.P.; VanBuren, R.; Colle, M.; Poorten, T.J.; Wai, C.M.; Niederhuth, C.E.; Alger, E.; Ou, S.; Acharya, C.B.; Wang, J.; et al. Single-molecule sequencing and optical mapping yields an improved genome of woodland strawberry (Fragaria vesca) with chromosome-scale contiguity. GigaScience 2017, 7, gix124. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Pi, M.; Gao, Q.; Liu, Z.; Kang, C. Updated annotation of the wild strawberry Fragaria vesca V4 genome. Hortic. Res. 2019, 6, 61. [Google Scholar] [CrossRef] [PubMed]
- Liu, T.; Li, M.; Liu, Z.; Ai, X.; Li, Y. Reannotation of the cultivated strawberry genome and establishment of a strawberry genome database. Hortic. Res. 2021, 8, 31. [Google Scholar] [CrossRef] [PubMed]
- Mao, J.; Wang, Y.; Wang, B.; Li, J.; Zhang, C.; Zhang, W.; Li, X.; Li, J.; Zhang, J.; Li, H.; et al. High-quality haplotype-resolved genome assembly of cultivated octoploid strawberry. Hortic. Res. 2023, 10, uhad002. [Google Scholar] [CrossRef] [PubMed]
- Zhou, Y.; Xiong, J.; Shu, Z.; Dong, C.; Gu, T.; Sun, P.; He, S.; Jiang, M.; Xia, Z.; Xue, J.; et al. The telomere-to-telomere genome of Fragaria vesca reveals the genomic evolution of Fragaria and the origin of cultivated octoploid strawberry. Hortic. Res. 2023, 10, uhad027. [Google Scholar] [CrossRef]
- Zhao, M.; Haxim, Y.; Liang, Y.; Qiao, S.; Gao, B.; Zhang, D.; Li, X. Genome-wide investigation of AP2/ERF gene family in the desert legume Eremosparton songoricum: Identification, classification, evolution, and expression profiling under drought stress. Front. Plant Sci. 2022, 13, 885694. [Google Scholar] [CrossRef]
- Yang, X.; Tuskan, G.A.; Cheng, M. Divergence of the Dof Gene Families in Poplar, Arabidopsis, and Rice Suggests Multiple Modes of Gene Evolution after Duplication. Plant Physiol. 2006, 142, 820–830. [Google Scholar] [CrossRef] [PubMed]
- Chen, Q.; Zhang, X.; Fang, Y.; Wang, B.; Xu, S.; Zhao, K.; Zhang, J.; Fang, J. Genome-Wide Identification and Expression Analysis of the R2R3-MYB Transcription Factor Family Revealed Their Potential Roles in the Flowering Process in Longan (Dimocarpus longan). Front. Plant Sci. 2022, 13, 820439. [Google Scholar] [CrossRef] [PubMed]
- Darwish, O.; Slovin, J.P.; Kang, C.; Hollender, C.A.; Geretz, A.; Houston, S.J.; Liu, Z.; Alkharouf, N.W. SGR: An online genomic resource for the woodland strawberry. BMC Plant Biol. 2013, 13, 223. [Google Scholar] [CrossRef] [PubMed]
- Lv, K.; Li, J.; Zhao, K.; Chen, S.; Nie, J.; Zhang, W.; Liu, G.; Wei, H. Overexpression of an AP2/ERF family gene, BpERF13, in birch enhances cold tolerance through upregulating CBF genes and mitigating reactive oxygen species. Plant Sci. 2020, 292, 110375. [Google Scholar] [CrossRef] [PubMed]
- Xie, W.; Ding, C.; Hu, H.; Dong, G.; Zhang, G.; Qian, Q.; Ren, D. Molecular Events of Rice AP2/ERF Transcription Factors. Int. J. Mol. Sci. 2022, 23, 12013. [Google Scholar] [CrossRef] [PubMed]
- Sakuma, Y.; Liu, Q.; Dubouzet, J.G.; Abé, H.; Shinozaki, K.; Yamaguchi-Shinozaki, K. DNA-binding specificity of the ERF/AP2 domain of Arabidopsis DREBs, transcription factors involved in dehydration- and cold-inducible gene expression. Biochem. Biophys. Res. Commun. 2002, 290, 998–1009. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Liao, J.; Ling, Q.; Xi, Y.; Qian, Y. Genome-wide identification and expression profiling analysis of maize AP2/ERF superfamily genes reveal essential roles in abiotic stress tolerance. BMC Genom. 2022, 23, 125. [Google Scholar] [CrossRef]
- Jia, H.; Song, J.; Yang, W.; Zhang, X.; Zhu, Y.; Zhao, D.; Wang, H. Genome-wide characterization of AP2/ERF genes and their potential roles in bulb and bolt development in Allium sativum. Sci. Hortic. 2023, 322, 112359. [Google Scholar] [CrossRef]
