Mulberry Leaf Compounds and Gut Microbiota in Alzheimer’s Disease and Diabetes: A Study Using Network Pharmacology, Molecular Dynamics Simulation, and Cellular Assays
Abstract
:1. Introduction
2. Results
2.1. Cluster Analysis of Mulberry Leaf Components
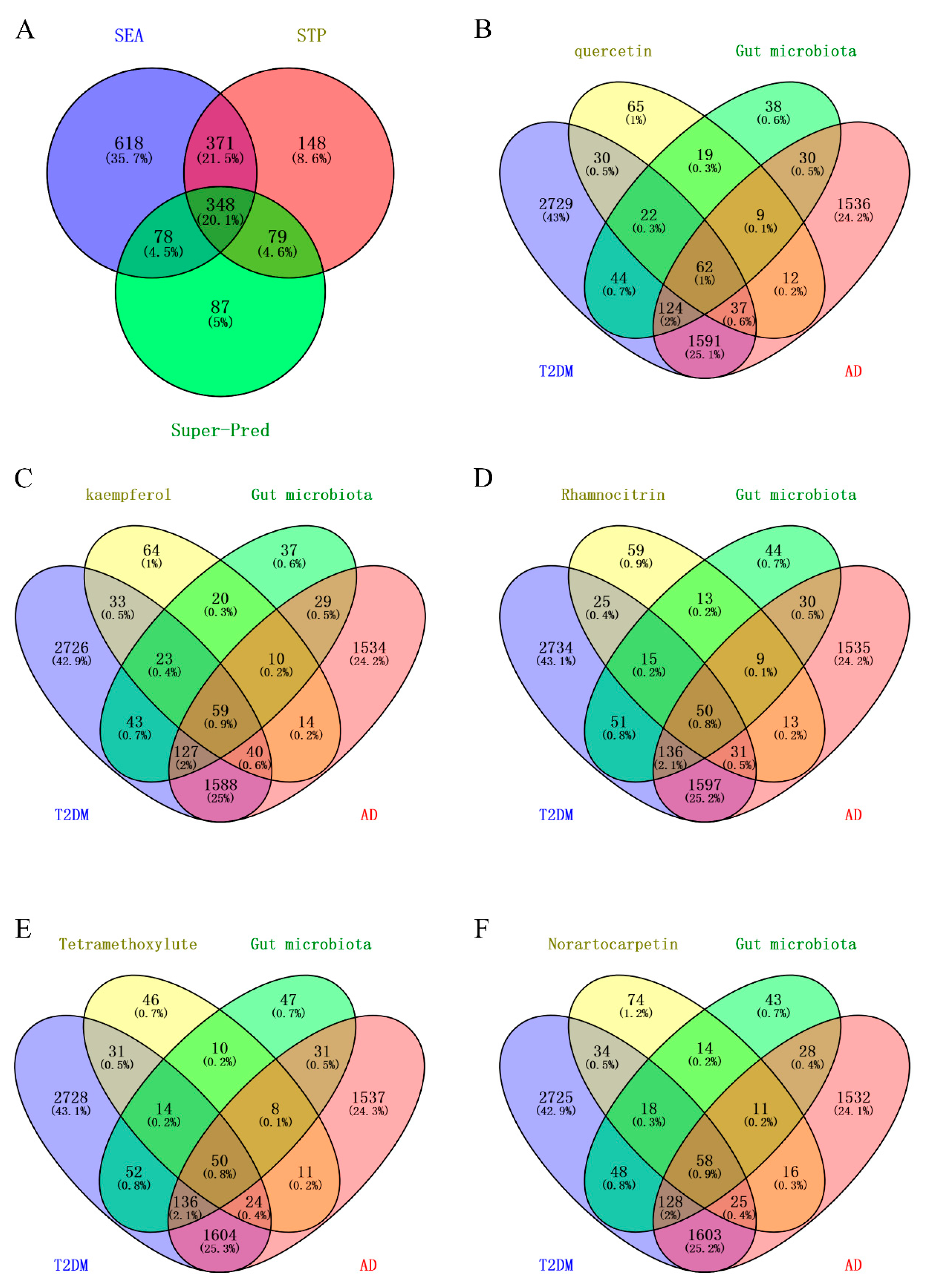
2.2. Targets of Mulberry Leaf Components Combined with Gut Microbiota to Intervene in AD and T2DM
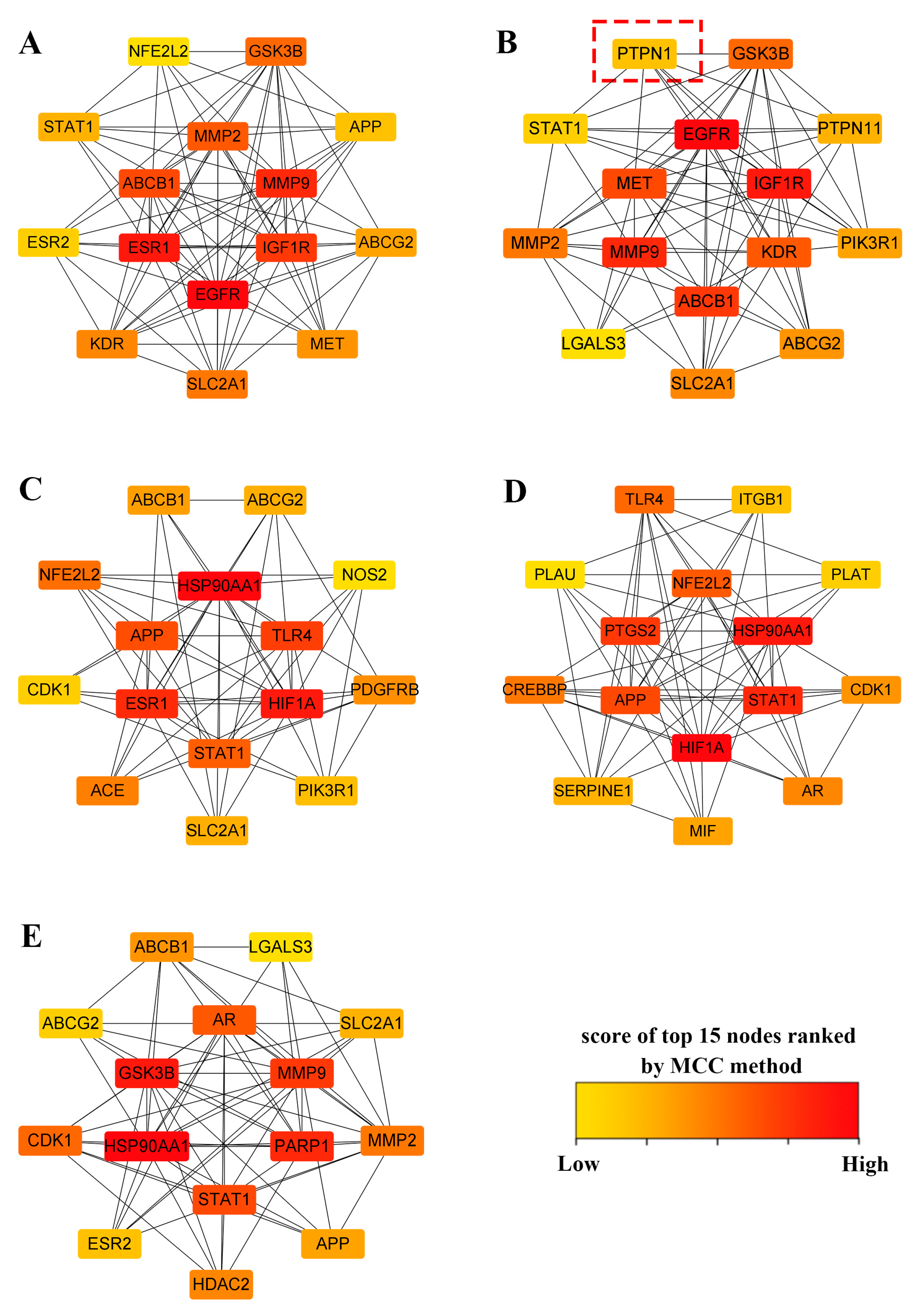
2.3. Construction of PPI Networks and Top 15 Targets’ Screening
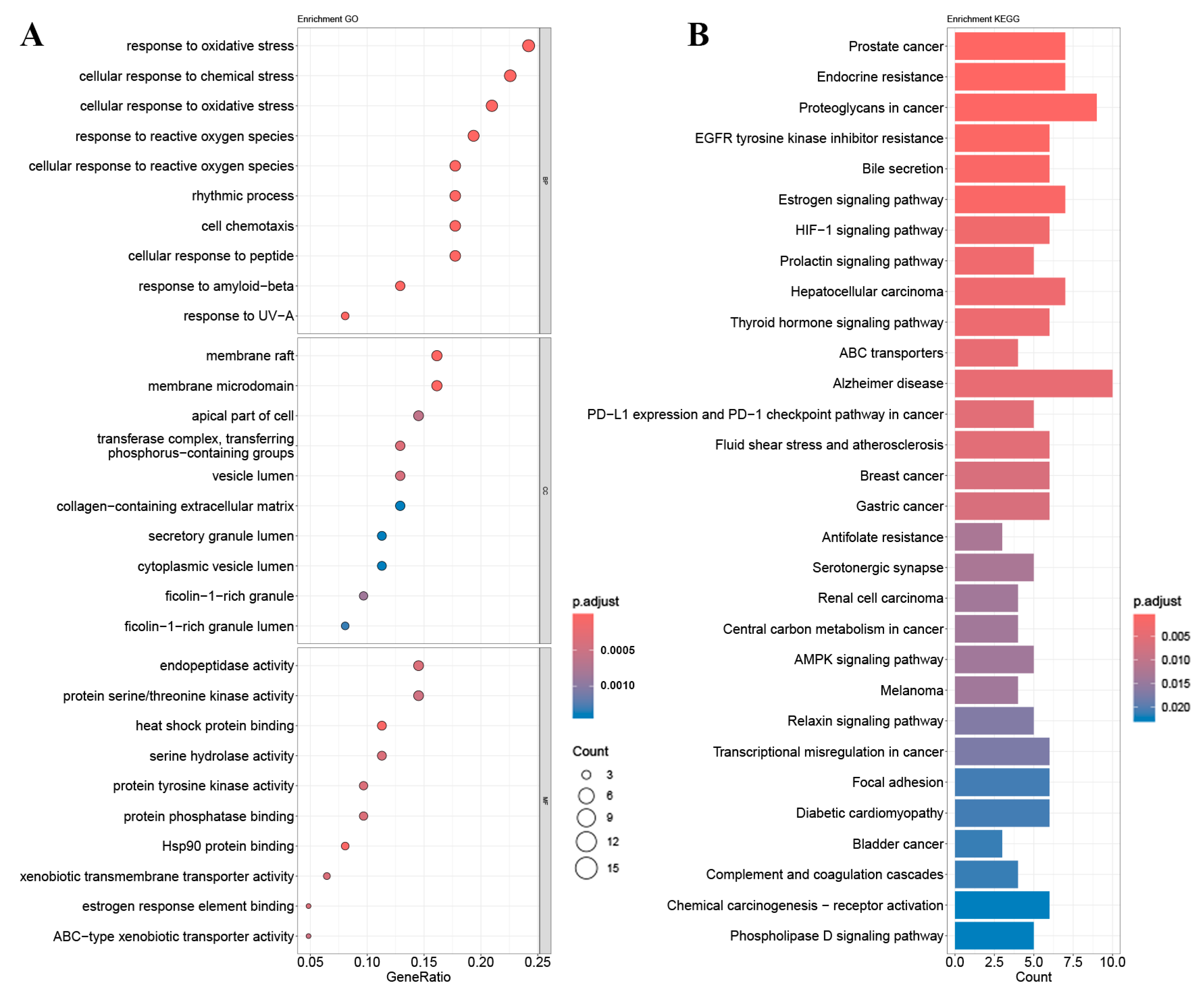
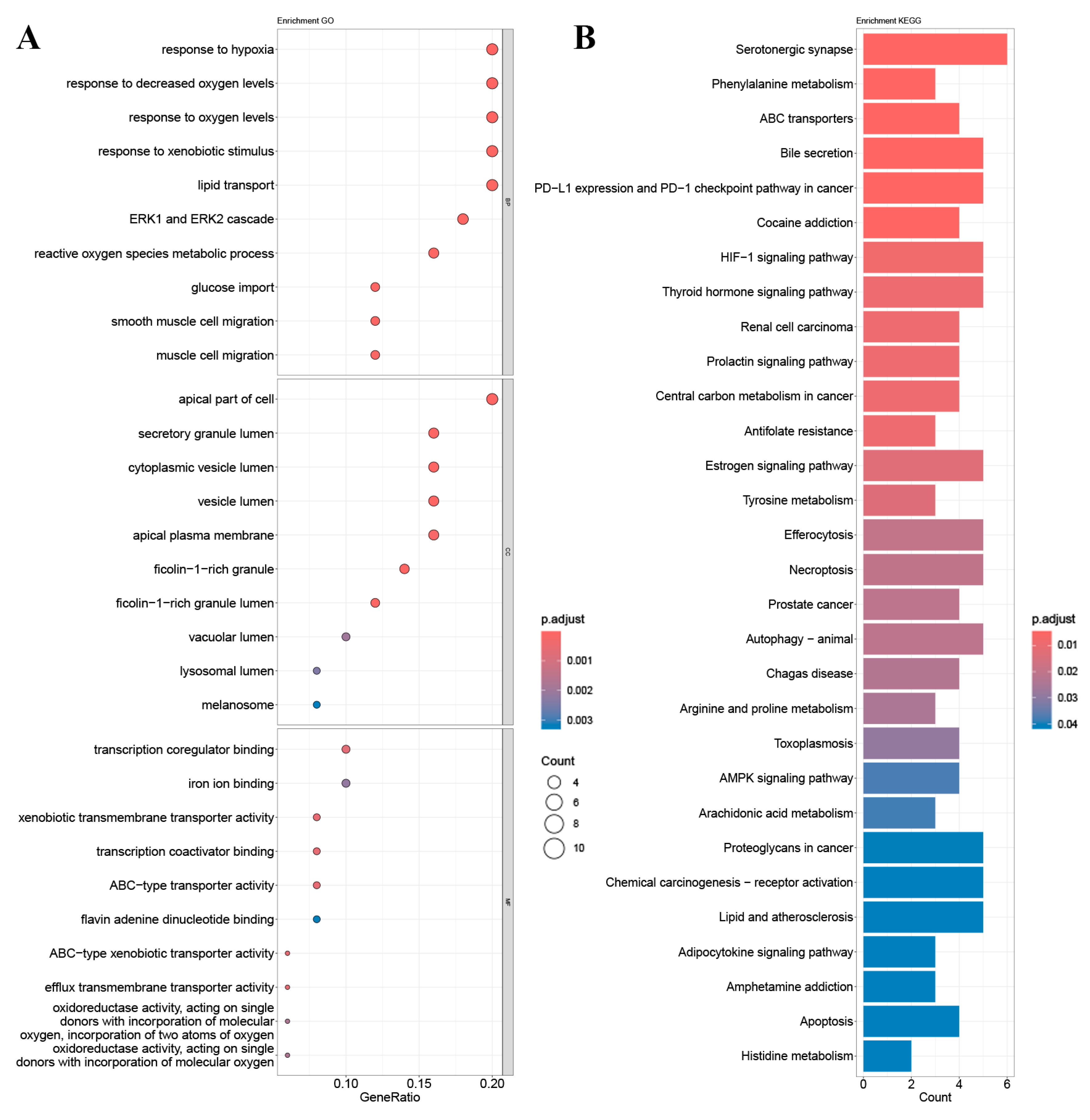
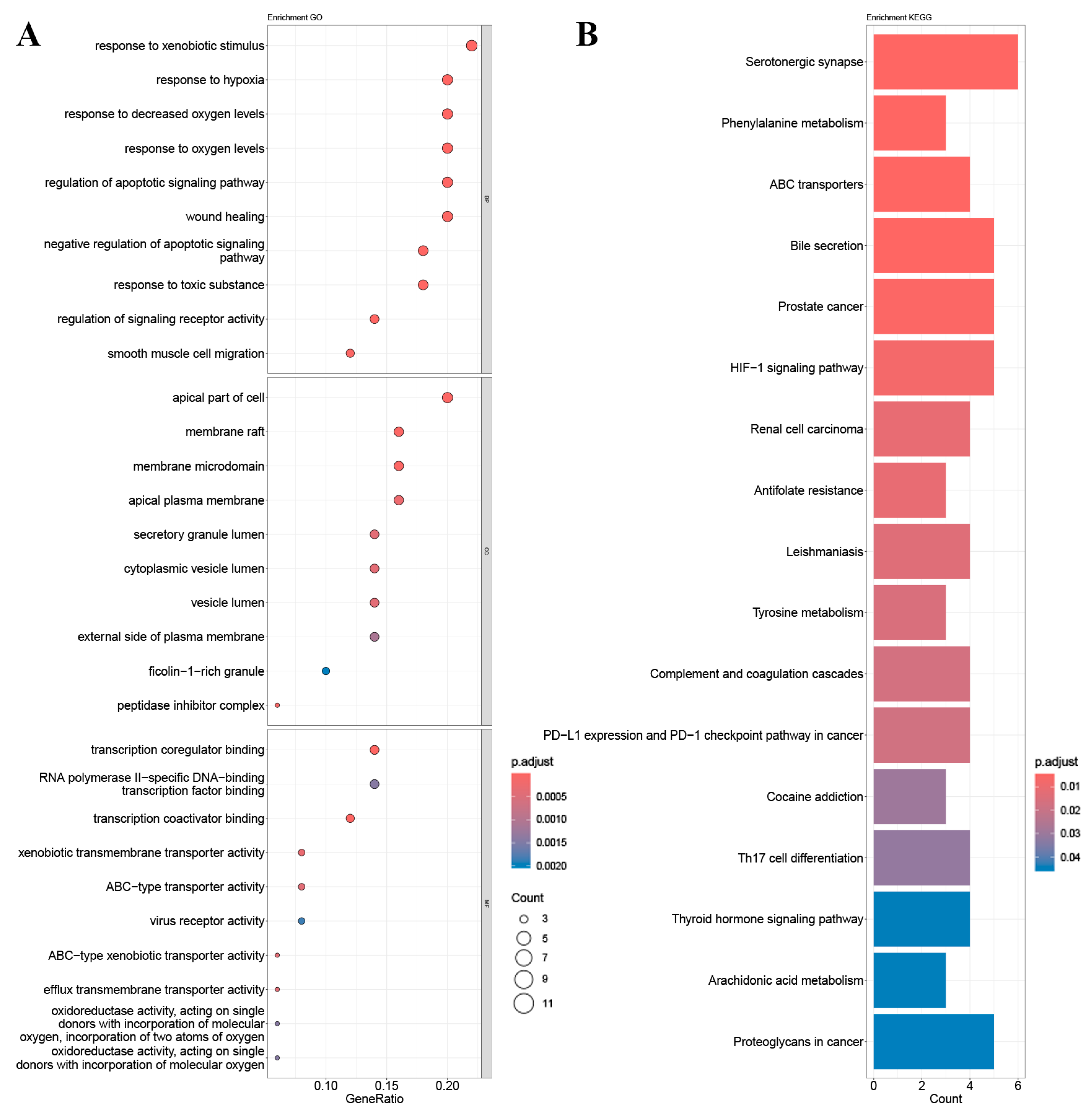
2.4. GO Gene Enrichment and KEGG Pathway Analysis
2.5. Quantum Chemical Calculation
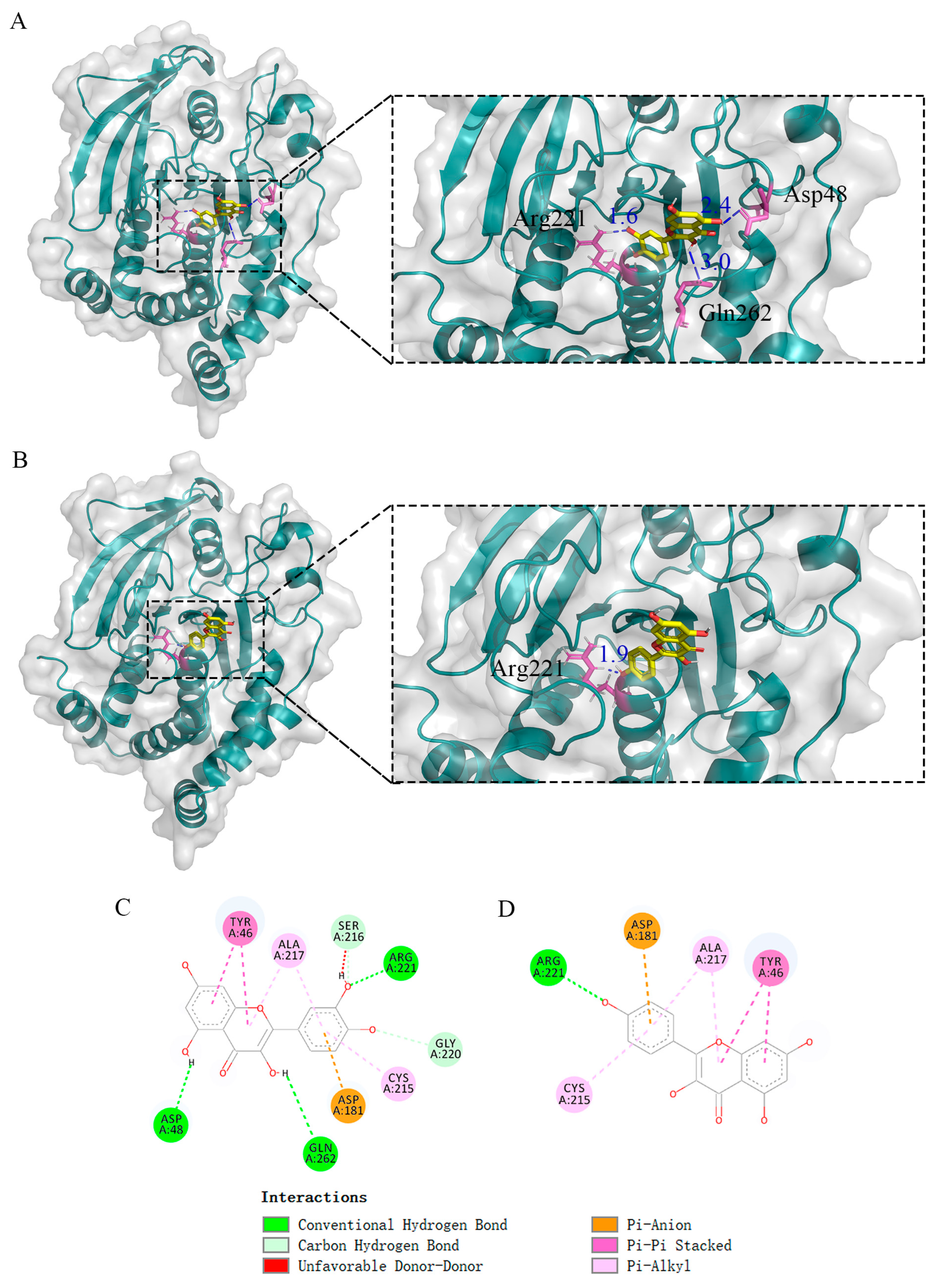
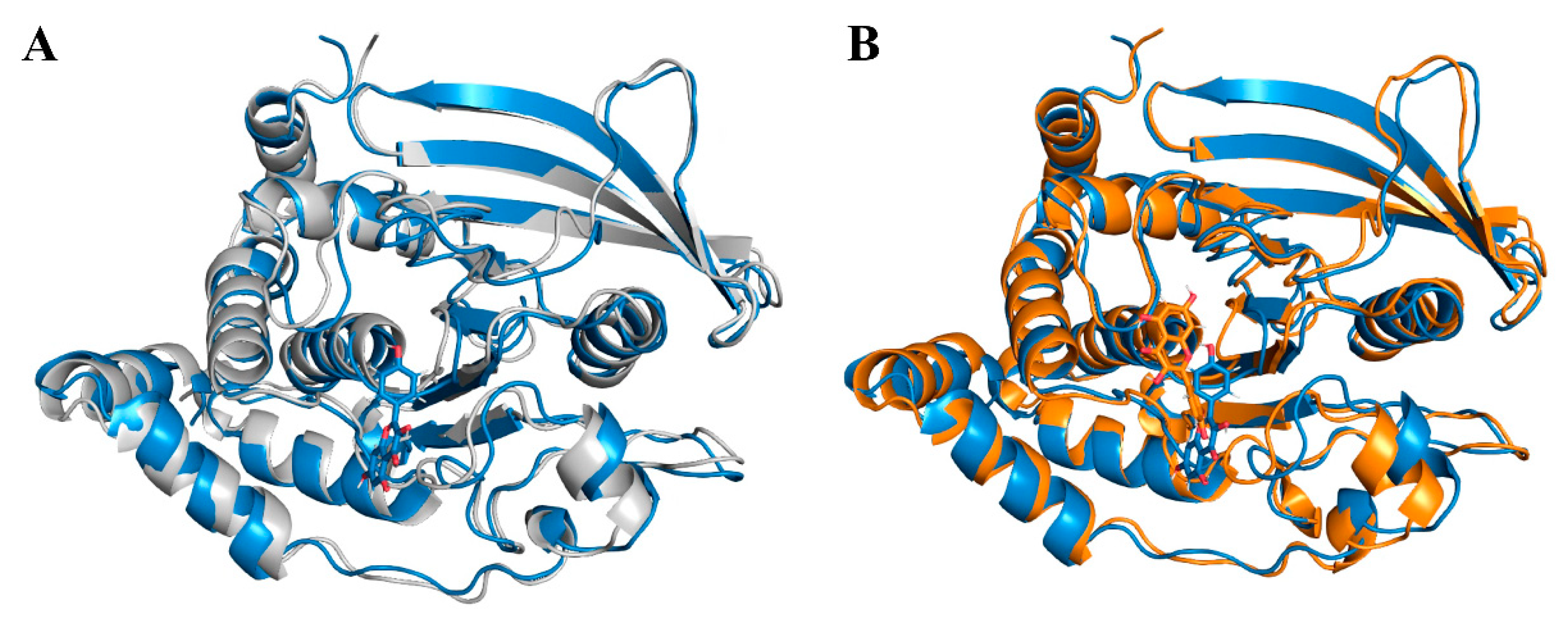
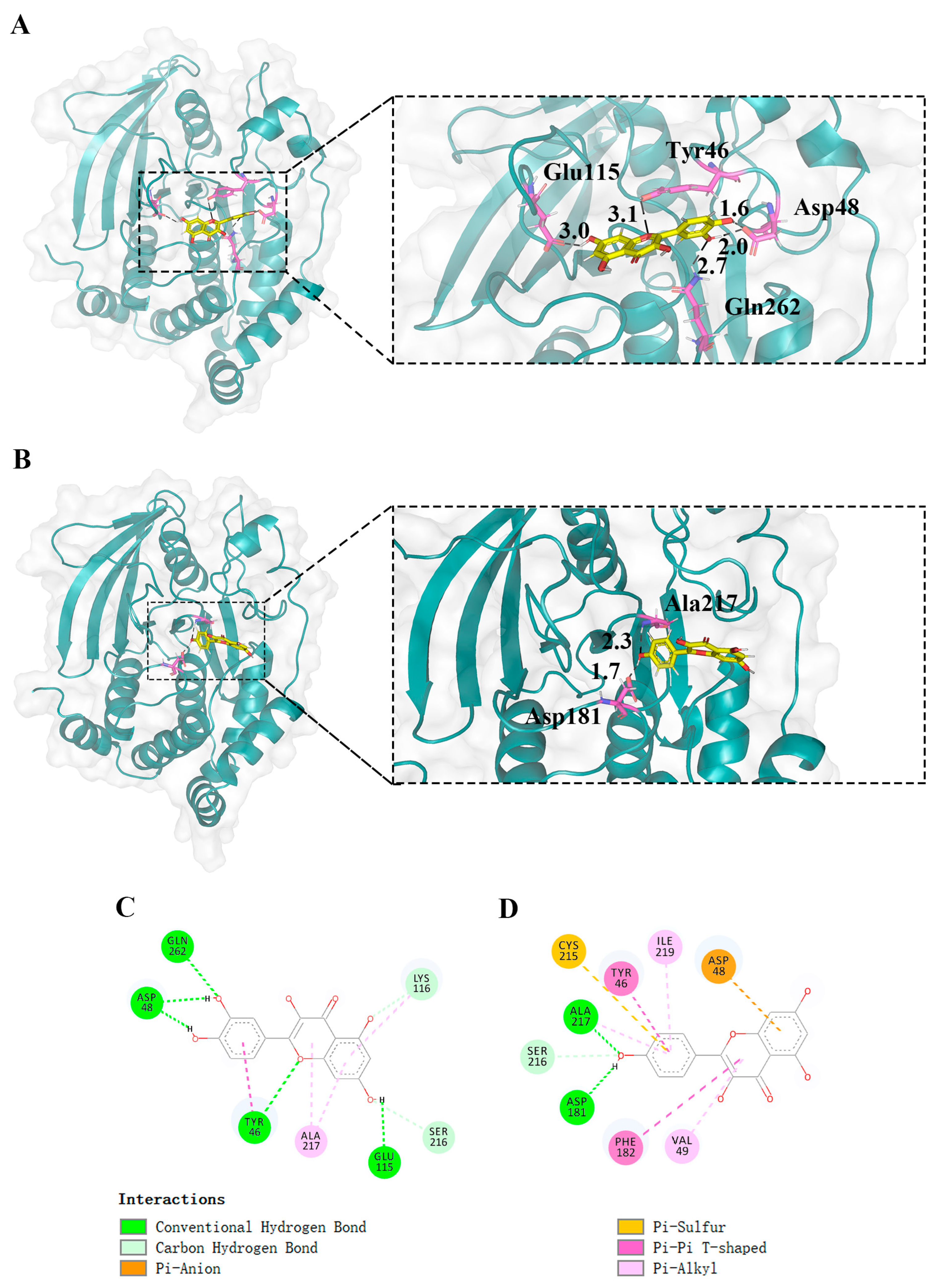
2.6. Molecular Docking
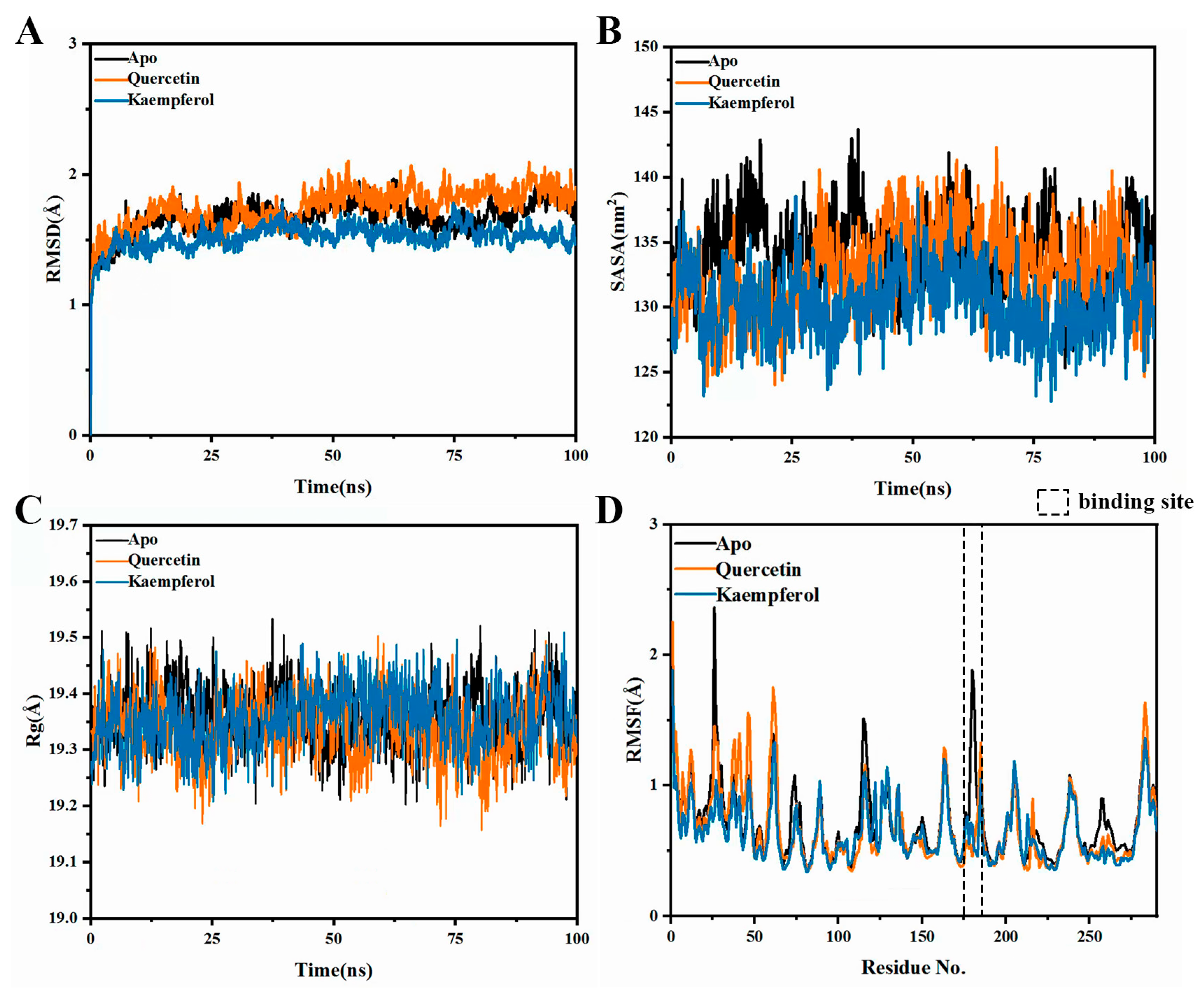
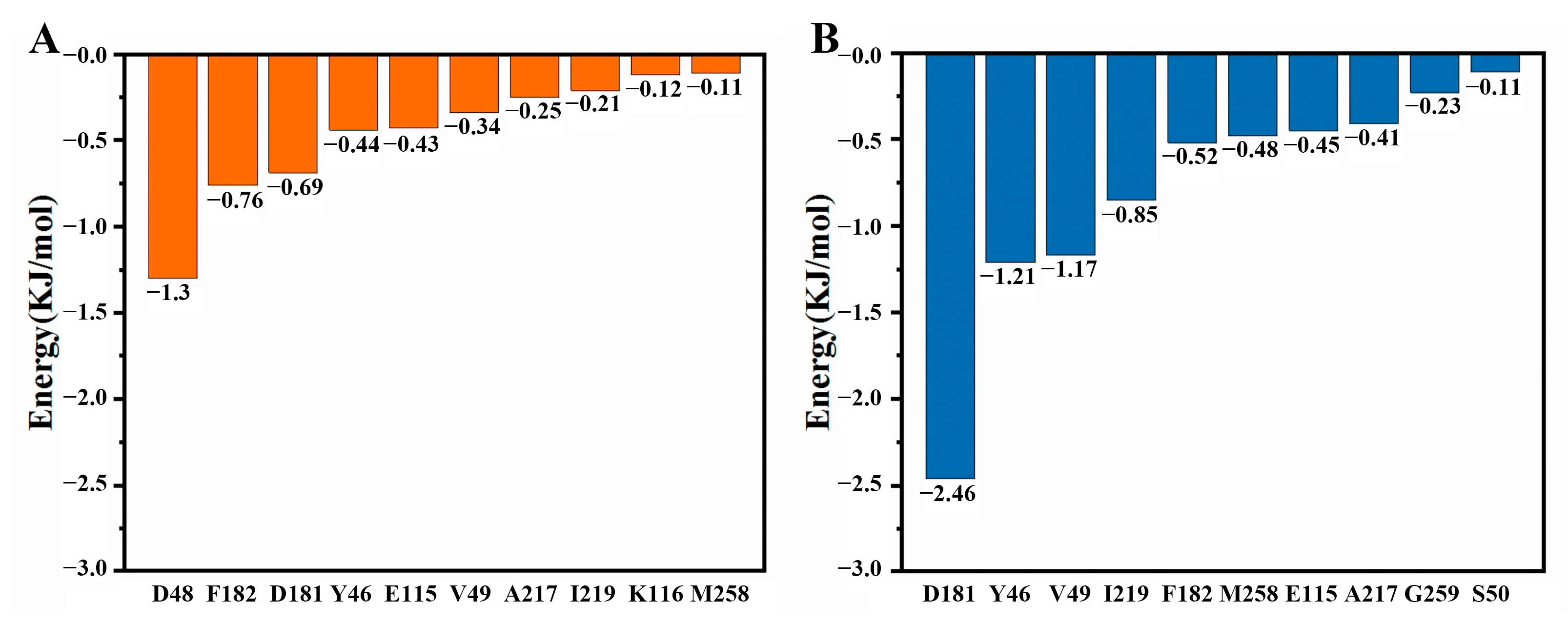
2.7. Molecular Dynamics Simulations
2.8. Half Maximal Inhibitory Concentration (IC50) and Tyrosine Phosphorylation of Kaempferol
2.9. Prediction of ADMET Properties
3. Discussion
4. Materials and Methods
4.1. Acquisition and Cluster Analysis of Components in Mulberry Leaf
4.2. Prediction of AD-Related Targets and T2DM-Related Targets
4.3. Prediction of Targets of Quercetin and Its Structural Analogs
4.4. Prediction of Gut Microbiota Metabolite Targets
4.5. Acquisition Targets for Mulberry Leaf Ingredients Combined with Gut Microbiota to Intervene in AD and T2DM
4.6. Construction of the Protein–Protein Interaction (PPI) Network
4.7. GO and KEGG Enrichment Analysis
4.8. Quantum Chemical Calculation
4.9. Molecular Docking
4.10. Molecular Dynamics Simulations
4.11. IC50 and Tyrosine Phosphorylation of Kaempferol
4.12. Prediction of ADMET Properties
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Athanasaki, A.; Melanis, K.; Tsantzali, I.; Stefanou, M.I.; Ntymenou, S.; Paraskevas, S.G.; Kalamatianos, T.; Boutati, E.; Lambadiari, V.; Voumvourakis, K.I.; et al. Type 2 Diabetes Mellitus as a Risk Factor for Alzheimer’s Disease: Review and Meta-Analysis. Biomedicines 2022, 10, 778. [Google Scholar] [CrossRef] [PubMed]
- Jeremic, D.; Jiménez-Díaz, L.; Navarro-López, J.D. Past, Present and Future of Therapeutic Strategies against Amyloid-β Peptides in Alzheimer’s Disease: A Systematic Review. Ageing Res. Rev. 2021, 72, 101496. [Google Scholar] [CrossRef] [PubMed]
- Khan, S.; Barve, K.H.; Kumar, M.S. Recent Advancements in Pathogenesis, Diagnostics and Treatment of Alzheimer’s Disease. Curr. Neuropharmacol. 2020, 18, 1106–1125. [Google Scholar] [CrossRef] [PubMed]
- Pleen, J.; Townley, R. Alzheimer’s Disease Clinical Trial Update 2019–2021. J. Neurol. 2022, 269, 1038–1051. [Google Scholar] [CrossRef] [PubMed]
- Tatulian, S.A. Challenges and Hopes for Alzheimer’s Disease. Drug Discov. Today 2022, 27, 1027–1043. [Google Scholar] [CrossRef] [PubMed]
