Functional Identification of HhUGT74AG11—A Key Glycosyltransferase Involved in Biosynthesis of Oleanane-Type Saponins in Hedera helix
Abstract
1. Introduction
2. Results
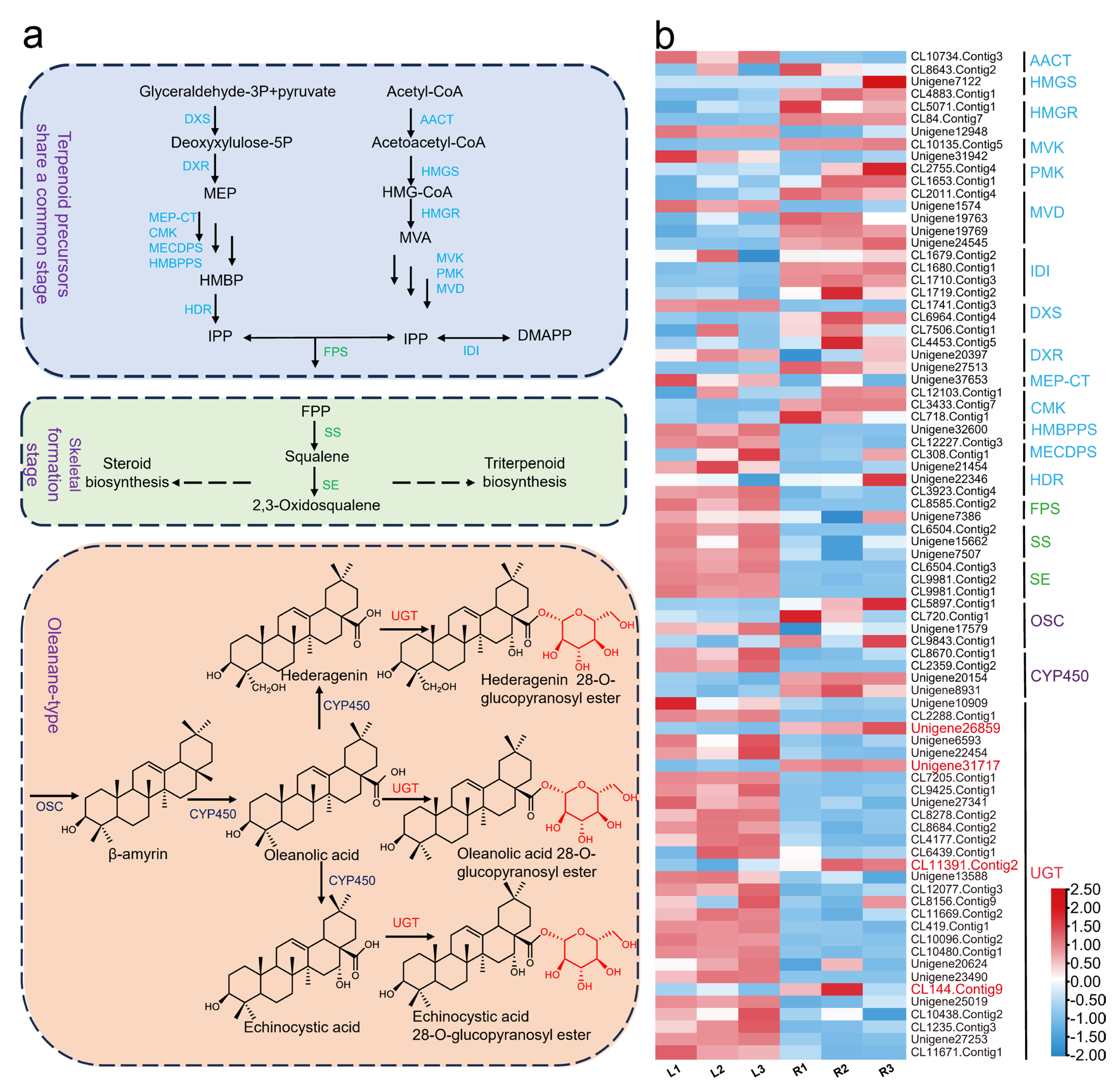
2.1. UGT Candidate Gene Screening
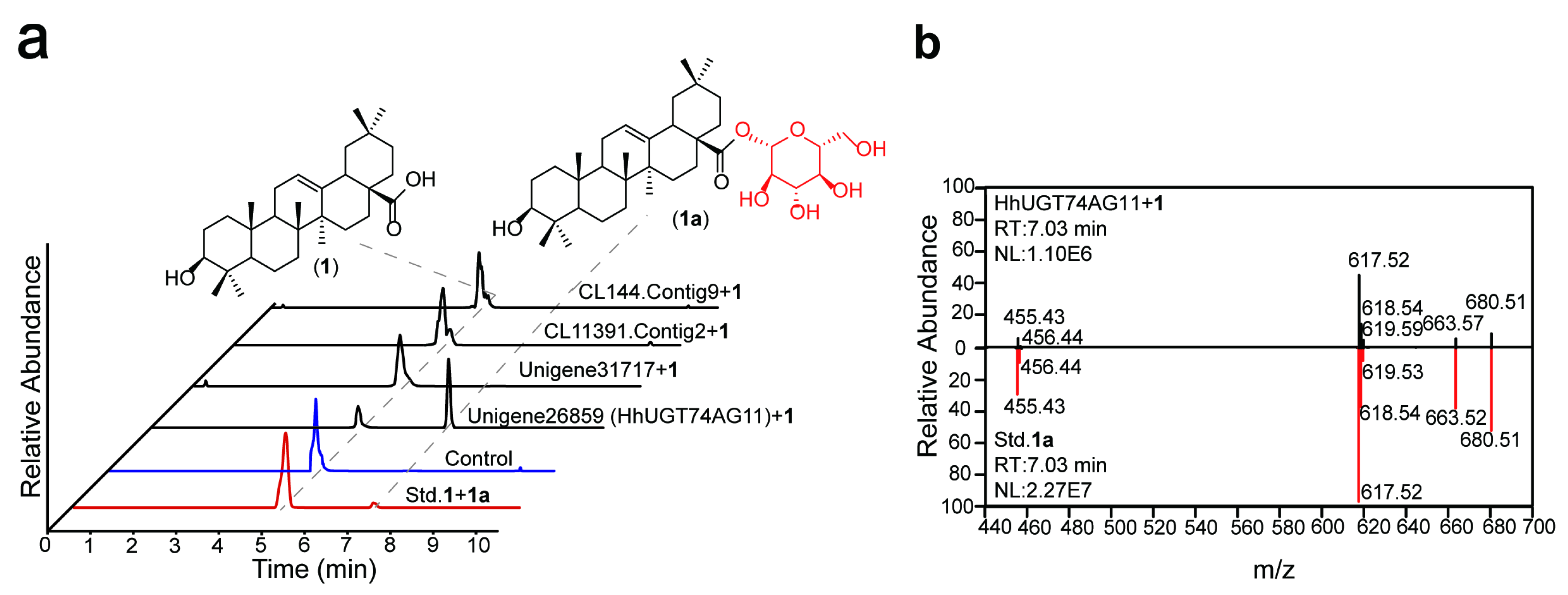
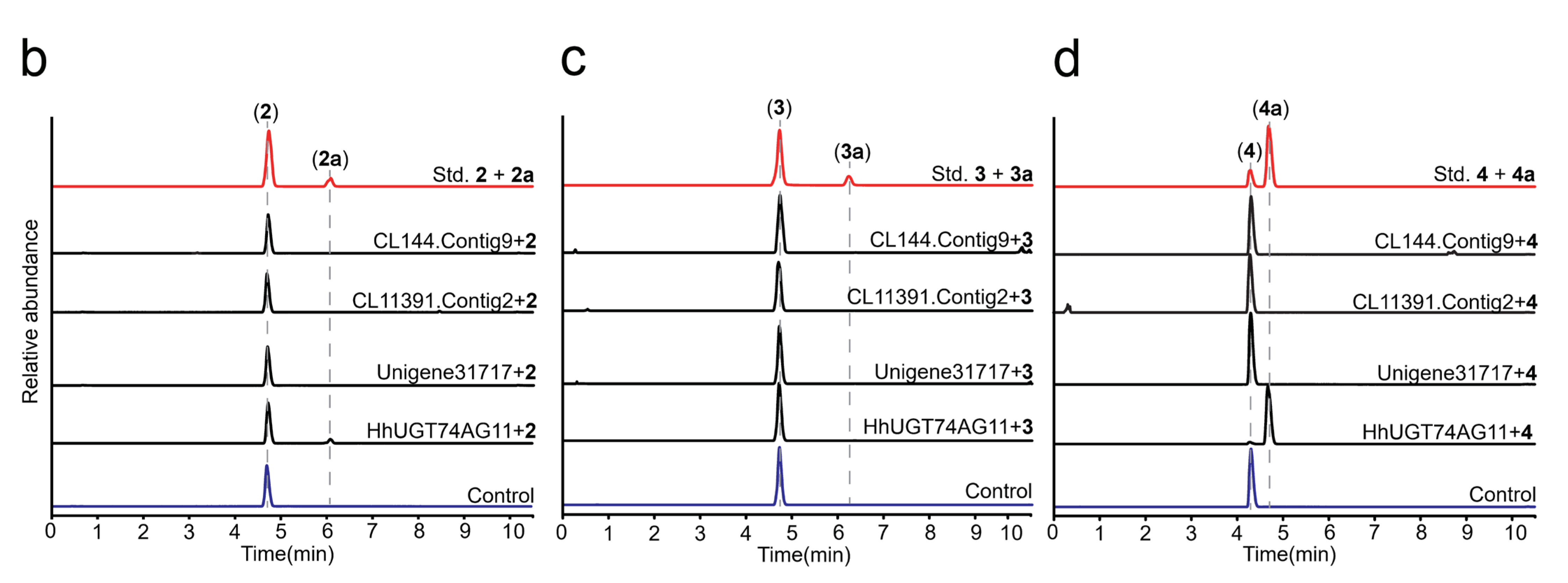
2.2. In Vitro Enzyme Activity Determination of UGTs
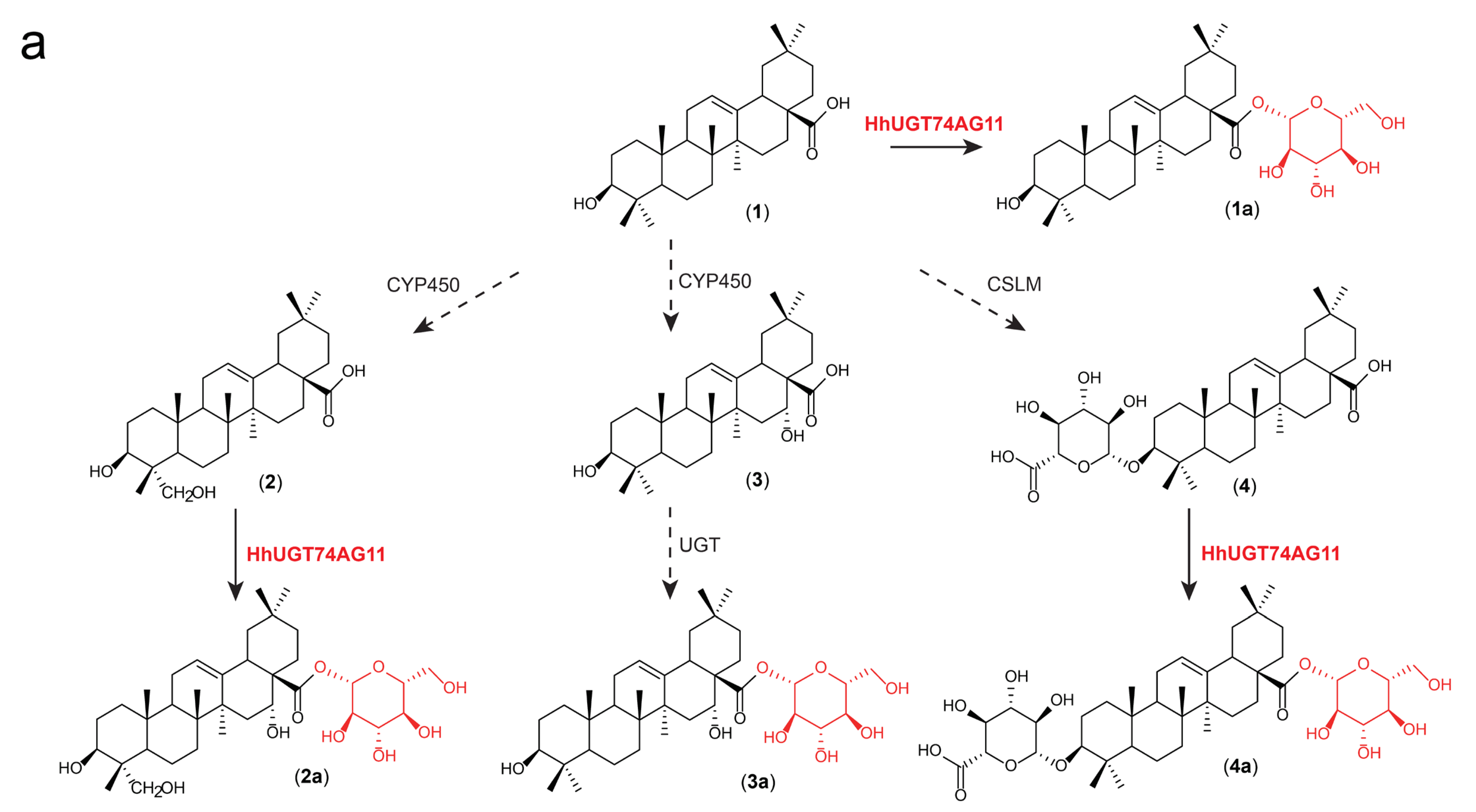
2.3. Substrate Hybridity of HhUGT74AG11
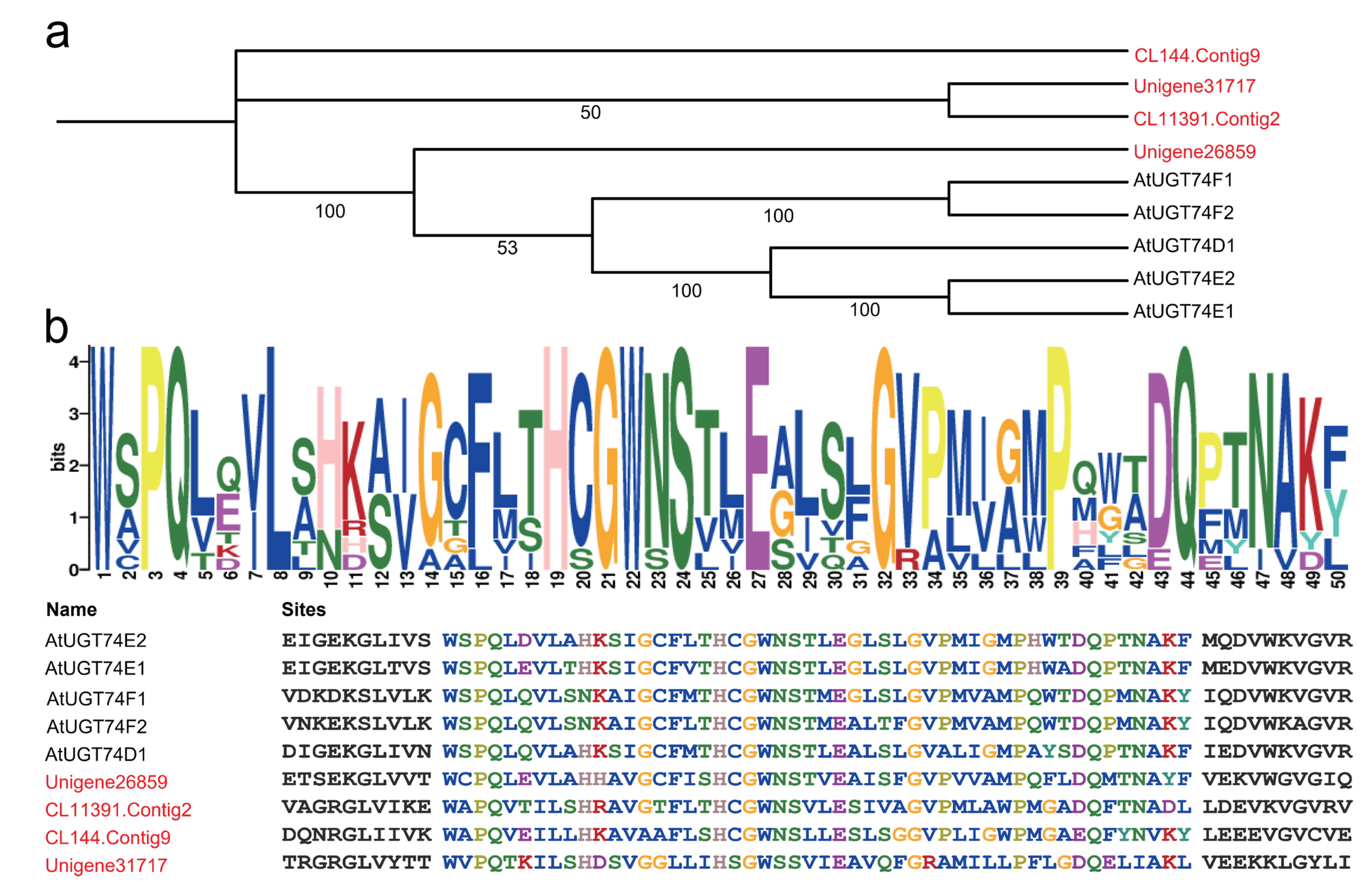
2.4. Cluster Analysis of HhUGT74AG11
3. Discussion
4. Materials and Methods
4.1. Botanical Substances and Compounds
4.2. Examining the Transcriptome and Discovering Genes Associated with Triterpenoid Synthesis
4.3. Bioinformatics Analysis
4.4. Total RNA Extraction, cDNA First-Strand Synthesis, and Gene Cloning
4.5. Expression of Escherichia coli and Protein Separation and Purification
4.6. In Vitro Enzyme Activity Determination
4.7. UPLC-ESI-MS Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Baharara, H.; Moghadam, A.T.; Sahebkar, A.; Emami, S.A.; Tayebi, T.; Mohammadpour, A.H. The Effects of Ivy (Hedera helix) on Respiratory Problems and Cough in Humans: A Review. Adv. Exp. Med. Biol. 2021, 1328, 361–376. [Google Scholar] [PubMed]
- Sun, H.-P.; Li, F.; Ruan, Q.-M.; Zhong, X.-H. Identification and Validation of Reference Genes for Quantitative Real-Time PCR Studies in Hedera helix L. Plant Physiol. Biochem. PPB 2016, 108, 286–294. [Google Scholar] [CrossRef] [PubMed]