- Xu, W.; Li, F.; Ling, L.; Liu, A. Genome-wide survey and expression profiles of the AP2/ERF family in castor bean (Ricinus communis L.). BMC Genom. 2013, 14, 785. [Google Scholar] [CrossRef]
- Mitsukawa, N.; Okumura, S.; Shirano, Y.; Sato, S.; Kato, T.; Harashima, S.; Shibata, D. Overexpression of an Arabidopsis thaliana high-affinity phosphate transporter gene in tobacco cultured cells enhances cell growth under phosphate-limited conditions. Proc. Natl. Acad. Sci. USA 1997, 94, 7098–7102. [Google Scholar] [CrossRef]
- Okamuro, J.K.; Caster, B.; Villarroel, R.; Van Montagu, M.; Jofuku, K.D. The AP2 domain of APETALA2 defines a large new family of DNA binding proteins in Arabidopsis. Proc. Natl. Acad. Sci. USA 1997, 94, 7076–7081. [Google Scholar] [CrossRef] [PubMed]
- Wu, D.; Zhang, K.; Li, C.; Xie, G.; Lu, M.; Qian, Y.; Shu, Y.; Shen, Q. Genome-wide comprehensive characterization and Transcriptomic analysis of AP2/ERF gene family revealed its role in seed oil and ALA formation in Perilla (Perilla frutescens). Gene 2023, 889, 147808. [Google Scholar] [CrossRef] [PubMed]
- Xu, S.; Yao, S.; Huang, R.; Tan, Y.H.; Huang, D. Transcriptome-wide analysis of the AP2/ERF transcription factor gene family involved in the regulation of gypenoside biosynthesis in Gynostemma pentaphyllum. Plant Physiol. Biochem. 2020, 154, 238–247. [Google Scholar] [CrossRef] [PubMed]
- Karanja, B.K.; Xu, L.; Wang, Y.; Tang, M.; M’mbone Muleke, E.; Dong, J.; Liu, L. Genome-wide characterization of the AP2/ERF gene family in radish (Raphanus sativus L.): Unveiling evolution and patterns in response to abiotic stresses. Gene 2019, 718, 144048. [Google Scholar] [CrossRef] [PubMed]
- Zhu, X.; Wang, B.; Liu, W.; Wei, X.; Wang, X.; Du, X.; Liu, H. Genome-wide analysis of AP2/ERF gene and functional analysis of CqERF24 gene in drought stress in quinoa. Int. J. Biol. Macromol. 2023, 253 Pt. 8, 127582. [Google Scholar] [CrossRef]
- Hughes, A.L. The evolution of functionally novel proteins after gene duplication. Proc. Biol. Sci. 1994, 256, 119–124. [Google Scholar] [PubMed]
- Zhu, Y.; Liu, X.; Gao, Y.; Li, K.; Guo, W. Transcriptome-based identification of AP2/ERF family genes and their cold-regulated expression during the dormancy phase transition of Chinese cherry flower buds. Sci. Hortic. 2021, 275, 109666. [Google Scholar] [CrossRef]
- Wan, R.; Song, J.; Lv, Z.; Qi, X.; Han, X.; Guo, Q.; Wang, S.S.; Shi, J.; Jian, Z.; Hu, Q.; et al. Genome-Wide Identification and Comprehensive Analysis of the AP2/ERF Gene Family in Pomegranate Fruit Development and Postharvest Preservation. Genes. 2022, 13, 895. [Google Scholar] [CrossRef] [PubMed]
- Wan, L.; Zhang, J.; Zhang, H.; Zhang, Z.; Quan, R.; Zhou, S.; Huang, R. Transcriptional activation of OsDERF1 in OsERF3 and OsAP2-39 negatively modulates ethylene synthesis and drought tolerance in rice. PLoS ONE 2011, 6, e25216. [Google Scholar] [CrossRef] [PubMed]
- Chen, K.; Li, G.; Bressan, R.A.; Song, C.; Zhu, J.; Zhao, Y. Abscisic acid dynamics, signaling and functions in plants. J. Integr. Plant Biol. 2019, 62, 25–54. [Google Scholar] [CrossRef]
- Li, B.; Wang, X.; Wang, X.; Xi, Z. An AP2/ERF transcription factor VvERF63 positively regulates cold tolerance in Arabidopsis and grape leaves. Environ. Exp. Bot. 2023, 205, 105124. [Google Scholar] [CrossRef]
- Sena, F.; Monza, J.; Signorelli, S. Determination of Free Proline in Plants. Methods Mol. Biol. 2024, 2798, 183–194. [Google Scholar] [PubMed]
- Yu, Y.; Ni, Y.; Qiao, T.; Ji, X.; Xu, J.; Li, B.; Sun, Q. Overexpression of VvASMT1 from grapevine enhanced salt and osmotic stress tolerance in Nicotiana benthamiana. PLoS ONE 2022, 17, e0269028. [Google Scholar] [CrossRef] [PubMed]