- Liu, G.; Li, Y.; Pan, A.; Hu, Y.; Chen, S.; Qian, F.; Rimm, E.B.; Manson, J.E.; Stampfer, M.J.; Giatsidis, G.; et al. Adherence to a Healthy Lifestyle in Association With Microvascular Complications Among Adults With Type 2 Diabetes. JAMA Netw. Open 2023, 6, e2252239. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Wang, D.; Liu, Y.-P. Metabolite Profiles of Diabetes Mellitus and Response to Intervention in Anti-Hyperglycemic Drugs. Front. Endocrinol. 2023, 14, 1237934. [Google Scholar] [CrossRef] [PubMed]
- Saeedi, P.; Petersohn, I.; Salpea, P.; Malanda, B.; Karuranga, S.; Unwin, N.; Colagiuri, S.; Guariguata, L.; Motala, A.A.; Ogurtsova, K.; et al. Global and Regional Diabetes Prevalence Estimates for 2019 and Projections for 2030 and 2045: Results from the International Diabetes Federation Diabetes Atlas, 9th Edition. Diabetes Res. Clin. Pract. 2019, 157, 107843. [Google Scholar] [CrossRef]
- Kahn, S.E. The Relative Contributions of Insulin Resistance and Beta-Cell Dysfunction to the Pathophysiology of Type 2 Diabetes. Diabetologia 2003, 46, 3–19. [Google Scholar] [CrossRef] [PubMed]
- Knop, F.K.; Vilsbøll, T.; Højberg, P.V.; Larsen, S.; Madsbad, S.; Vølund, A.; Holst, J.J.; Krarup, T. Reduced Incretin Effect in Type 2 Diabetes: Cause or Consequence of the Diabetic State? Diabetes 2007, 56, 1951–1959. [Google Scholar] [CrossRef] [PubMed]
- Kaur, R.; Kaur, M.; Singh, J. Endothelial Dysfunction and Platelet Hyperactivity in Type 2 Diabetes Mellitus: Molecular Insights and Therapeutic Strategies. Cardiovasc. Diabetol. 2018, 17, 121. [Google Scholar] [CrossRef] [PubMed]
- Zheng, Y.; Ley, S.H.; Hu, F.B. Global Aetiology and Epidemiology of Type 2 Diabetes Mellitus and Its Complications. Nat. Rev. Endocrinol. 2018, 14, 88–98. [Google Scholar] [CrossRef] [PubMed]
- Haan, M.N. Therapy Insight: Type 2 Diabetes Mellitus and the Risk of Late-Onset Alzheimer’s Disease. Nat. Clin. Pract. Neurol. 2006, 2, 159–166. [Google Scholar] [CrossRef] [PubMed]
- Hu, Z.; Jiao, R.; Wang, P.; Zhu, Y.; Zhao, J.; De Jager, P.; Bennett, D.A.; Jin, L.; Xiong, M. Shared Causal Paths Underlying Alzheimer’s Dementia and Type 2 Diabetes. Sci. Rep. 2020, 10, 4107. [Google Scholar] [CrossRef] [PubMed]
- Nuthikattu, S.; Milenkovic, D.; Norman, J.E.; Villablanca, A.C. Single Nuclei Transcriptomics in Diabetic Mice Reveals Altered Brain Hippocampal Endothelial Cell Function, Permeability, and Behavior. Biochim. Biophys. Acta Mol. Basis Dis. 2023, 1870, 166970. [Google Scholar] [CrossRef]
- Profenno, L.A.; Porsteinsson, A.P.; Faraone, S.V. Meta-Analysis of Alzheimer’s Disease Risk with Obesity, Diabetes, and Related Disorders. Biol. Psychiatry 2010, 67, 505–512. [Google Scholar] [CrossRef] [PubMed]
