Overexpression of Potato PYL16 Gene in Tobacco Enhances the Transgenic Plant Tolerance to Drought Stress
Abstract
1. Introduction
2. Results
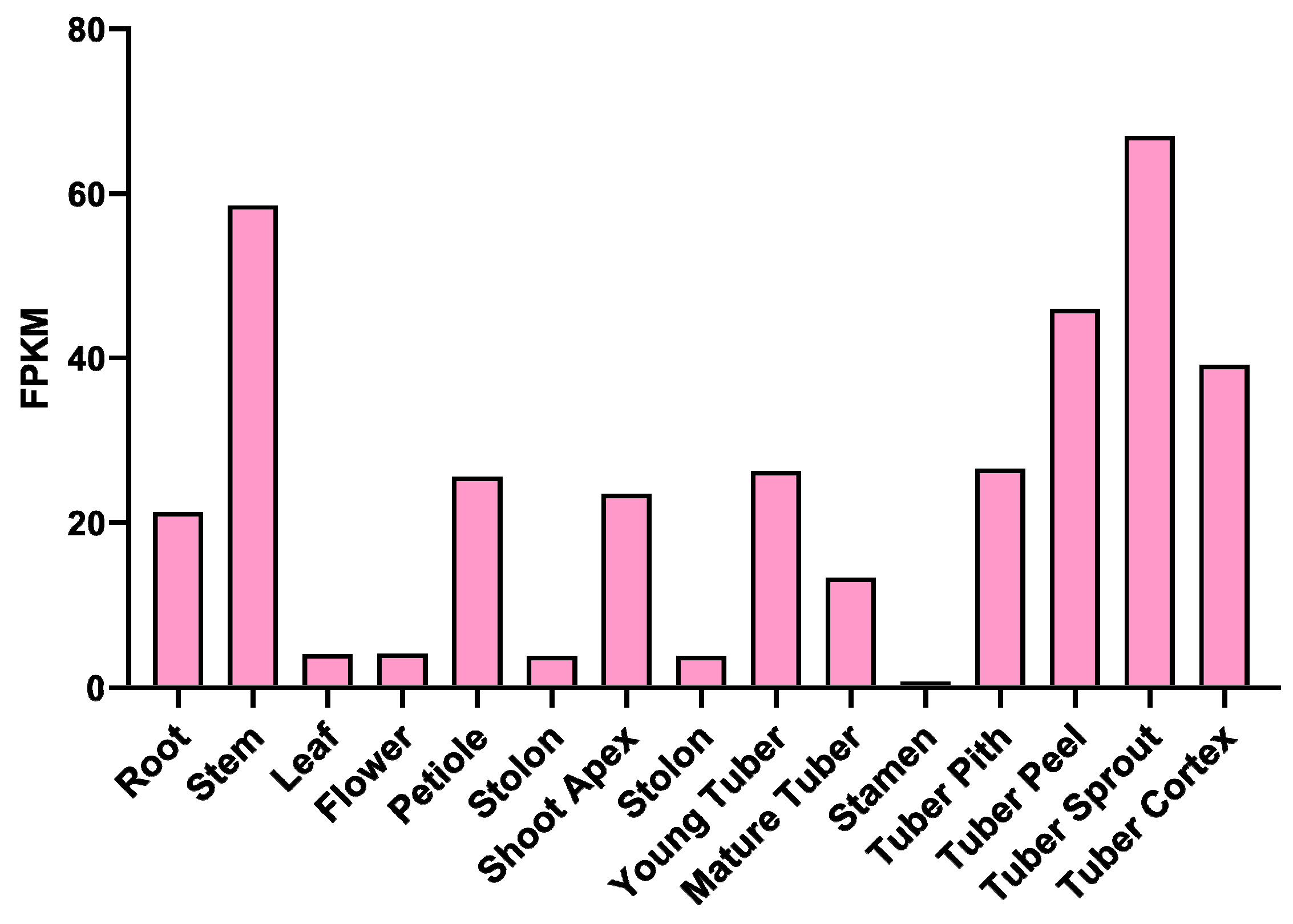
2.1. Screening and Molecular Characterization Analysis of StPYL16
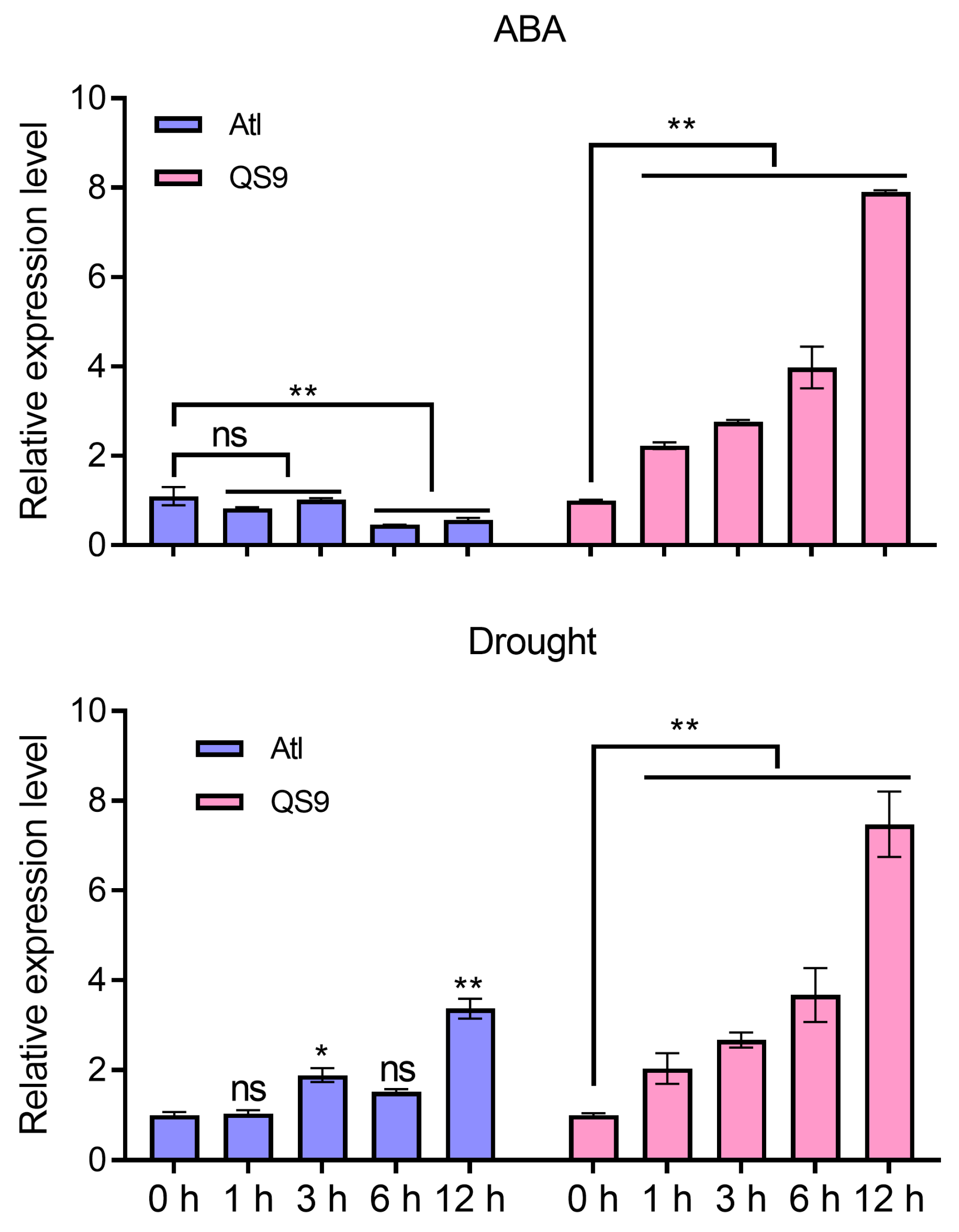
2.2. StPYL16 Is Strongly Induced by Drought Treatment
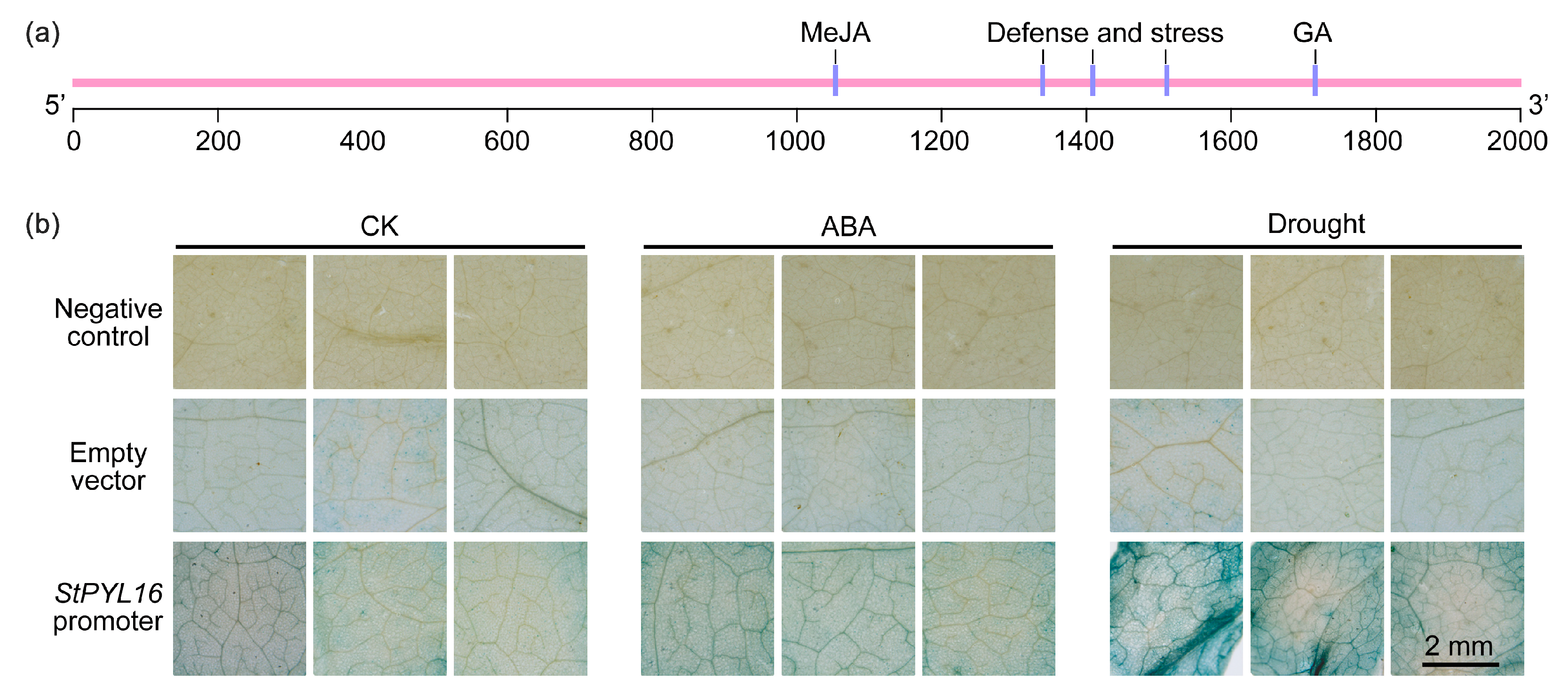
2.3. Drought Enhances the Activity of StPYL16 Promoter
2.4. Transient Transformation of StPYL16 Improves Drought Tolerance of Transgenic Tobacco
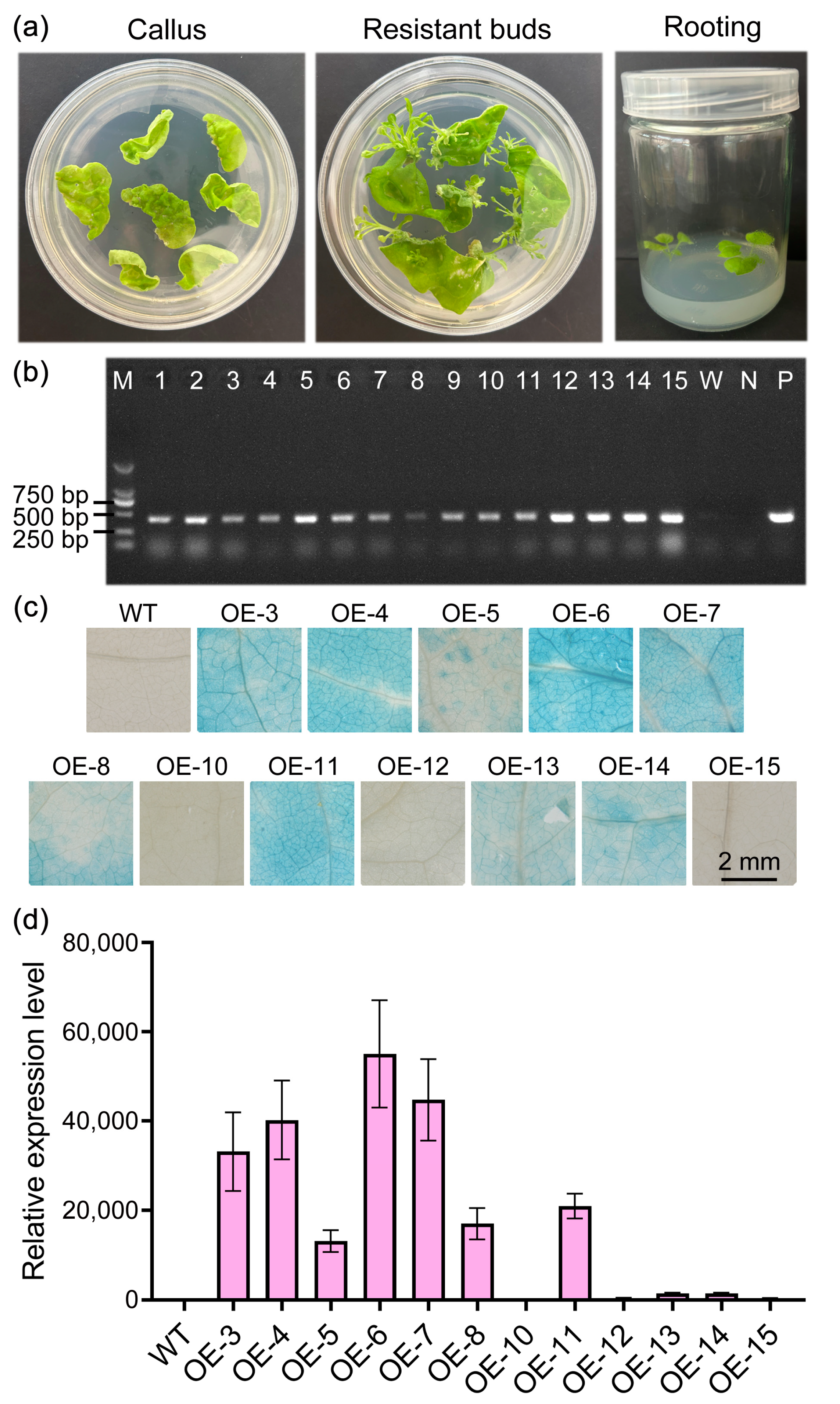
2.5. Generation of Stable Transgenic Tobacco
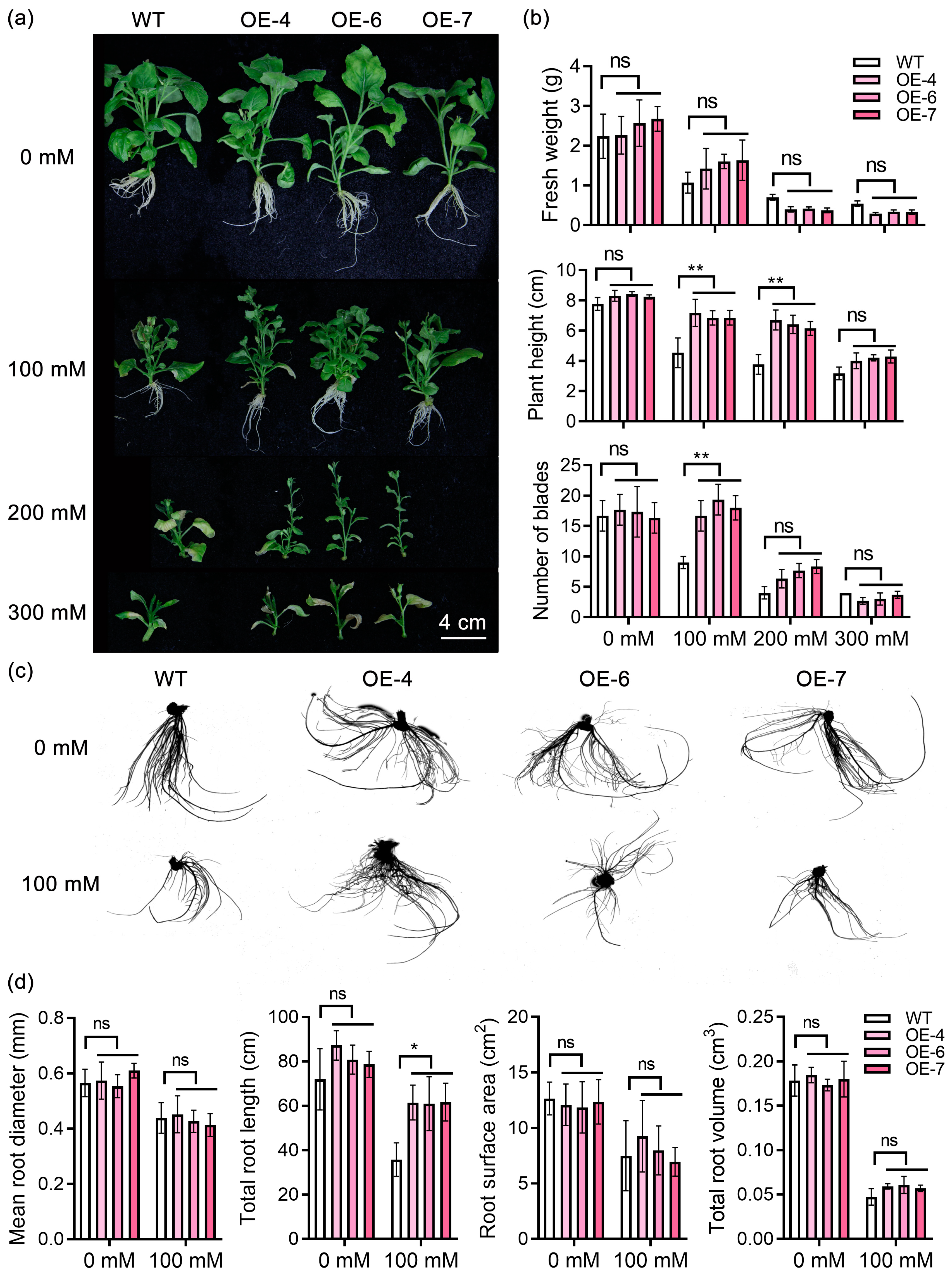
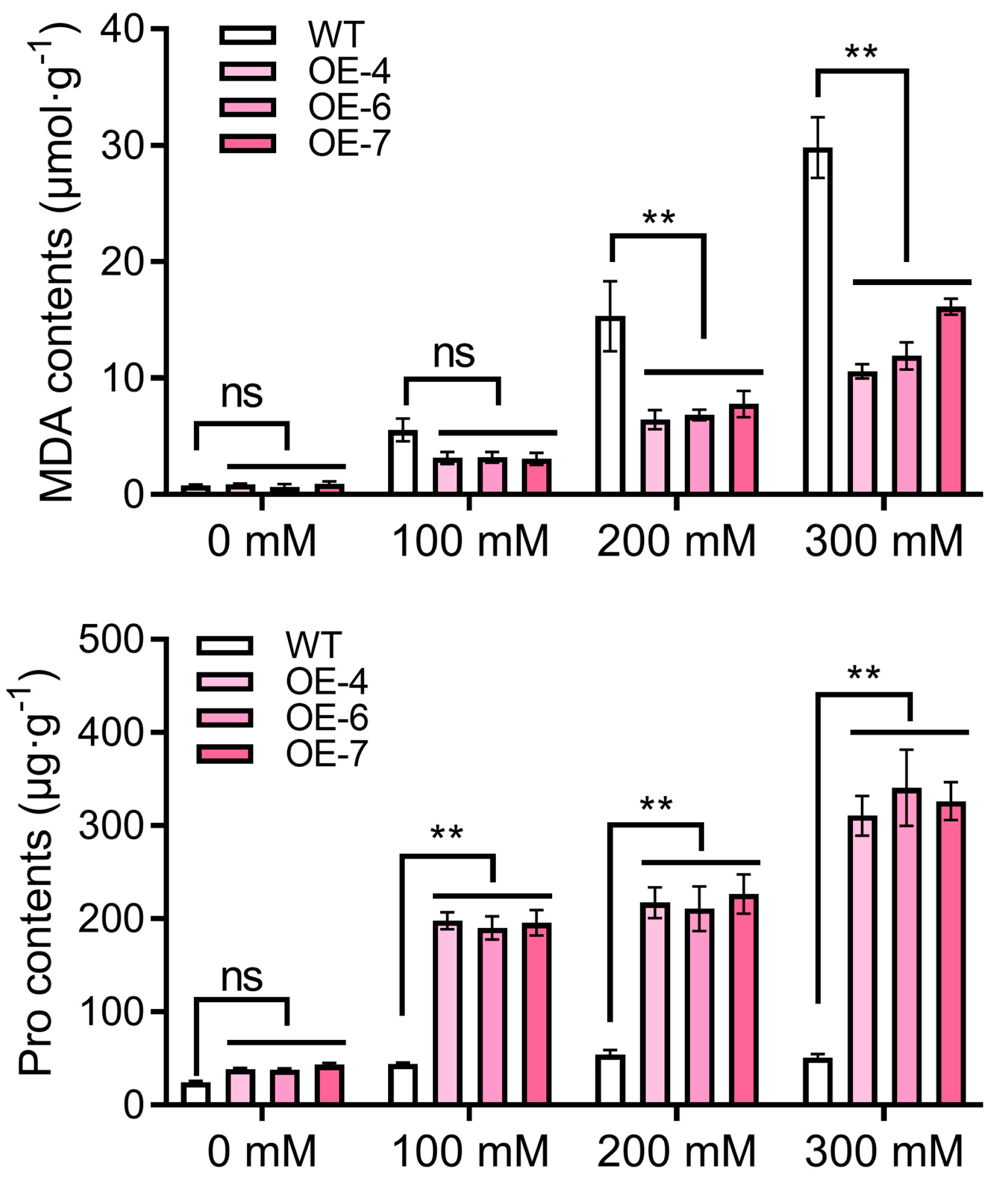
2.6. Overexpression of StPYL16 Increases Tobacco Drought Tolerance
2.7. Stress-Related Gene Expression in Transgenic Tobacco Plants under Drought Stress Mediated by StPYL16
3. Discussion
4. Materials and Methods
4.1. Plant Material and Growth Conditions
4.2. Isolation and Characterization of StPYL16
4.3. Expression Analysis of StPYL16
4.4. Promoter Cis-eElement Analysis
4.5. Evaluation of Drought Resistance after Transient Transformation of Tobacco
4.6. Evaluation of Drought Resistance after Stable Transformation of Tobacco
4.7. β-Glucuronidase (GUS) Staining
4.8. Determination of Phenotypic and Physiological Indicators
4.9. Expression Measurement of Stress-rResponsive Genes
4.10. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Bansal, K.C.; Lenka, S.K.; Mondal, T.K. Genomic resources for breeding crops with enhanced abiotic stress tolerance. Plant Breed. 2013, 133, 1–11. [Google Scholar] [CrossRef]
- Bartels, D.; Sunkar, R. Drought and Salt Tolerance in Plants. Crit. Rev. Plant Sci. 2005, 24, 23–58. [Google Scholar] [CrossRef]
- Lee, S.C.; Luan, S. ABA signal transduction at the crossroad of biotic and abiotic stress responses. Plant Cell Environ. 2011, 35, 53–60. [Google Scholar] [CrossRef] [PubMed]