- Akhtar, M.; Shaukat, A.; Zahoor, A.; Chen, Y.; Deng, G. Anti-Inflammatory Effects of Hederacoside-C on Staphylococcus Aureus Induced Inflammation via TLRs and Their Downstream Signal Pathway In Vivo and In Vitro. Microb. Pathog. 2019, 137, 103767. [Google Scholar] [CrossRef] [PubMed]
- Matsuda, H.; Morikawa, T.; Nakamura, S.; Muraoka, O.; Yoshikawa, M. New Biofunctional Effects of Oleanane-Type Triterpene Saponins. J. Nat. Med. 2023, 77, 644–664. [Google Scholar] [CrossRef] [PubMed]
- Xu, X.-H.; Li, T.; Fong, C.M.V.; Chen, X.; Chen, X.-J.; Wang, Y.-T.; Huang, M.-Q.; Lu, J.-J. Saponins from Chinese Medicines as Anticancer Agents. Molecules 2016, 21, 1326. [Google Scholar] [CrossRef] [PubMed]
- Yu, M.; Si, L.; Wang, Y.; Wu, Y.; Yu, F.; Jiao, P.; Shi, Y.; Wang, H.; Xiao, S.; Fu, G.; et al. Discovery of Pentacyclic Triterpenoids as Potential Entry Inhibitors of Influenza Viruses. J. Med. Chem. 2014, 57, 10058–10071. [Google Scholar] [CrossRef] [PubMed]
- Lang, C.; Röttger-Lüer, P.; Staiger, C. A valuable option for the treatment of respiratory diseases: Review on the clinical evidence of the ivy leaves dry extract EA 575®. Planta Medica 2015, 81, 968–974. [Google Scholar] [CrossRef] [PubMed]
- Barnes, L.A.; Leach, M.; Anheyer, D.; Brown, D.; Steel, A. The Effects of Hedera helix on Viral Respiratory Infections in Humans: A Rapid Review. Adv. Integr. Med. 2020, 7, 222–226. [Google Scholar] [CrossRef] [PubMed]