- de la Monte, S.M. Type 3 Diabetes Is Sporadic Alzheimer’s Disease: Mini-Review. Eur. Neuropsychopharmacol. 2014, 24, 1954–1960. [Google Scholar] [CrossRef]
- de la Monte, S.M.; Wands, J.R. Alzheimer’s Disease Is Type 3 Diabetes-Evidence Reviewed. J. Diabetes Sci. Technol. 2008, 2, 1101–1113. [Google Scholar] [CrossRef]
- Janoutová, J.; Machaczka, O.; Zatloukalová, A.; Janout, V. Is Alzheimer’s Disease a Type 3 Diabetes? A Review. Cent. Eur. J. Public Health 2022, 30, 139–143. [Google Scholar] [CrossRef] [PubMed]
- Kumkoon, T.; Srisaisap, M.; Boonserm, P. Biosynthesized Silver Nanoparticles Using Morus alba (White Mulberry) Leaf Extract as Potential Antibacterial and Anticancer Agents. Molecules 2023, 28, 1213. [Google Scholar] [CrossRef] [PubMed]
- Liu, T.; Li, Y.; Wang, L.; Zhang, X.; Zhang, Y.; Gai, X.; Chen, L.; Liu, L.; Yang, L.; Wang, B. Network Pharmacology-Based Exploration Identified the Antiviral Efficacy of Quercetin Isolated from Mulberry Leaves against Enterovirus 71 via the NF-κB Signaling Pathway. Front. Pharmacol. 2023, 14, 1260288. [Google Scholar] [CrossRef] [PubMed]
- Ma, G.; Chai, X.; Hou, G.; Zhao, F.; Meng, Q. Phytochemistry, Bioactivities and Future Prospects of Mulberry Leaves: A Review. Food Chem. 2022, 372, 131335. [Google Scholar] [CrossRef] [PubMed]
- Naowaboot, J.; Pannangpetch, P.; Kukongviriyapan, V.; Kukongviriyapan, U.; Nakmareong, S.; Itharat, A. Mulberry Leaf Extract Restores Arterial Pressure in Streptozotocin-Induced Chronic Diabetic Rats. Nutr. Res. 2009, 29, 602–608. [Google Scholar] [CrossRef] [PubMed]
- Ramadan, E.M.; Abou-Taleb, K.A.; Galal, G.F.; Abdel-Hamid, N.S. Antibacterial, Antibiofilm and Antitumor Activities of Grape and Mulberry Leaves Ethanolic Extracts towards Bacterial Clinical Strains. Ann. Agric. Sci. 2017, 62, 151–159. [Google Scholar] [CrossRef]
- Suriyaprom, S.; Srisai, P.; Intachaisri, V.; Kaewkod, T.; Pekkoh, J.; Desvaux, M.; Tragoolpua, Y. Antioxidant and Anti-Inflammatory Activity on LPS-Stimulated RAW 264.7 Macrophage Cells of White Mulberry (Morus alba L.) Leaf Extracts. Molecules 2023, 28, 4395. [Google Scholar] [CrossRef] [PubMed]
- Thaipitakwong, T.; Numhom, S.; Aramwit, P. Mulberry Leaves and Their Potential Effects against Cardiometabolic Risks: A Review of Chemical Compositions, Biological Properties and Clinical Efficacy. Pharm. Biol. 2018, 56, 109–118. [Google Scholar] [CrossRef] [PubMed]
- Wei, J.; Zhu, Y.; Lin, T.; Tao, H.; Chen, L.; Xu, Z.; Lv, Z.; Liu, P. Preliminary Comparisons of Tender Shoots and Young Leaves of 12 Mulberry Varieties as Vegetables and Constituents Relevant for Their Potential Use as Functional Food for Blood Sugar Control. Plants 2023, 12, 3748. [Google Scholar] [CrossRef] [PubMed]
- Yu, Y.; Chen, Y.; Shi, X.; Ye, C.; Wang, J.; Huang, J.; Zhang, B.; Deng, Z. Hepatoprotective Effects of Different Mulberry Leaf Extracts against Acute Liver Injury in Rats by Alleviating Oxidative Stress and Inflammatory Response. Food Funct. 2022, 13, 8593–8604. [Google Scholar] [CrossRef] [PubMed]