- Dong, T.; Park, Y.; Hwang, I. Abscisic acid: Biosynthesis, inactivation, homoeostasis and signalling. Essays Biochem. 2015, 58, 29–48. [Google Scholar] [CrossRef] [PubMed]
- Cutler, S.R.; Rodriguez, P.L.; Finkelstein, R.R.; Abrams, S.R. Abscisic Acid: Emergence of a Core Signaling Network. Annu. Rev. Plant Biol. 2010, 61, 651–679. [Google Scholar] [CrossRef]
- Chen, K.; Li, G.J.; Bressan, R.A.; Song, C.P.; Zhu, J.K.; Zhao, Y. Abscisic acid dynamics, signaling, and functions in plants. J. Integr. Plant. Biol. 2020, 62, 25–54. [Google Scholar] [CrossRef]
- Xue, T.; Wang, D.; Zhang, S.; Ehlting, J.; Ni, F.; Jakab, S.; Zheng, C.; Zhong, Y. Genome-wide and expression analysis of protein phosphatase 2C in rice and Arabidopsis. BMC Genom. 2008, 9, 550. [Google Scholar] [CrossRef] [PubMed]
- Park, S.-Y.; Fung, P.; Nishimura, N.; Jensen, D.R.; Fujii, H.; Zhao, Y.; Lumba, S.; Santiago, J.; Rodrigues, A.; Chow, T.-F.F.; et al. Abscisic Acid Inhibits Type 2C Protein Phosphatases via the PYR/PYL Family of START Proteins. Science 2009, 324, 1068–1071. [Google Scholar] [CrossRef] [PubMed]
- Saha, J.; Chatterjee, C.; Sengupta, A.; Gupta, K.; Gupta, B. Genome-wide analysis and evolutionary study of sucrose non-fermenting 1-related protein kinase 2 (SnRK2) gene family members in Arabidopsis and Oryza. Comput. Biol. Chem. 2014, 49, 59–70. [Google Scholar] [CrossRef]
- Wei, K.; Pan, S. Maize protein phosphatase gene family: Identification and molecular characterization. BMC Genom. 2014, 15, 773. [Google Scholar] [CrossRef]
- Fan, W.; Zhao, M.; Li, S.; Bai, X.; Li, J.; Meng, H.; Mu, Z. Contrasting transcriptional responses of PYR1/PYL/RCAR ABA receptors to ABA or dehydration stress between maize seedling leaves and roots. BMC Plant Biol. 2016, 16, 99. [Google Scholar] [CrossRef] [PubMed]
