Abstract
Streptomyces are well-known for producing bioactive secondary metabolites, with numerous antimicrobials essential to fight against infectious diseases. Globally, multidrug-resistant (MDR) microorganisms significantly challenge human and veterinary diseases. To tackle this issue, there is an urgent need for alternative antimicrobials. In the search for potent agents, we have isolated four Streptomyces species PC1, BT1, BT2, and BT3 from soils collected from various geographical regions of the Himalayan country Nepal, which were then identified based on morphology and 16S rRNA gene sequencing. The relationship of soil microbes with different Streptomyces species has been shown in phylogenetic trees. Antimicrobial potency of isolates was carried out against Staphylococcus aureus American Type Culture Collection (ATCC) 43300, Shigella sonnei ATCC 25931, Salmonella typhi ATCC 14028, Klebsiella pneumoniae ATCC 700603, and Escherichia coli ATCC 25922. Among them, Streptomyces species PC1 showed the highest zone of inhibition against tested pathogens. Furthermore, ethyl acetate extracts of shake flask fermentation of these Streptomyces strains were subjected to liquid chromatography-tandem mass spectrometric (LC-MS/MS) analysis for their metabolic comparison and Global Natural Products Social Molecular Networking (GNPS) web-based molecular networking. We found very similar metabolite composition in four strains, despite their geographical variation. In addition, we have identified thirty-seven metabolites using LC-MS/MS analysis, with the majority belonging to the diketopiperazine class. Among these, to the best of our knowledge, four metabolites, namely cyclo-(Ile-Ser), 2-n-hexyl-5-n-propylresorcinol, 3-[(6-methylpyrazin-2-yl) methyl]-1H-indole, and cyclo-(d-Leu-l-Trp), were detected for the first time in Streptomyces species. Besides these, other 23 metabolites including surfactin B, surfactin C, surfactin D, and valinomycin were identified with the help of GNPS-based molecular networking.
1. Introduction
Pathogens, which are causes of infectious diseases, acquire resistance to commonly used antibiotics due to excessive and improper application, leading to a growing necessity to combat the escalating issue of drug resistance. In 2015, the World Health Organization (WHO) officially recognized a worldwide health crisis, stating that 700,000 deaths occur annually due to infections caused by MDR. The WHO also projected a staggering 10 million deaths by the year 2050 if the issue is not effectively addressed [1].
For that reason, researchers around the world are currently focusing on the search for potent antimicrobials from natural sources with the help of different tools and techniques. Currently, metabolomics is emerging as a promising avenue for the analysis of the metabolites present in extracts under definite conditions and can facilitate and speed up the search for alternative bioactive molecules [2]. Metabolomics based on liquid chromatography-high resolution mass spectrometry (LC-HRMS) is an effective analytical approach for evaluating mixtures of metabolites in a biological system to aid in the dereplication and discovery of novel natural products [3]. In addition, a more recent approach in drug discovery programs is molecular networking, a tandem mass spectrometry (MS/MS) data organizational approach. It has been demonstrated that molecular networking can be used to analyze complex MS/MS data utilizing GNPS web-based platform, which can aid in the identification of various compounds because of the network formed due to similarity in collision-induced dissociation-tandem mass spectrometry (CID-MS/MS) fragmentation pattern [4]. This, in turn, contributes to the identification of precursor ions or molecules towards the discovery of lead compounds.
Streptomyces, the largest genus within the phylum Actinomycetota, is renowned for being a major source of a broad spectrum of bioactive secondary metabolites including antibiotics, anticancer and antiviral agents, and various enzymes [5]. These are usually isolated from the soil and marine environment using a variety of growth media, such as a series of International Streptomyces Project (ISP) media, casein starch agar (CSA) media, chitin agar, and starch nitrate agar. Earlier studies have reported that the genus Streptomyces has contributed to the production of more than 74% of the antibiotics currently available such as streptomycin, tetracycline, chloramphenicol, erythromycin, neomycin, nystatin, etc. [6,7].