- Dajas, F. Life or Death: Neuroprotective and Anticancer Effects of Quercetin. J. Ethnopharmacol. 2012, 143, 383–396. [Google Scholar] [CrossRef] [PubMed]
- Jiang, H.; Yamashita, Y.; Nakamura, A.; Croft, K.; Ashida, H. Quercetin and Its Metabolite Isorhamnetin Promote Glucose Uptake through Different Signalling Pathways in Myotubes. Sci. Rep. 2019, 9, 2690. [Google Scholar] [CrossRef] [PubMed]
- Khan, H.; Ullah, H.; Aschner, M.; Cheang, W.S.; Akkol, E.K. Neuroprotective Effects of Quercetin in Alzheimer’s Disease. Biomolecules 2019, 10, 59. [Google Scholar] [CrossRef] [PubMed]
- Vessal, M.; Hemmati, M.; Vasei, M. Antidiabetic Effects of Quercetin in Streptozocin-Induced Diabetic Rats. Comp. Biochem. Physiol. Toxicol. Part C Pharmacol. 2003, 135, 357–364. [Google Scholar] [CrossRef] [PubMed]
- Grabrucker, S.; Marizzoni, M.; Silajdžić, E.; Lopizzo, N.; Mombelli, E.; Nicolas, S.; Dohm-Hansen, S.; Scassellati, C.; Moretti, D.V.; Rosa, M.; et al. Microbiota from Alzheimer’s Patients Induce Deficits in Cognition and Hippocampal Neurogenesis. Brain J. Neurol. 2023, 146, 4916–4934. [Google Scholar] [CrossRef] [PubMed]
- Jiang, C.; Li, G.; Huang, P.; Liu, Z.; Zhao, B. The Gut Microbiota and Alzheimer’s Disease. J. Alzheimers Dis. 2017, 58, 1–15. [Google Scholar] [CrossRef] [PubMed]
- Buckley, M.M.; O’Mahony, S.M.; O’Malley, D. Convergence of Neuro-Endocrine-Immune Pathways in the Pathophysiology of Irritable Bowel Syndrome. World J. Gastroenterol. 2014, 20, 8846–8858. [Google Scholar]
- El Aidy, S.; Dinan, T.G.; Cryan, J.F. Gut Microbiota: The Conductor in the Orchestra of Immune–Neuroendocrine Communication. Clin. Ther. 2015, 37, 954–967. [Google Scholar] [CrossRef] [PubMed]
- Duan, H.; Khan, G.J.; Shang, L.-J.; Peng, H.; Hu, W.-C.; Zhang, J.-Y.; Hua, J.; Cassandra, A.; Rashed, M.M.A.; Zhai, K.-F. Computational Pharmacology and Bioinformatics to Explore the Potential Mechanism of Schisandra against Atherosclerosis. Food Chem. Toxicol. 2021, 150, 112058. [Google Scholar] [CrossRef] [PubMed]
- Nguyen, N.H.; Fumery, M.; Dulai, P.S.; Prokop, L.J.; Sandborn, W.J.; Murad, M.H.; Singh, S. Comparative Efficacy and Tolerability of Pharmacological Agents for Management of Mild to Moderate Ulcerative Colitis: A Systematic Review and Network Meta-Analyses. Lancet Gastroenterol. Hepatol. 2018, 3, 742–753. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.-S.; Gong, X.-F.; Feng, J.-L.; Li, H.-S.; Li, X.; Deng, S.; Ren, P.-Z.; Wang, J.-M.; Lv, M.-S.; Jin, R.-F.; et al. Study on the Mechanism of Jiawei Shengjiang Powder in Improving Male Asthma-Induced Asthenospermia Based on Network Pharmacology and Bioinformatics. Drug Des. Devel. Ther. 2021, 15, 1245–1259. [Google Scholar] [CrossRef] [PubMed]