- Sun, L.; Wang, Y.-P.; Chen, P.; Ren, J.; Ji, K.; Li, Q.; Li, P.; Dai, S.-J.; Leng, P. Transcriptional regulation of SlPYL, SlPP2C, and SlSnRK2 gene families encoding ABA signal core components during tomato fruit development and drought stress. J. Exp. Bot. 2011, 62, 5659–5669. [Google Scholar] [CrossRef] [PubMed]
- Gomez-Cadenas, A.; Vives, V.; I Zandalinas, S.; Manzi, M.; M Sanchez-Perez, A.; M Perez-Clemente, R.; Arbona, V. Abscisic acid: A versatile phytohormone in plant signaling and beyond. Curr. Protein. Pept. Sc. 2015, 16, 413–434. [Google Scholar] [CrossRef] [PubMed]
- Shi, H.; Ye, T.; Zhu, J.-K.; Chan, Z. Constitutive production of nitric oxide leads to enhanced drought stress resistance and extensive transcriptional reprogramming in Arabidopsis. J. Exp. Bot. 2014, 65, 4119–4131. [Google Scholar] [CrossRef] [PubMed]
- Zhao, Y.; Chan, Z.; Gao, J.; Xing, L.; Cao, M.; Yu, C.; Hu, Y.; You, J.; Shi, H.; Zhu, Y.; et al. ABA receptor PYL9 promotes drought resistance and leaf senescence. Proc. Natl. Acad. Sci. USA 2016, 113, 1949–1954. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Q.; Kong, X.; Yu, Q.; Ding, Y.; Li, X.; Yang, Y. Responses of PYR/PYL/RCAR ABA Receptors to Contrasting stresses, Heat and Cold in Arabidopsis. Plant Signal. Behav. 2019, 14, 1670596. [Google Scholar] [CrossRef]
- Tian, X.; Wang, Z.; Li, X.; Lv, T.; Liu, H.; Wang, L.; Niu, H.; Bu, Q. Characterization and functional analysis of pyrab-actin resistance-like abscisic acid receptor family in rice. Rice 2015, 8, 28. [Google Scholar] [CrossRef] [PubMed]
- Verma, R.K.; Santosh Kumar, V.V.; Yadav, S.K.; Pushkar, S.; Rao, M.V.; Chinnusamy, V. Overexpression of ABA recep-tor PYL10 gene confers drought and cold tolerance to indica rice. Front Plant Sci. 2019, 10, 1488. [Google Scholar] [CrossRef]
- Kim, H.; Lee, K.; Hwang, H.; Bhatnagar, N.; Kim, D.Y.; Yoon, I.S.; Byun, M.O.; Kim, S.T.; Jung, K.H.; Kim, B.G. Over-expression of PYL5 in rice enhances drought tolerance, inhibits growth, and modulates gene expression. J. Exp. Bot. 2014, 65, 453–464. [Google Scholar] [CrossRef]