The secondary metabolites produced from the Streptomyces species were mostly well known for their structural diversity and novelty. The search for novel lead compounds from conventional soil-isolated Streptomyces has experienced a downturn due to the rediscovery of the same or known compounds [8]. Despite this fact, some researchers have been reporting novel lead compounds from soil-derived Streptomyces species. Yang et al. isolated and identified eight new fasamycin-type antibiotics and streptovertimycins A–H from Streptomyces morookaense strain derived from soil [9]. Two new cyclic thiopeptides, geninthiocins E and F with antiviral activities were isolated from soil-derived Streptomyces sp. CPCC 200267 by Fang et al. [10]. Similarly, dipimprinine E and dipimprinine F two alkaloids showing significant anticancer activity were isolated from soil-derived Streptomyces sp. 44414B [11]. Picolinamycin, a novel antibiotic showing antibacterial activity against multi-drug resistant bacterial strains was isolated from soil-derived Streptomyces sp. SM01 [12]. Moreover, Hu et al. isolated and identified two new phenazines with antimicrobial activity; 6-hydroxyphenazine-1-carboxamide and methyl 6-carbamoylphenazine-1-carboxylate from soil-derived Streptomyces species [13]. In addition, a novel pranonaphthoquinone antibiotic, xiakemycin A, was isolated from Streptomyces sp. CC8-201 [14]. Thus, the above evidence indicates that there are still chances of finding new bioactive molecules from soil microbes.
Nepal’s varied ecological landscape spans a spectrum of climatic zones, stretching from tropical conditions in the lowlands to alpine environments in the lofty Himalayan peaks. This broad diversity of habitats provides a range of niches for microbial communities, including the genus Streptomyces. The capacity of Streptomyces species to generate a variety of physiologically active secondary metabolites, which are very powerful against microbial pathogens, makes them one of the most influential groups of bacteria [15]. In this study, we have collected four soil samples from different niches of Nepal with varying altitudes to isolate Streptomyces species. The primary goal of this study is to explore and analyze the potent antimicrobial metabolites present in Streptomyces isolated from soil by tandem mass spectrometry and GNPS-based molecular networking.
2. Results
2.1. Isolation of Streptomyces Species
Four soil samples PC1, BT1, BT2, and BT3 were collected from different ecosystems in Nepal, ranging from 1010 m to 2743 m above sea level (Table S1). As suggested by Bergey’s Manual of Systematic Bacteriology, bacterial strains PC1, BT1, BT2, and BT3 were initially identified based on biochemical assays, morphology, growth pattern, and Gram staining [16]. They were grown on ISP4 media, and their secondary metabolites harvesting was determined by the growth curve, which varied in individual bacterial strains. The color of aerial mycelia of the isolates BT1, BT2, and BT3 were greyish-white in appearance, while isolate PC1 was found to be whitish, which is presented in Figure S1. The Gram-staining revealed that the isolates were composed of hair-like mycelium and flagellated Gram-positive bacteria. We isolated more than twenty actinomycetes strains from each soil sample PC1, BT1, BT2, and BT3, respectively, but most of them were discarded based on their similar morphological characters, antimicrobial properties, and molecular sequencing. Finally, only one peculiar actinomycetes strain was selected from each soil sample. Further research on those soils may lead to the isolation of additional actinomycetes in the future.
2.2. PCR of 16S rRNA and Molecular Sequencing
About 1.5 kb 16S rRNA gene was amplified using universal oligonucleotides (27F and 1492R) from the genomic DNA of soil microbes (PC1, BT1, BT2, and BT3). Then, it was subjected to molecular sequencing as methods described earlier. The homology search using the BLAST tool in NCBI for the 16S rRNA gene sequences of these actinomycetes revealed that all their sequences exhibited high similarity (greater than 99.0%) with several sequences of Streptomyces species in the GenBank. Their 16S rRNA gene sequences were deposited in the GenBank (PC1, accession number OR577614; BT1, accession number OR578351; BT2, accession number PP106255; BT3, accession number OR905603). Multiple sequence alignment was performed for our sequences with twelve 16S rRNA genes showing the highest sequence similarity from NCBI, and a phylogenetic tree was generated using the MEGA software (version 11.0.13) (https://www.megasoftware.net/) [17]. All four were assigned as Streptomyces species considering their closeness to many sequences of different Streptomyces. Figure S3 displays the link between the isolates and the nearest phylogenetic neighbors as well as a comparison of the sequencing findings of Streptomyces isolates.
2.3. Antimicrobial Assays
The antimicrobial activity of Streptomyces species was tested against various Gram-positive and Gram-negative bacteria. The zone of inhibition of respective bacterial fermented extracts against tested pathogenic bacteria is listed below in Table S2. Among four isolates, Streptomyces sp. PC1 showed the highest zone of inhibition against all tested pathogenic bacteria. Thus, the MIC and MBC values (Table 1) of the extract of Streptomyces sp. PC1 was determined against S. aureus and E. coli along with Streptomyces species BT1, BT2, and BT3. The MICs of Streptomyces sp. PC1 against S. aureus and E. coli were 0.65 mg/mL and 1.5 mg/mL, respectively, while positive control neomycin exhibited MICs of 0.62 µg/mL and 0.78 µg/mL against S. aureus and E. coli, respectively. The MBCs of Streptomyces sp. PC1 against S. aureus and E. coli were 2.63 mg/mL and 3.0 mg/mL, respectively, compared to positive control (neomycin), which had MBCs of 1.25 µg/mL and 1.56 µg/mL against S. aureus and E. coli, respectively (Figure S2).