- Hou, M.; Wang, R.; Zhao, S.; Wang, Z. Ginsenosides in Panax genus and Their Biosynthesis. Acta Pharm. Sin. B 2021, 11, 1813–1834. [Google Scholar] [CrossRef]
- Shan, C.; Wang, C.; Zhang, S.; Shi, Y.; Ma, K.; Yang, Q.; Wu, J. Transcriptome Analysis of Clinopodium Gracile (Benth.) Matsum and Identification of Genes Related to Triterpenoid Saponin Biosynthesis. BMC Genomics 2020, 21, 49. [Google Scholar] [CrossRef]
- Wang, J.; Guo, Y.; Yin, X.; Wang, X.; Qi, X.; Xue, Z. Diverse Triterpene Skeletons Are Derived from the Expansion and Divergent Evolution of 2,3-Oxidosqualene Cyclases in Plants. Crit. Rev. Biochem. Mol. Biol. 2022, 57, 113–132. [Google Scholar] [CrossRef]
- Seki, H.; Tamura, K.; Muranaka, T. P450s and UGTs: Key Players in the Structural Diversity of Triterpenoid Saponins. Plant Cell Physiol. 2015, 56, 1463–1471. [Google Scholar] [CrossRef] [PubMed]
- He, B.; Bai, X.; Tan, Y.; Xie, W.; Feng, Y.; Yang, G.-Y. Glycosyltransferases: Mining, Engineering and Applications in Biosynthesis of Glycosylated Plant Natural Products. Synth. Syst. Biotechnol. 2022, 7, 602–620. [Google Scholar] [CrossRef]
- Jung, S.-C.; Kim, W.; Park, S.C.; Jeong, J.; Park, M.K.; Lim, S.; Lee, Y.; Im, W.-T.; Lee, J.H.; Choi, G.; et al. Two Ginseng UDP-Glycosyltransferases Synthesize Ginsenoside Rg3 and Rd. Plant Cell Physiol. 2014, 55, 2177–2188. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Zhang, H.; Ri, H.C.; An, Z.; Wang, X.; Zhou, J.-N.; Zheng, D.; Wu, H.; Wang, P.; Yang, J.; et al. Deletion and Tandem Duplications of Biosynthetic Genes Drive the Diversity of Triterpenoids in Aralia elata. Nat. Commun. 2022, 13, 2224. [Google Scholar] [CrossRef]