- Mega, R.; Abe, F.; Kim, J.-S.; Tsuboi, Y.; Tanaka, K.; Kobayashi, H.; Sakata, Y.; Hanada, K.; Tsujimoto, H.; Kikuchi, J.; et al. Tuning water-use efficiency and drought tolerance in wheat using abscisic acid receptors. Nat. Plants 2019, 5, 153–159. [Google Scholar] [CrossRef]
- Zhao, Y.; Qi, G.; Ren, F.; Wang, Y.; Wang, P.; Wu, X. Analysis of PYL genes and their potential relevance to stress tol-erance and berry ripening in grape. J. Am. Soc. Hortic. Sci. 2020, 145, 308–317. [Google Scholar] [CrossRef]
- Romero, P.; Lafuente, M.T.; Rodrigo, M.J. The Citrus ABA signalosome: Identification and transcriptional regulation during sweet orange fruit ripening and leaf dehydration. J. Exp. Bot. 2012, 63, 4931–4945. [Google Scholar] [CrossRef] [PubMed]
- Hu, W.; Yan, Y.; Shi, H.; Liu, J.; Miao, H.; Tie, W.; Ding, Z.; Ding, X.; Wu, C.; Liu, Y.; et al. The core regulatory network of the abscisic acid pathway in banana: Genome-wide identification and expression analyses during development, ripening, and abiotic stress. BMC Plant Biol. 2017, 17, 145. [Google Scholar] [CrossRef] [PubMed]
- Hou, H.; Lv, L.; Huo, H.; Dai, H.; Zhang, Y. Genome-wide identification of the ABA receptors genes and their re-sponse to abiotic stress in apple. Plants 2020, 9, 1028. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Ren, W.; An, P.; Pan, Z.; Wang, L.; Dong, Z.; He, D.; Yang, J.; Pan, S.; Tian, H. Responses of Crop Water Use Efficiency to Climate Change and Agronomic Measures in the Semiarid Area of Northern China. PLoS ONE 2015, 10, e0137409. [Google Scholar] [CrossRef] [PubMed]
- Ali, A.; Pardo, J.M.; Yun, D.-J. Desensitization of ABA-Signaling: The Swing From Activation to Degradation. Front. Plant Sci. 2020, 11, 379. [Google Scholar] [CrossRef] [PubMed]