- Proença, C.; Ribeiro, D.; Freitas, M.; Carvalho, F.; Fernandes, E. A Comprehensive Review on the Antidiabetic Activity of Flavonoids Targeting PTP1B and DPP-4: A Structure-Activity Relationship Analysis. Crit. Rev. Food Sci. Nutr. 2022, 62, 4095–4151. [Google Scholar] [CrossRef]
- Olloquequi, J.; Cano, A.; Sanchez-López, E.; Carrasco, M.; Verdaguer, E.; Fortuna, A.; Folch, J.; Bulló, M.; Auladell, C.; Camins, A.; et al. Protein Tyrosine Phosphatase 1B (PTP1B) as a Potential Therapeutic Target for Neurological Disorders. Biomed. Pharmacother. Biomed. Pharmacother. 2022, 155, 113709. [Google Scholar] [CrossRef] [PubMed]
- Vieira, M.N.N.; Lyra ESilva, N.M.; Ferreira, S.T.; De Felice, F.G. Protein Tyrosine Phosphatase 1B (PTP1B): A Potential Target for Alzheimer’s Therapy? Front. Aging Neurosci. 2017, 9, 7. [Google Scholar] [CrossRef] [PubMed]
- Yip, S.-C.; Saha, S.; Chernoff, J. PTP1B: A Double Agent in Metabolism and Oncogenesis. Trends Biochem. Sci. 2010, 35, 442–449. [Google Scholar] [CrossRef] [PubMed]
- Stuible, M.; Doody, K.M.; Tremblay, M.L. PTP1B and TC-PTP: Regulators of Transformation and Tumorigenesis. Cancer Metastasis Rev. 2008, 27, 215–230. [Google Scholar] [CrossRef] [PubMed]
- Nickel, J.; Gohlke, B.-O.; Erehman, J.; Banerjee, P.; Rong, W.W.; Goede, A.; Dunkel, M.; Preissner, R. SuperPred: Update on Drug Classification and Target Prediction. Nucleic Acids Res. 2014, 42, W26–W31. [Google Scholar] [CrossRef] [PubMed]
- Daina, A.; Michielin, O.; Zoete, V. SwissTargetPrediction: Updated Data and New Features for Efficient Prediction of Protein Targets of Small Molecules. Nucleic Acids Res. 2019, 47, W357–W364. [Google Scholar] [CrossRef] [PubMed]
- Keiser, M.J.; Roth, B.L.; Armbruster, B.N.; Ernsberger, P.; Irwin, J.J.; Shoichet, B.K. Relating Protein Pharmacology by Ligand Chemistry. Nat. Biotechnol. 2007, 25, 197–206. [Google Scholar] [CrossRef] [PubMed]
- Szklarczyk, D.; Gable, A.L.; Lyon, D.; Junge, A.; Wyder, S.; Huerta-Cepas, J.; Simonovic, M.; Doncheva, N.T.; Morris, J.H.; Bork, P.; et al. STRING V11: Protein-Protein Association Networks with Increased Coverage, Supporting Functional Discovery in Genome-Wide Experimental Datasets. Nucleic Acids Res. 2019, 47, D607–D613. [Google Scholar] [CrossRef] [PubMed]
- Shannon, P.; Markiel, A.; Ozier, O.; Baliga, N.S.; Wang, J.T.; Ramage, D.; Amin, N.; Schwikowski, B.; Ideker, T. Cytoscape: A Software Environment for Integrated Models of Biomolecular Interaction Networks. Genome Res. 2003, 13, 2498–2504. [Google Scholar] [CrossRef] [PubMed]
- Xu, X.; Liu, Y.; Fu, X.; Li, G.; Han, W.; Li, W. Network Pharmacology and Experiment Indicated That Medicinal Food Homologous Components Play Important Roles in Insomnia. Food Front. 2023, 4, 1859–1877. [Google Scholar] [CrossRef]