- Li, Y.; Baldauf, S.; Lim, E.K.; Bowles, D.J. Phylogenetic Analysis of the UDP-Glycosyltransferase Multigene Family of Arabidopsis thaliana. J. Biol. Chem. 2001, 276, 4338–4343. [Google Scholar] [CrossRef] [PubMed]
- Yang, Q.; Zhang, Y.; Qu, X.; Wu, F.; Li, X.; Ren, M.; Tong, Y.; Wu, X.; Yang, A.; Chen, Y.; et al. Genome-Wide Analysis of UDP-Glycosyltransferases Family and Identification of UGT Genes Involved in Abiotic Stress and Flavonol Biosynthesis in Nicotiana tabacum. BMC Plant Biol. 2023, 23, 204. [Google Scholar] [CrossRef]
- Sayama, T.; Ono, E.; Takagi, K.; Takada, Y.; Horikawa, M.; Nakamoto, Y.; Hirose, A.; Sasama, H.; Ohashi, M.; Hasegawa, H.; et al. The Sg-1 Glycosyltransferase Locus Regulates Structural Diversity of Triterpenoid Saponins of Soybean. Plant Cell 2012, 24, 2123–2138. [Google Scholar] [CrossRef]
- Wei, W.; Wang, P.; Wei, Y.; Liu, Q.; Yang, C.; Zhao, G.; Yue, J.; Yan, X.; Zhou, Z. Characterization of Panax ginseng UDP-Glycosyltransferases Catalyzing Protopanaxatriol and Biosyntheses of Bioactive Ginsenosides F1 and Rh1 in Metabolically Engineered Yeasts. Mol. Plant 2015, 8, 1412–1424. [Google Scholar] [CrossRef]
- Wang, D.; Wang, J.; Shi, Y.; Li, R.; Fan, F.; Huang, Y.; Li, W.; Chen, N.; Huang, L.; Dai, Z.; et al. Elucidation of the Complete Biosynthetic Pathway of the Main Triterpene Glycosylation Products of Panax notoginseng Using a Synthetic Biology Platform. Metab. Eng. 2020, 61, 131–140. [Google Scholar] [CrossRef]