- Sun, Y.; Pri-Tal, O.; Michaeli, D.; Mosquna, A. Evolution of Abscisic Acid Signaling Module and Its Perception. Front. Plant Sci. 2020, 11, 934. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.L.; Jiang, L.; Xin, Q.; Liu, Y.; Tan, J.X.; Chen, Z.Z. Structural basis and functions of abscisic acid receptors PYLs. Front. Plant Sci. 2015, 6, 88. [Google Scholar] [CrossRef] [PubMed]
- Dittrich, M.; Mueller, H.M.; Bauer, H.; Peirats-Llobet, M.; Rodriguez, P.L.; Geilfus, C.M.; Carpentier, S.C.; Al Rasheid, K.A.; Kollist, H.; Merilo, E. The role of Arabidopsis ABA receptors from the PYR/PYL/RCAR family in stomatal ac-climation and closure signal integration. Nat. Plants 2019, 5, 1002–1011. [Google Scholar] [CrossRef] [PubMed]
- González-Guzmán, M.; Rodríguez, L.; Lorenzo-Orts, L.; Pons, C.; Sarrión-Perdigones, A.; Fernández, M.A.; Peirats-Llobet, M.; Forment, J.; Moreno-Alvero, M.; Cutler, S.R.; et al. Tomato PYR/PYL/RCAR abscisic acid receptors show high expression in root, differential sensitivity to the abscisic acid agonist quinabactin, and the capability to enhance plant drought resistance. J. Exp. Bot. 2014, 65, 4451–4464. [Google Scholar] [CrossRef]
- Bai, G.; Xie, H.; Yao, H.; Li, F.; Chen, X.; Zhang, Y.; Xiao, B.; Yang, J.; Li, Y.; Yang, D.H. Genome-wide identification and characterization of ABA receptor PYL/RCAR gene family reveals evolution and roles in drought stress in Nico-tiana tabacum. BMC Genom. 2019, 20, 575. [Google Scholar] [CrossRef] [PubMed]
- Zhang, G.; Lu, T.; Miao, W.; Sun, L.; Tian, M.; Wang, J.; Hao, F. Genome-wide identification of ABA receptor PYL family and expression analysis of PYLs in response to ABA and osmotic stress in Gossypium. PeerJ 2017, 5, e4126. [Google Scholar] [CrossRef] [PubMed]
- Yadav, S.K.; Kumar, V.V.S.; Verma, R.K.; Yadav, P.; Saroha, A.; Wankhede, D.P.; Chaudhary, B.; Chinnusamy, V. Genome-wide identification and characterization of ABA receptor PYL gene family in rice. BMC Genom. 2020, 21, 676. [Google Scholar] [CrossRef] [PubMed]