Table 1.
MIC and MBC of various extracts against Staphylococcus aureus and Escherichia coli.
2.4. Metabolic Comparision in Streptomyces Species
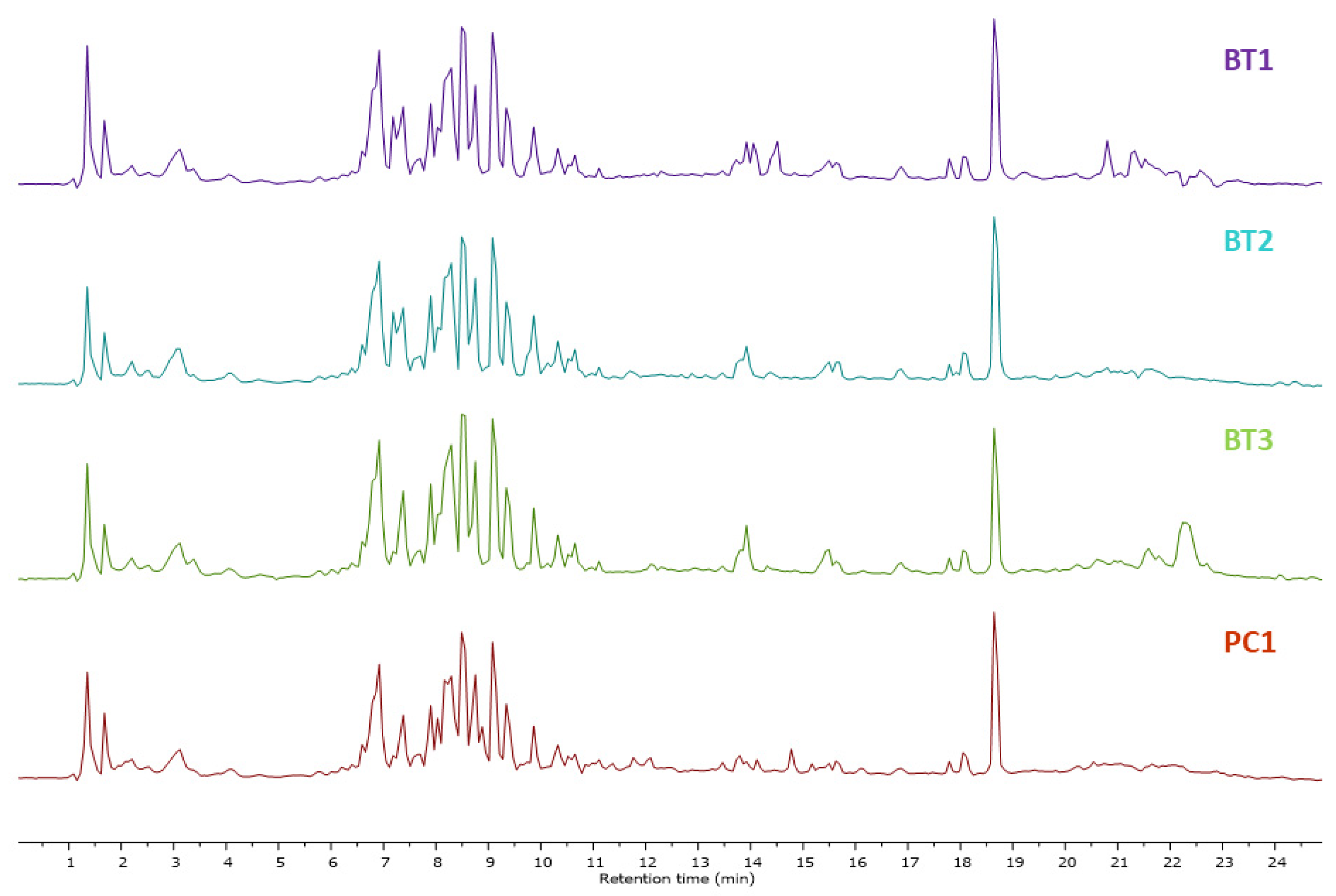
Mass spectrometry generated raw data were analyzed through MestReNova software 12.0.0, Spain (https://mestrelab.com/, accessed on 10–30 December 2023). Each peak was analyzed, detected, aligned, and annotated. To visualize any difference in metabolites, the total ion chromatograms (TIC) of ethyl acetate (EA) extracts of Streptomyces species BT1, BT2, BT3, and PC1 were stacked together as shown in Figure 1. Finally, the results were compared with literature and database libraries. Thirty-seven metabolites were detected in all samples of classes such as diketopiperazine and alkyl resorcinol. The secondary metabolites annotated through LC-HRMS/MS analysis are shown in Table 2 and Figure S4. The base peak chromatograms (BPC) and MS profiles of identified metabolites are displayed in Supplementary Figures S5–S41.