- Bin Kang, K.; Jayakodi, M.; Lee, Y.S.; Nguyen, V.B.; Park, H.-S.; Koo, H.J.; Choi, I.Y.; Kim, D.H.; Chung, Y.J.; Ryu, B.; et al. Identification of Candidate UDP-Glycosyltransferases Involved in Protopanaxadiol-Type Ginsenoside Biosynthesis in Panax Ginseng. Sci. Rep. 2018, 8, 11744. [Google Scholar] [CrossRef] [PubMed]
- Chen, K.; Zhang, M.; Xu, L.; Yi, Y.; Wang, L.; Wang, H.; Wang, Z.; Xing, J.; Li, P.; Zhang, X. Identification of Oxidosqualene Cyclases Associated with Saponin Biosynthesis from Astragalus Membranaceus Reveals a Conserved Motif Important for Catalytic Function. J. Adv. Res. 2023, 43, 247–257. [Google Scholar] [CrossRef]
- Bezruk, I.; Materiienko, A.; Gubar, S.; Proskurina, K.; Georgiyants, V. Estimation of the Influence of the Environmental Factors on the Accumulation of Phytochemicals and Antioxidant Capacity in the Ivy Leaves (Hedera helix L.). Nat. Prod. Res. 2020, 36, 1014–1019. [Google Scholar] [CrossRef]
- Meesapyodsuk, D.; Balsevich, J.; Reed, D.W.; Covello, P.S. Saponin Biosynthesis in Saponaria Vaccaria. CDNAs Encoding Beta-Amyrin Synthase and a Triterpene Carboxylic Acid Glucosyltransferase. Plant Physiol. 2007, 143, 959–969. [Google Scholar] [CrossRef]