- Lei, P.; Wei, X.; Gao, R.; Huo, F.; Nie, X.; Tong, W.; Song, W. Genome-wide identification of PYL gene family in wheat: Evolution, expression and 3D structure analysis. Genomics 2021, 113, 854–866. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Li, G.; Li, Y.; Kong, X.; Zhang, L.; Wang, J.; Li, X.; Yang, Y. ABA Receptor Subfamily III Enhances Abscisic Acid Sensitivity and Improves the Drought Tolerance of Arabidopsis. Int. J. Mol. Sci. 2018, 19, 1938. [Google Scholar] [CrossRef] [PubMed]
- Lenka, S.K.; Muthusamy, S.K.; Chinnusamy, V.; Bansal, K.C. Ectopic Expression of Rice PYL3 Enhances Cold and Drought Tolerance in Arabidopsis thaliana. Mol. Biotechnol. 2018, 60, 350–361. [Google Scholar] [CrossRef] [PubMed]
- He, Z.; Zhong, J.; Sun, X.; Wang, B.; Terzaghi, W.; Dai, M. The Maize ABA Receptors ZmPYL8, 9, and 12 Facilitate Plant Drought Resistance. Front. Plant Sci. 2018, 9, 422. [Google Scholar] [CrossRef] [PubMed]
- Yu, J.; Yang, L.; Liu, X.; Tang, R.; Wang, Y.; Ge, H.; Wu, M.; Zhang, J.; Zhao, F.; Luan, S.; et al. Overexpression of Poplar Pyrabactin Resistance-Like Abscisic Acid Receptors Promotes Abscisic Acid Sensitivity and Drought Resistance in Transgenic Arabidopsis. PLoS ONE 2016, 11, e0168040. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.; Feng, L.; Wei, N.; Liu, Z.-H.; Hu, S.; Li, X.-B. Overexpression of cotton PYL genes in Arabidopsis enhances the transgenic plant tolerance to drought stress. Plant Physiol. Biochem. 2017, 115, 229–238. [Google Scholar] [CrossRef]
- Liang, C.; Liu, Y.; Li, Y.; Meng, Z.; Yan, R.; Zhu, T.; Wang, Y.; Kang, S.; Ali Abid, M.; Malik, W. Activation of ABA re-ceptors gene GhPYL9-11A is positively correlated with cotton drought tolerance in transgenic Arabidopsis. Front Plant Sci. 2017, 8, 1453. [Google Scholar] [CrossRef]