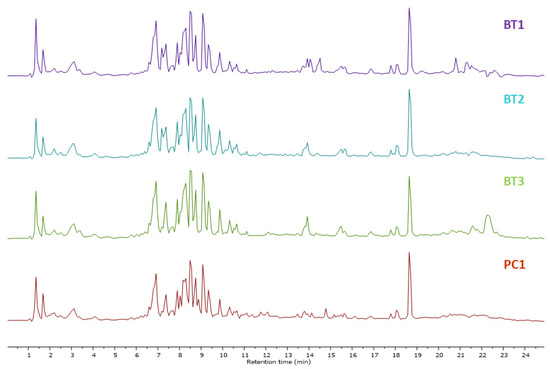
Figure 1.
Total ion chromatograms obtained in EA extracts of Streptomyces species BT1, BT2, BT3, and PC1 in a stacked format.

Table 2.
The list of annotated compounds in ethyl acetate fermentation extracts of Streptomyces species BT1, BT2, BT3, and PC1.
A molecular ion at m/z 284.139 [M+H]+ detected at retention time 9.41 min was identified as brevianamide F, isolated previously from Streptomyces sp.TN262 [40]. Cyclo-(Phenylalanyl-Prolyl), previously isolated and identified in Streptomyces sp. [39], was also identified in this study at m/z 245.129 [M+H]+ as a protonated ion. Likewise, a molecular ion at m/z 261.123 [M+H]+ detected at retention time 6.59 min was annotated as maculosin, which was identified previously in Streptomyces sp. KTM18 [20]. We detected a molecular ion at m/z 171.113 [M+H]+ in retention time 6.46 min and identified it as cyclo-(Gly-Leu), already reported in Streptomyces xanthophaeus [49]. Cyclo-(d-Ala-l-Pro) was detected at a retention time of 2.94 min with m/z 169.097 [M+H]+, already identified in mangrove-derived Streptomyces sp. by Tan et al. [47]. Another cyclic dipeptide compound, cyclo-(Tyr-Val), was detected at m/z 263.141 [M+H]+ and reported previously in Streptomyces sp. [45]. Moreover, a precursor ion detected at m/z 295.147 [M+H]+ was identified as cyclo-(Phenylalanyl-Phenylalanyl), reported previously in Streptomyces chrestomyceticus [44]. Cyclo-(l-Leucyl-l-Leucyl), previously identified in Streptomyces sp. [42], was also identified in our study at m/z 227.176 [M+H]+ as a precursor ion. A molecular ion detected at m/z 219.114 [M+H]+ at retention time 7.97 min was identified as cyclo-(l-Phe-l-Ala), previously detected in Streptomyces sp. [38]. Moreover, a protonated ion detected at m/z 277.155 [M+H]+ was identified as cyclo(Tyr-Leu), previously identified from soil-derived Streptomyces kunmingensis [37]. In addition, N-phenethylacetamide was annotated for a molecular ion at m/z 164.107 [M+H]+. A precursor ion at m/z 425.233 [M+H]+ detected at a retention time of 14.02 min was identified as neomarinone [21]. Similarly, a molecular ion at m/z 245.129 [M+H]+ was identified as cyclo-(d-Pro-d-Phe), previously reported by Alshaibani et al. in Streptomyces sp. SUK 25 [54]. Based on the literature survey, cyclo-(l-Val-l-Leu) was identified at m/z 213.160 [M+H]+ as a protonated ion, already reported in Streptomyces xiamenensis MCC A01570 [19]. In addition, dibutyl phthalate was annotated for a molecular ion at m/z 279.160 [M+H]+. A precursor ion [M+H]+ at m/z 269.093 detected at retention time 14.38 min was putatively identified as 1-acetyl-3-methoxycarbonyl-β-carboline. This compound was previously isolated from the fungus Ophiocordyceps sphecocephala BCC 2661 [46].