- Hussain, M.S.; Altamimi, A.S.A.; Afzal, M.; Almalki, W.H.; Kazmi, I.; Alzarea, S.I.; Gupta, G.; Shahwan, M.; Kukreti, N.; Wong, L.S.; et al. Kaempferol: Paving the Path for Advanced Treatments in Aging-Related Diseases. Exp. Gerontol. 2024, 188, 112389. [Google Scholar] [CrossRef] [PubMed]
- Jin, S.; Zhang, L.; Wang, L. Kaempferol, a Potential Neuroprotective Agent in Neurodegenerative Diseases: From Chemistry to Medicine. Biomed. Pharmacother. 2023, 165, 115215. [Google Scholar] [CrossRef] [PubMed]
- Zhou, Z.; Wang, M.; Guo, Z.; Zhang, X. Pharmacokinetic Evaluation of the Interaction between Oral Kaempferol and Ethanol in Rats. Acta Pharm. 2016, 66, 563–568. [Google Scholar] [CrossRef] [PubMed]
- Liang, S.; Tran, E.; Du, X.; Dong, J.; Sudholz, H.; Chen, H.; Qu, Z.; Huntington, N.D.; Babon, J.J.; Kershaw, N.J.; et al. A Small Molecule Inhibitor of PTP1B and PTPN2 Enhances T Cell Anti-Tumor Immunity. Nat. Commun. 2023, 14, 4524. [Google Scholar] [CrossRef] [PubMed]
- Douty, B.; Wayland, B.; Ala, P.J.; Bower, M.J.; Pruitt, J.; Bostrom, L.; Wei, M.; Klabe, R.; Gonneville, L.; Wynn, R.; et al. Isothiazolidinone Inhibitors of PTP1B Containing Imidazoles and Imidazolines. Bioorg. Med. Chem. Lett. 2008, 18, 66–71. [Google Scholar] [CrossRef] [PubMed]
- Halayal, R.Y.; Bagewadi, Z.K.; Maliger, R.B.; Al Jadidi, S.; Deshpande, S.H. Network Pharmacology Based Anti-Diabetic Attributes of Bioactive Compounds from Ocimum gratissimum L. through Computational Approach. Saudi J. Biol. Sci. 2023, 30, 103766. [Google Scholar] [CrossRef] [PubMed]
- Yang, L.; Zhao, Y.; Qu, R.; Fu, Y.; Zhou, C.; Yu, J. A Network Pharmacology and Molecular Docking Approach to Reveal the Mechanism of Chaihu Anxin Capsule in Depression. Front. Endocrinol. 2023, 14, 1256045. [Google Scholar] [CrossRef] [PubMed]
- Gan, L.; Inamura, Y.; Shimizu, Y.; Yokoi, Y.; Ohnishi, Y.; Song, Z.; Kumaki, Y.; Kikukawa, T.; Demura, M.; Ito, M.; et al. A Basic Study of the Effects of Mulberry Leaf Administration to Healthy C57BL/6 Mice on Gut Microbiota and Metabolites. Metabolites 2023, 13, 1003. [Google Scholar] [CrossRef] [PubMed]
- Ru, J.; Li, P.; Wang, J.; Zhou, W.; Li, B.; Huang, C.; Li, P.; Guo, Z.; Tao, W.; Yang, Y.; et al. TCMSP: A Database of Systems Pharmacology for Drug Discovery from Herbal Medicines. J. Cheminform. 2014, 6, 13. [Google Scholar] [CrossRef] [PubMed]
- Whirl-Carrillo, M.; Huddart, R.; Gong, L.; Sangkuhl, K.; Thorn, C.F.; Whaley, R.; Klein, T.E. An Evidence-Based Framework for Evaluating Pharmacogenomics Knowledge for Personalized Medicine. Clin. Pharmacol. Ther. 2021, 110, 563–572. [Google Scholar] [CrossRef] [PubMed]
- Cheng, L.; Qi, C.; Yang, H.; Lu, M.; Cai, Y.; Fu, T.; Ren, J.; Jin, Q.; Zhang, X. gutMGene: A Comprehensive Database for Target Genes of Gut Microbes and Microbial Metabolites. Nucleic Acids Res. 2022, 50, D795–D800. [Google Scholar] [CrossRef] [PubMed]