- Chen, C.; Wu, Y.; Li, J.; Wang, X.; Zeng, Z.; Xu, J.; Liu, Y.; Feng, J.; Chen, H.; He, Y.; et al. TBtools-II: A “One for All, All for One” Bioinformatics Platform for Biological Big-data Mining. Mol. Plant 2023, 16, 1733–1742. [Google Scholar] [CrossRef] [PubMed]
- Zhao, Y.; Ma, R.; Xu, D.; Bi, H.; Xia, Z.; Peng, H. Genome-Wide Identification and Analysis of the AP2 Transcription Factor Gene Family in Wheat (Triticum aestivum L.). Front. Plant Sci. 2019, 10, 1286. [Google Scholar] [CrossRef] [PubMed]
- Han, D.; Zhang, Z.; Ni, B.; Ding, H.; Liu, W.; Li, W.; Chai, L.; Yang, G. Isolation and functional analysis of MxNAS3 involved in enhanced iron stress tolerance and abnormal flower in transgenic Arabidopsis. J. Plant Interact. 2018, 13, 433–441. [Google Scholar] [CrossRef]
- Han, D.; Han, J.; Xu, T.; Li, X.; Yao, C.; Li, T.; Sun, X.; Wang, X.; Yang, G. Overexpression of MbERF12, an ERF gene from Malus baccata (L.) Borkh, increases cold and salt tolerance in Arabidopsis thaliana associated with ROS scavenging through ethylene signal transduction. In Vitro Cell Dev. Biol. Plant. 2021, 57, 760–770. [Google Scholar] [CrossRef]
- Lee, H.Y.; Back, K. Melatonin induction and its role in high light stress tolerance in Arabidopsis thaliana. J. Pineal Res. 2018, 65, e12504. [Google Scholar] [CrossRef]
- Li, X.; Liang, X.; Li, W.; Yao, A.; Liu, W.; Wang, Y.; Yang, G.; Han, D. Isolation and functional analysis of MbCBF2, a Malus baccata (L.) Borkh CBF transcription factor gene, with functions in tolerance to cold and salt stress in transgenic Arabidopsis thaliana. Int. J. Mol. Sci. 2022, 23, 9827. [Google Scholar] [CrossRef]
- Lin, K.H.; Sei, S.C.; Su, Y.H.; Chiang, C.M. Overexpression of the Arabidopsis and winter squash superoxide dismutase genes enhances chilling tolerance via ABA-sensitive transcriptional regulation in transgenic Arabidopsis. Plant Signal. Behav. 2019, 14, 1685728. [Google Scholar] [CrossRef]
- Maren, N.A.; Duduit, J.R.; Huang, D.; Zhao, F.; Ranney, T.G.; Liu, W. Stepwise Optimization of Real-Time RT-PCR Analysis. Methods Mol. Biol. 2023, 2653, 317–332. [Google Scholar] [PubMed]
- Wang, W.; Shi, S.; Kang, W.; He, L. Enriched endogenous free Spd and Spm in alfalfa (Medicago sativa L.) under drought stress enhance drought tolerance by inhibiting H2O2 production to increase antioxidant enzyme activity. J. Plant Physiol. 2023, 291, 154139. [Google Scholar] [CrossRef] [PubMed]
- Tao, J.; Wu, F.; Wen, H.; Liu, X.; Luo, W.; Gao, L.; Jiang, Z.; Mo, B.; Chen, X.; Kong, W.; et al. RCD1 Promotes Salt Stress Tolerance in Arabidopsis by Repressing ANAC017 Activity. Int. J. Mol. Sci. 2023, 24, 9793. [Google Scholar] [CrossRef] [PubMed]

Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wei, Y.; Kong, Y.; Li, H.; Yao, A.; Han, J.; Zhang, W.; Li, X.; Li, W.; Han, D. Genome-Wide Characterization and Expression Profiling of the AP2/ERF Gene Family in Fragaria vesca L. Int. J. Mol. Sci. 2024, 25, 7614. https://doi.org/10.3390/ijms25147614
Wei Y, Kong Y, Li H, Yao A, Han J, Zhang W, Li X, Li W, Han D. Genome-Wide Characterization and Expression Profiling of the AP2/ERF Gene Family in Fragaria vesca L. International Journal of Molecular Sciences. 2024; 25(14):7614. https://doi.org/10.3390/ijms25147614
Chicago/Turabian StyleWei, Yangfan, Yihan Kong, Huiwen Li, Anqi Yao, Jiaxin Han, Wenhao Zhang, Xingguo Li, Wenhui Li, and Deguo Han. 2024. "Genome-Wide Characterization and Expression Profiling of the AP2/ERF Gene Family in Fragaria vesca L." International Journal of Molecular Sciences 25, no. 14: 7614. https://doi.org/10.3390/ijms25147614
APA StyleWei, Y., Kong, Y., Li, H., Yao, A., Han, J., Zhang, W., Li, X., Li, W., & Han, D. (2024). Genome-Wide Characterization and Expression Profiling of the AP2/ERF Gene Family in Fragaria vesca L. International Journal of Molecular Sciences, 25(14), 7614. https://doi.org/10.3390/ijms25147614