- Apel, K.; Hirt, H. Reactive oxygen species: Metabolism, oxidative stress, and signal transduction. Annu. Rev. Plant Biol. 2004, 55, 373–399. [Google Scholar] [CrossRef] [PubMed]
- Shi, H.; Chen, L.; Ye, T.; Liu, X.; Ding, K.; Chan, Z. Modulation of auxin content in Arabidopsis confers improved drought stress resistance. Plant Physiol. Biochem. 2014, 82, 209–217. [Google Scholar] [CrossRef] [PubMed]
- Ashraf, M. Biotechnological approach of improving plant salt tolerance using antioxidants as markers. Biotechnol. Adv. 2008, 27, 84–93. [Google Scholar] [CrossRef] [PubMed]
- Sachdev, S.; Ansari, S.A.; Ansari, M.I.; Fujita, M.; Hasanuzzaman, M. Abiotic Stress and Reactive Oxygen Species: Generation, Signaling, and Defense Mechanisms. Antioxidants 2021, 10, 277. [Google Scholar] [CrossRef] [PubMed]
- Gill, S.S.; Tuteja, N. Reactive oxygen species and antioxidant machinery in abiotic stress tolerance in crop plants. Plant Physiol. Biochem. 2010, 48, 909–930. [Google Scholar] [CrossRef] [PubMed]
- Sharma, P.; Jha, A.B.; Dubey, R.S.; Pessarakli, M. Reactive oxygen species, oxidative damage, and antioxidative de-fense mechanism in plants under stressful conditions. J. Exp. Bot. 2012, 2012, 217037. [Google Scholar] [CrossRef]
- Jaleel, C.A.; Gopi, R.; Sankar, B.; Manivannan, P.; Kishorekumar, A.; Sridharan, R.; Panneerselvam, R. Studies on germination, seedling vigour, lipid peroxidation and proline metabolism in Catharanthus roseus seedlings under salt stress. South Afr. J. Bot. 2007, 73, 190–195. [Google Scholar] [CrossRef]
- Meena, M.; Divyanshu, K.; Kumar, S.; Swapnil, P.; Zehra, A.; Shukla, V.; Yadav, M.; Upadhyay, R. Regulation of L-proline biosynthesis, signal transduction, transport, accumulation and its vital role in plants during variable en-vironmental conditions. Heliyon. 2019, 5, 12–19. [Google Scholar] [CrossRef]