A compound exhibiting a molecular ion peak at m/z 170.081 [M+H]+ was annotated as pyridoxine [23]. Furthermore, the precursor ion at m/z 245.10 [M+H]+ was annotated as cyclo-(l-Pro-l-OMet), detected at a retention time of 2.53 min, as per the analysis of spectral data of Yang et al. [24]. A molecular ion at m/z 1008.660 [M+H]+ detected at retention time 2.53 min was identified as surfactin C13 [25]. Likewise, a molecular ion at m/z 149.023 [M+H]+ detected at a retention time of 18.64 min was annotated as phthalic anhydride. Another molecular ion at m/z 556.531 [M+H]+ detected at a retention time of 20.68 min was identified as phytoceramide. Similarly, the [M+H]+ at m/z 155.081 [M+H]+ detected at retention time 2.20 min was identified as cyclo-(Pro-Gly), based on the existing literature [30]. A molecular ion at m/z 211.144 [M+H]+ detected at retention time 8.03 min was identified as cyclo-(l-Leu-l-Pro), previously reported by Zin et al. [31]. Furthermore, the molecular ion at m/z 261.119 [M+H]+ detected as a protonated ion at retention time 6.79 min was previously isolated from Streptomyces asenjonii by Abdelkader et al. and was annotated as cyclo-(l-Phenylalanyl-trans-4-hydroxy-l-Proline) [55]. As per the literature, the sodium adduct formed at m/z 344.183 [M+Na]+ detected at retention time 10.85 min was identified as coronafacoyl-l-isoleucine. This metabolite was previously isolated from Streptomyces scabies [56]. Another molecular ion peak detected at retention time 6.79 min having m/z 261.119 [M+H]+ was annotated as cyclo-(d-Pro-l-Tyr), previously detected at Streptomyces sp. strain 22-4 [32]. In addition, a metabolite was detected at m/z 197.12 [M+H]+ as a protonated ion in the retention time of 6.85 min, and identified as cyclo-(Pro-Val) as per the existing literature [34]. Similarly, the molecular ion detected at a retention time of 7.18 min with m/z 180.102 [M+H]+ was annotated as N-acetyl tyramine, from the spectral analysis carried out by Driche et al. [35]. The metabolite cyclo-(l-Ala-l-Leu), previously isolated from Streptomyces species from the soil environment, was detected at m/z 185.129 [M+H]+ as a protonated ion at the retention time of 7.57 min [57]. We identified another compound detected at m/z 274.275 [M+H]+ as N-lauryl diethanolamine. Likewise, a protonated ion detected at m/z 237.185 [M+H]+ at retention time 18.05 min was tentatively identified as 2-hexyl-5-methylresorcinol which was reported already in S. clavuligerus [53].
Identification of Metabolites New to Streptomyces Species
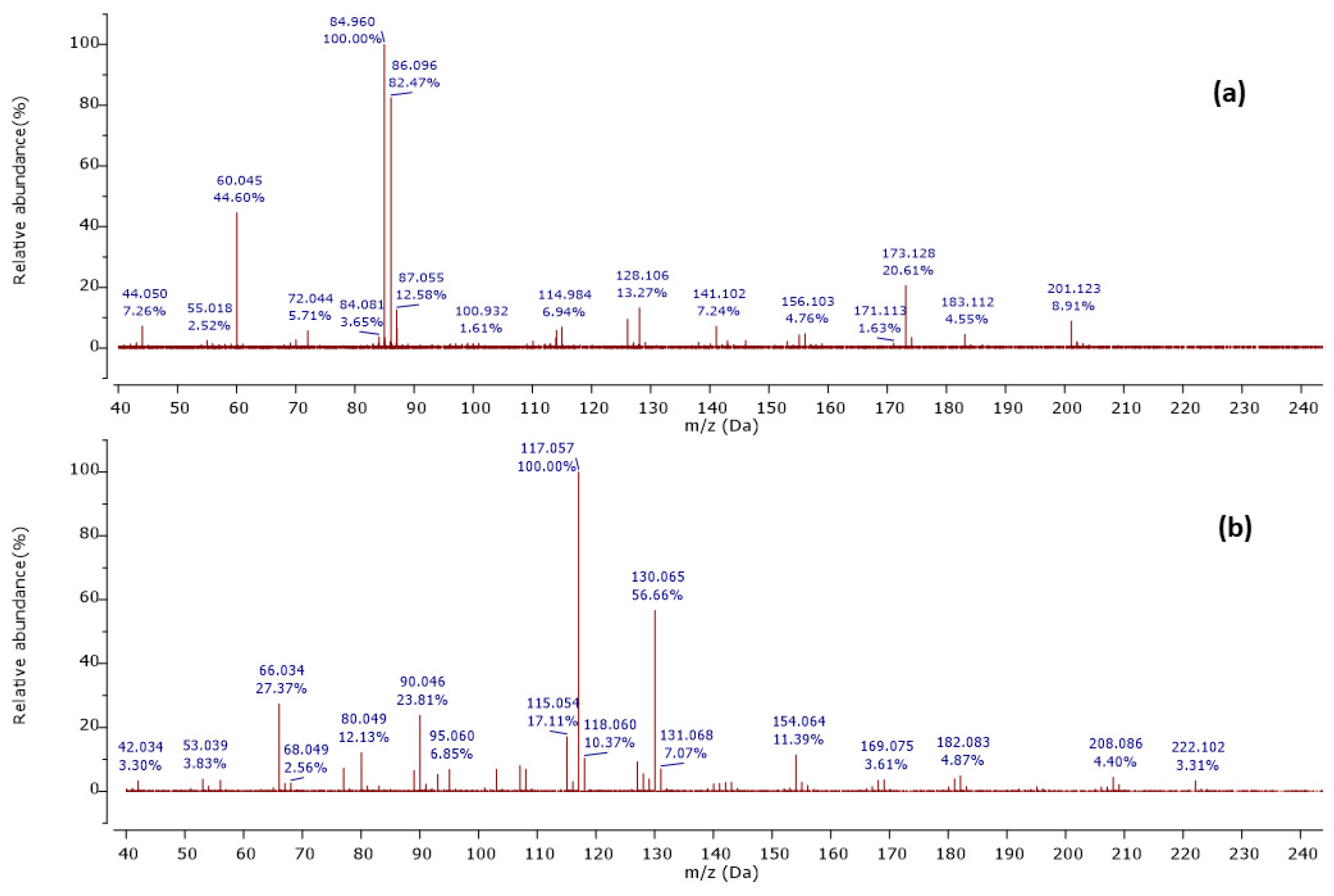
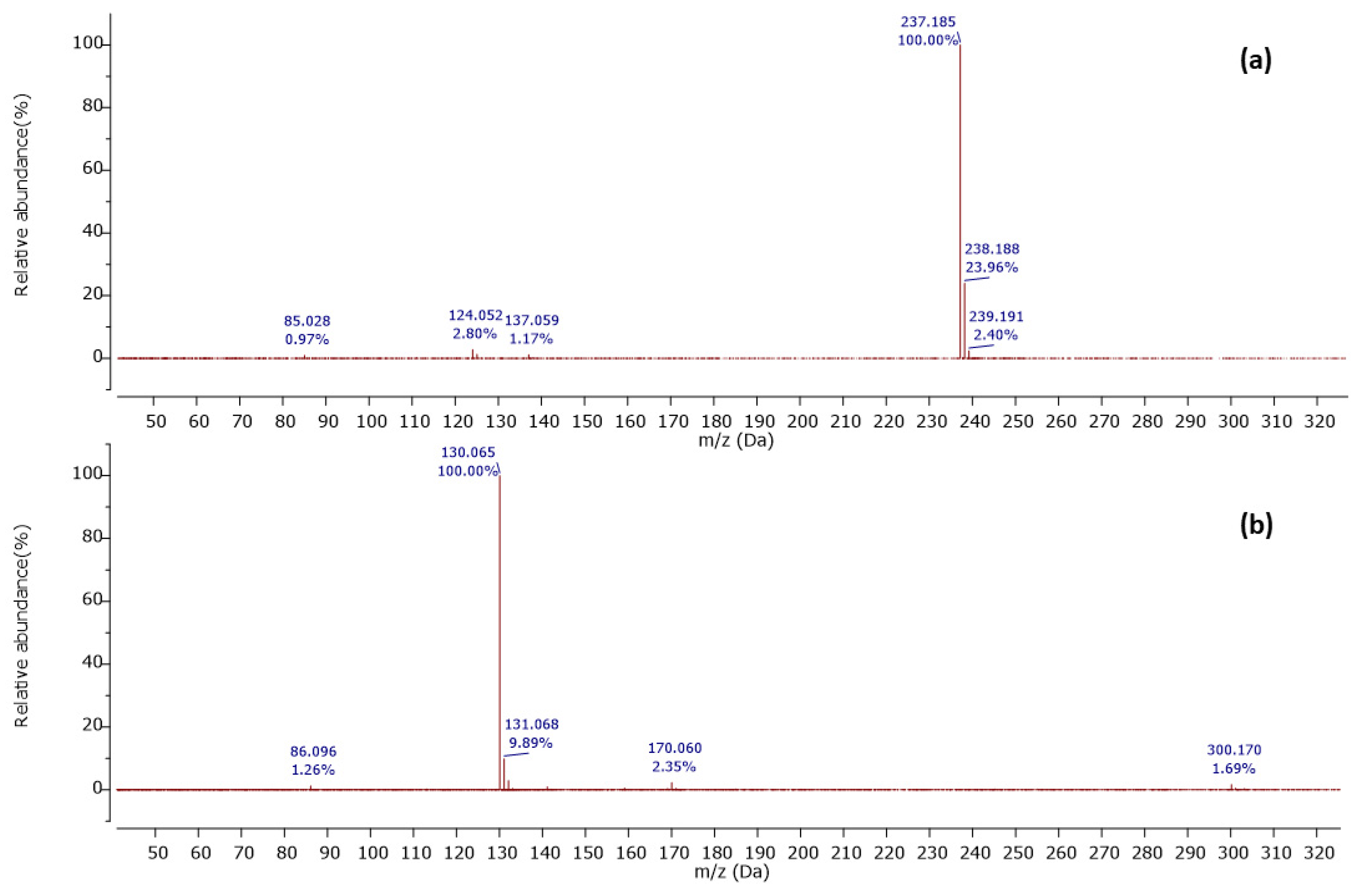
Among 37 metabolites, we identified four metabolites for the first time in Streptomyces species. A molecular ion [M+H]+ was detected at m/z 201.124 at a retention time of 6.20 min, with its MS2 peaks shown in Figure 2a.