- Dai, L.; Liu, C.; Zhu, Y.; Zhang, J.; Men, Y.; Zeng, Y.; Sun, Y. Functional Characterization of Cucurbitadienol Synthase and Triterpene Glycosyltransferase Involved in Biosynthesis of Mogrosides from Siraitia grosvenorii. Plant Cell Physiol. 2015, 56, 1172–1182. [Google Scholar] [CrossRef]
- Rahimi, S.; Kim, J.; Mijakovic, I.; Jung, K.-H.; Choi, G.; Kim, S.-C.; Kim, Y.-J. Triterpenoid-Biosynthetic UDP-Glycosyltransferases from Plants. Biotechnol. Adv. 2019, 37, 107394. [Google Scholar] [CrossRef] [PubMed]
- Wang, M.; Ji, Q.; Lai, B.; Liu, Y.; Mei, K. Structure-Function and Engineering of Plant UDP-Glycosyltransferase. Comput. Struct. Biotechnol. J. 2023, 21, 5358–5371. [Google Scholar] [CrossRef] [PubMed]
- Rehman, H.M.; Khan, U.M.; Nawaz, S.; Saleem, F.; Ahmed, N.; Rana, I.A.; Atif, R.M.; Shaheen, N.; Seo, H. Genome Wide Analysis of Family-1 UDP Glycosyltransferases in Populus Trichocarpa Specifies Abiotic Stress Responsive Glycosylation Mechanisms. Genes 2022, 13, 1640. [Google Scholar] [CrossRef]
- Caputi, L.; Malnoy, M.; Goremykin, V.; Nikiforova, S.; Martens, S. A Genome-Wide Phylogenetic Reconstruction of Family 1 UDP-Glycosyltransferases Revealed the Expansion of the Family during the Adaptation of Plants to Life on Land. Plant J. 2012, 69, 1030–1042. [Google Scholar] [CrossRef]
- Shibuya, M.; Nishimura, K.; Yasuyama, N.; Ebizuka, Y. Identification and Characterization of Glycosyltransferases Involved in the Biosynthesis of Soyasaponin I in Glycine max. FEBS Lett. 2010, 584, 2258–2264. [Google Scholar] [CrossRef]