- Sun, B.; Zhao, Y.; Shi, S.; Yang, M.; Xiao, K. TaZFP1, a C2H2 type-ZFP gene of T. aestivum, mediates salt stress toler-ance of plants by modulating diverse stress-defensive physiological processes. Plant Physiol. Bioch. 2019, 136, 127–142. [Google Scholar] [CrossRef]
- Jefferson, R.A.; Kavanagh, T.A.; Bevan, M.W. GUS fusions: Beta-glucuronidase as a sensitive and versatile gene fu-sion marker in higher plants. EMBO J. 1987, 6, 3901–3907. [Google Scholar] [CrossRef]
- Magné, C.; Larher, F. High sugar content of extracts interferes with colorimetric determination of amino acids and free proline. Anal. Biochem. 1992, 200, 115–118. [Google Scholar] [CrossRef] [PubMed]


Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yao, P.; Zhang, C.; Bi, Z.; Liu, Y.; Liu, Z.; Wei, J.; Su, X.; Bai, J.; Cui, J.; Sun, C. Overexpression of Potato PYL16 Gene in Tobacco Enhances the Transgenic Plant Tolerance to Drought Stress. Int. J. Mol. Sci. 2024, 25, 8644. https://doi.org/10.3390/ijms25168644
Yao P, Zhang C, Bi Z, Liu Y, Liu Z, Wei J, Su X, Bai J, Cui J, Sun C. Overexpression of Potato PYL16 Gene in Tobacco Enhances the Transgenic Plant Tolerance to Drought Stress. International Journal of Molecular Sciences. 2024; 25(16):8644. https://doi.org/10.3390/ijms25168644
Chicago/Turabian StyleYao, Panfeng, Chunli Zhang, Zhenzhen Bi, Yuhui Liu, Zhen Liu, Jia Wei, Xinglong Su, Jiangping Bai, Junmei Cui, and Chao Sun. 2024. "Overexpression of Potato PYL16 Gene in Tobacco Enhances the Transgenic Plant Tolerance to Drought Stress" International Journal of Molecular Sciences 25, no. 16: 8644. https://doi.org/10.3390/ijms25168644
APA StyleYao, P., Zhang, C., Bi, Z., Liu, Y., Liu, Z., Wei, J., Su, X., Bai, J., Cui, J., & Sun, C. (2024). Overexpression of Potato PYL16 Gene in Tobacco Enhances the Transgenic Plant Tolerance to Drought Stress. International Journal of Molecular Sciences, 25(16), 8644. https://doi.org/10.3390/ijms25168644