- UniProt Consortium. UniProt: The Universal Protein Knowledgebase in 2021. Nucleic Acids Res. 2021, 49, D480–D489. [Google Scholar] [CrossRef] [PubMed]
- Ashburner, M.; Ball, C.A.; Blake, J.A.; Botstein, D.; Butler, H.; Cherry, J.M.; Davis, A.P.; Dolinski, K.; Dwight, S.S.; Eppig, J.T.; et al. Gene Ontology: Tool for the Unification of Biology. The Gene Ontology Consortium. Nat. Genet. 2000, 25, 25–29. [Google Scholar] [CrossRef] [PubMed]
- Gene Ontology Consortium. The Gene Ontology Resource: Enriching a GOld Mine. Nucleic Acids Res. 2021, 49, D325–D334. [Google Scholar] [CrossRef] [PubMed]
- Hosseini, S.; Ketabi, S.; Hasheminasab, G. QSAR Study of Antituberculosis Activity of Oxadiazole Derivatives Using DFT Calculations. J. Recept. Signal Transduct. Res. 2022, 42, 503–511. [Google Scholar] [CrossRef] [PubMed]
- Lu, T.; Chen, F. Multiwfn: A Multifunctional Wavefunction Analyzer. J. Comput. Chem. 2012, 33, 580–592. [Google Scholar] [CrossRef] [PubMed]
- Eberhardt, J.; Santos-Martins, D.; Tillack, A.F.; Forli, S. AutoDock Vina 1.2.0: New Docking Methods, Expanded Force Field, and Python Bindings. J. Chem. Inf. Model. 2021, 61, 3891–3898. [Google Scholar] [CrossRef] [PubMed]
- Seeliger, D.; de Groot, B.L. Ligand Docking and Binding Site Analysis with PyMOL and Autodock/Vina. J. Comput.-Aided Mol. Des. 2010, 24, 417–422. [Google Scholar] [CrossRef] [PubMed]
- Maier, J.A.; Martinez, C.; Kasavajhala, K.; Wickstrom, L.; Hauser, K.E.; Simmerling, C. ff14SB: Improving the Accuracy of Protein Side Chain and Backbone Parameters from ff99SB. J. Chem. Theory Comput. 2015, 11, 3696–3713. [Google Scholar] [CrossRef]
- Davidchack, R.L.; Handel, R.; Tretyakov, M.V. Langevin Thermostat for Rigid Body Dynamics. J. Chem. Phys. 2009, 130, 234101. [Google Scholar] [CrossRef]
- Roe, D.R.; Cheatham, T.E. PTRAJ and CPPTRAJ: Software for Processing and Analysis of Molecular Dynamics Trajectory Data. J. Chem. Theory Comput. 2013, 9, 3084–3095. [Google Scholar] [CrossRef]
- Liu, K.; Guo, F.; Ma, Y.; Yu, X.; Fu, X.; Li, W.; Han, W. Functionalized Fullerene Potentially Inhibits SARS-CoV-2 Infection by Modulating Spike Protein Conformational Changes. Int. J. Mol. Sci. 2023, 24, 14471. [Google Scholar] [CrossRef] [PubMed]
- Shi, D.; Dong, H.; Zhu, Z.; Sun, M.; Su, J.; Zhang, X.; Xu, Y.; Fu, X. Expression of Catalytic Domain of Protein Tyrosine Phosphatase 1B and Preparation of Its Polyclonal Antibody. Chem. Res. Chin. Univ. 2007, 23, 204–207. [Google Scholar] [CrossRef]
- Xiong, G.; Wu, Z.; Yi, J.; Fu, L.; Yang, Z.; Hsieh, C.; Yin, M.; Zeng, X.; Wu, C.; Lu, A.; et al. ADMETlab 2.0: An Integrated Online Platform for Accurate and Comprehensive Predictions of ADMET Properties. Nucleic Acids Res. 2021, 49, W5–W14. [Google Scholar] [CrossRef] [PubMed]







| Name | MOL ID | Dimension 1 | Dimension 2 | Dimension 3 |
|---|---|---|---|---|