- Song, C.; Zhao, S.; Hong, X.; Liu, J.; Schulenburg, K.; Schwab, W. A UDP-Glucosyltransferase Functions in Both Acylphloroglucinol Glucoside and Anthocyanin Biosynthesis in Strawberry (Fragaria × ananassa). Plant J. 2016, 85, 730–742. [Google Scholar] [CrossRef] [PubMed]
- Richman, A.; Swanson, A.; Humphrey, T.; Chapman, R.; McGarvey, B.; Pocs, R.; Brandle, J. Functional Genomics Uncovers Three Glucosyltransferases Involved in the Synthesis of the Major Sweet Glucosides of Stevia rebaudiana. Plant J. 2005, 41, 56–67. [Google Scholar] [CrossRef] [PubMed]
- Kim, O.T.; Jin, M.L.; Lee, D.Y.; Jetter, R. Characterization of the Asiatic Acid Glucosyltransferase, UGT73AH1, Involved in Asiaticoside Biosynthesis in Centella asiatica (L.) Urban. Int. J. Mol. Sci. 2017, 18, 2630. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.-T.; Wang, Z.-L.; Chen, K.; Yao, M.-J.; Zhang, M.; Wang, R.-S.; Zhang, J.-H.; Ågren, H.; Li, F.-D.; Li, J.; et al. Insights into the Missing Apiosylation Step in Flavonoid Apiosides Biosynthesis of Leguminosae Plants. Nat. Commun. 2023, 14, 6658. [Google Scholar] [CrossRef] [PubMed]
- Sun, H.; Li, F.; Xu, Z.; Sun, M.; Cong, H.; Qiao, F.; Zhong, X. De Novo Leaf and Root Transcriptome Analysis to Identify Putative Genes Involved in Triterpenoid Saponins Biosynthesis in Hedera helix L. PLoS ONE 2017, 12, e0182243. [Google Scholar] [CrossRef]
- Kumar, S.; Stecher, G.; Li, M.; Knyaz, C.; Tamura, K. MEGA X: Molecular Evolutionary Genetics Analysis across Computing Platforms. Mol. Biol. Evol. 2018, 35, 1547–1549. [Google Scholar] [CrossRef]

Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yu, H.; Zhou, J.; Zhang, J.; He, X.; Peng, S.; Ling, H.; Dong, Z.; Lu, X.; Tian, Y.; Guan, G.; et al. Functional Identification of HhUGT74AG11—A Key Glycosyltransferase Involved in Biosynthesis of Oleanane-Type Saponins in Hedera helix. Int. J. Mol. Sci. 2024, 25, 4067. https://doi.org/10.3390/ijms25074067
Yu H, Zhou J, Zhang J, He X, Peng S, Ling H, Dong Z, Lu X, Tian Y, Guan G, et al. Functional Identification of HhUGT74AG11—A Key Glycosyltransferase Involved in Biosynthesis of Oleanane-Type Saponins in Hedera helix. International Journal of Molecular Sciences. 2024; 25(7):4067. https://doi.org/10.3390/ijms25074067
Chicago/Turabian StyleYu, Han, Jun Zhou, Jing Zhang, Xinyi He, Siqing Peng, Hao Ling, Zhuang Dong, Xiangyang Lu, Yun Tian, Guiping Guan, and et al. 2024. "Functional Identification of HhUGT74AG11—A Key Glycosyltransferase Involved in Biosynthesis of Oleanane-Type Saponins in Hedera helix" International Journal of Molecular Sciences 25, no. 7: 4067. https://doi.org/10.3390/ijms25074067
APA StyleYu, H., Zhou, J., Zhang, J., He, X., Peng, S., Ling, H., Dong, Z., Lu, X., Tian, Y., Guan, G., Tang, Q., Zhong, X., & He, Y. (2024). Functional Identification of HhUGT74AG11—A Key Glycosyltransferase Involved in Biosynthesis of Oleanane-Type Saponins in Hedera helix. International Journal of Molecular Sciences, 25(7), 4067. https://doi.org/10.3390/ijms25074067