| quercetin | MOL000098 | 13.98971333 | −15.4718551 | −12.91115122 |
| kaempferol | MOL000422 | 14.93055973 | −15.56046554 | −13.2378193 |
| rhamnocitrin | MOL000251 | 14.27188684 | −15.25028232 | −13.87688149 |
| tetramethoxyluteolin | MOL007879 | 14.28316604 | −14.20247304 | −12.73193365 |
| norartocarpetin | MOL006630 | 15.14767912 | −14.11761894 | −13.39243877 |
| Name | Number of Nodes | Number of Edges | Average Node Degree | Avg. Local Clustering Coefficient |
|---|---|---|---|---|
| quercetin | 62 | 316 | 10.20 | 0.46 |
| kaempferol | 59 | 233 | 7.90 | 0.42 |
| rhamnocitrin | 50 | 194 | 7.76 | 0.52 |
| tetramethoxyluteolin | 50 | 192 | 7.68 | 0.55 |
| norartocarpetin | 58 | 227 | 7.83 | 0.50 |
| Energy | Value (kcal/mol) |
|---|---|
| ΔEvdW (van der Waals energy) | −19.06 ± 0.44 |
| ΔEele (electrostatic energy) | −24.19 ± 1.34 |
| ΔGgas (gas-phase free energy change) | 31.89 ± 0.99 |
| ΔGsolv (solvation free energy change) | −43.25 ± 1.19 |
| binding free energy | −11.36 ± 0.51 |
| System Components | Concentration |
|---|---|
| MOPS-NaOH buffer (pH 7.0) | 25 mM |
| EDTA | 1 mM |
| DTT | 1 mM |
| BSA | 1 mg/mL |
| NaCl | 0.1 M |
| p-NPP | 10 mM |
| PTP1B | 40 ng |
| Reagent Name | Manufacturers |
|---|---|
| BL21 with PT7-ΔPTP1B recombinant plasmid | Laboratory preservation (Fisher Laboratory, College of Life Sciences, Jilin University, Changchun, China) |
| Kaempferol | Aladdin (Shanghai, China) |
| HepG2 cell | Nanjing Keygen Company (Nanjing, China) |
| SDS-PAGE | SIGMA Corporation of America (Ronkonkoma, NY, USA) |
| Phosphotyrosine antibody (PY99) | Santa Cruz Biotechnology (Santa Cruz, CA, USA) (RRID: AB_628123) |
| β-Actin antibodies | Santa Cruz Biotechnology (Santa Cruz, CA, USA) (RRID: AB_626632) |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Bai, X.; Zhao, X.; Liu, K.; Yang, X.; He, Q.; Gao, Y.; Li, W.; Han, W. Mulberry Leaf Compounds and Gut Microbiota in Alzheimer’s Disease and Diabetes: A Study Using Network Pharmacology, Molecular Dynamics Simulation, and Cellular Assays. Int. J. Mol. Sci. 2024, 25, 4062. https://doi.org/10.3390/ijms25074062
Bai X, Zhao X, Liu K, Yang X, He Q, Gao Y, Li W, Han W. Mulberry Leaf Compounds and Gut Microbiota in Alzheimer’s Disease and Diabetes: A Study Using Network Pharmacology, Molecular Dynamics Simulation, and Cellular Assays. International Journal of Molecular Sciences. 2024; 25(7):4062. https://doi.org/10.3390/ijms25074062
Chicago/Turabian StyleBai, Xue, Xinyi Zhao, Kaifeng Liu, Xiaotang Yang, Qizheng He, Yilin Gao, Wannan Li, and Weiwei Han. 2024. "Mulberry Leaf Compounds and Gut Microbiota in Alzheimer’s Disease and Diabetes: A Study Using Network Pharmacology, Molecular Dynamics Simulation, and Cellular Assays" International Journal of Molecular Sciences 25, no. 7: 4062. https://doi.org/10.3390/ijms25074062