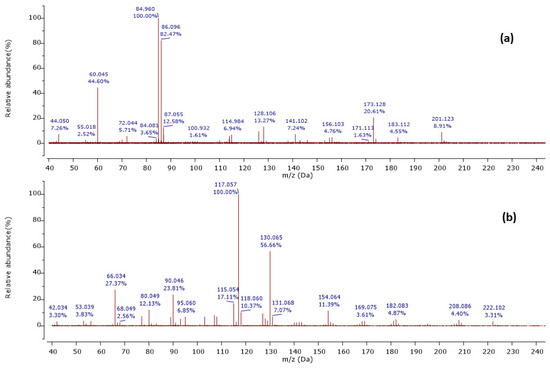
Figure 2.
Observed MS/MS profiles of the precursor protonated molecules at m/z 201.124 (a) and m/z 224.118 (b).
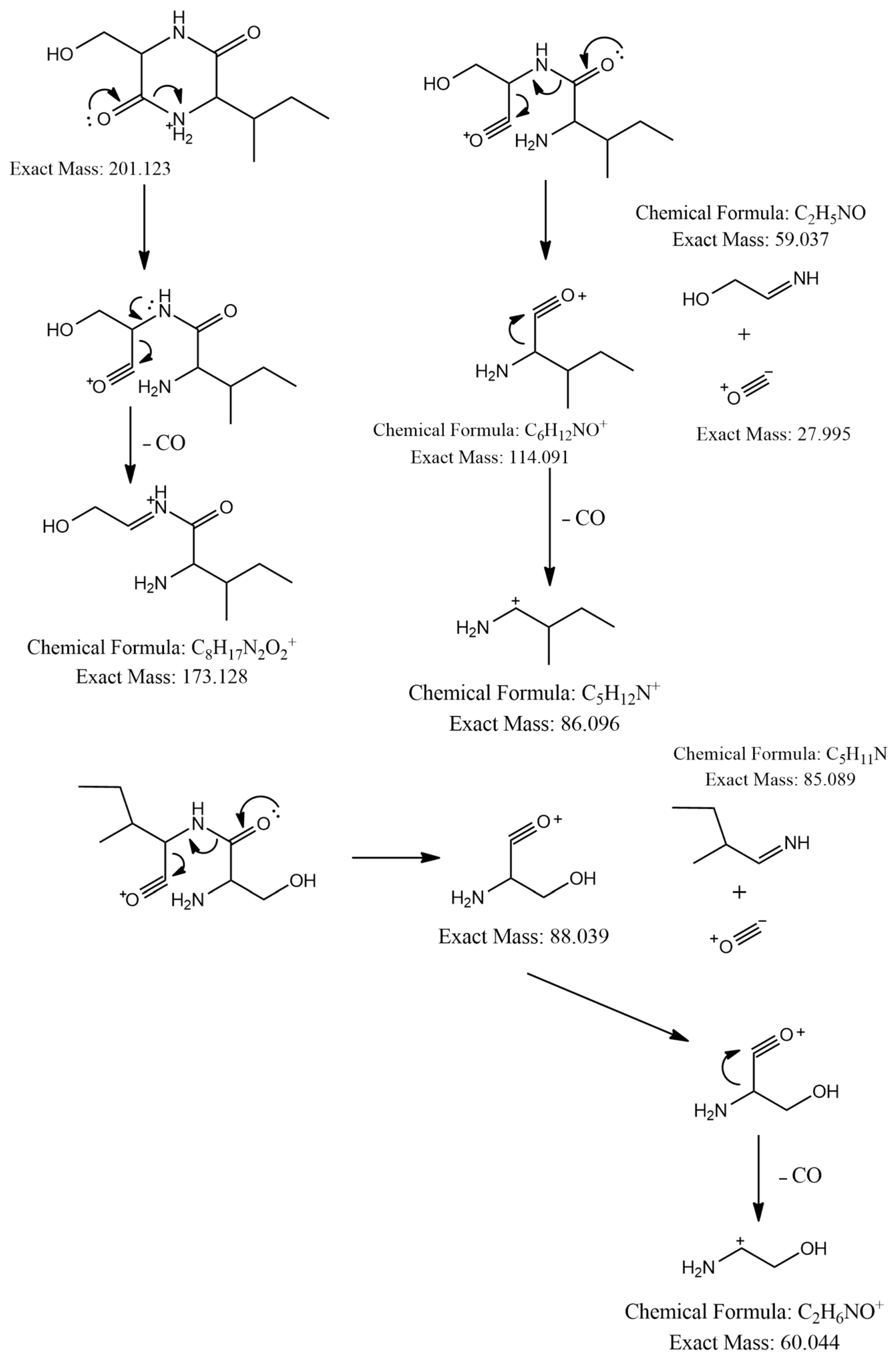
The MS2 spectrum showed the fragment ions at m/z 173 [M+H-28]+ due to [C8H17N2O2]+ ion formed by loss of a CO molecule from precursor ion, m/z 114 [M+H-59-28]+ owing to [C6H12NO]+ ion formed by departure of a neutral C2H5NO unit and a CO molecule simultaneously, and [C6H12NO]+ ion further lose a CO molecule to give a distinct peak at m/z 86 attributed to C5H12N+ ion. Furthermore, an ion at m/z 60 [M+H-85-28-28]+ was observed due to the loss of a neutral C5H11N unit and two CO molecules as shown in Figure 3. Thus, we identified this compound as cyclo-(Ile-Ser), isolated previously from Ophiocordyceps sobolifera [48].
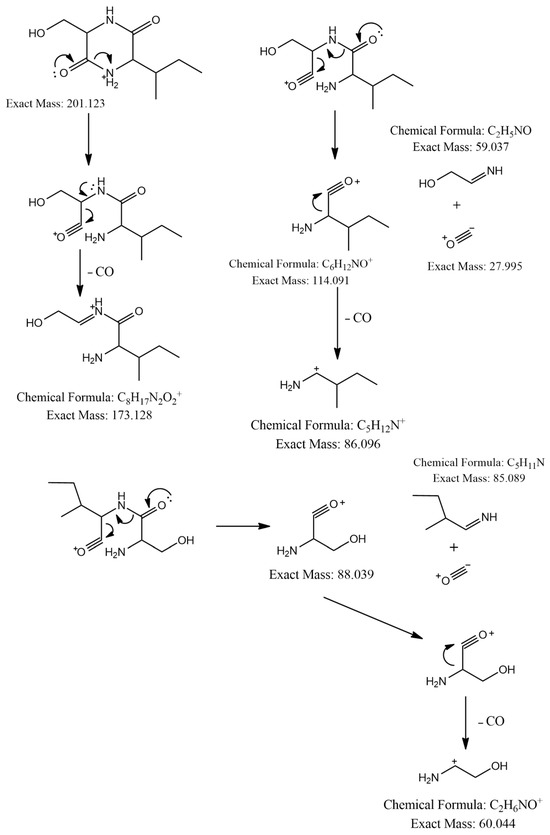
Figure 3.
Observed fragmentation pattern of cyclo-(Ile-Ser) in (+)-ESI mode.
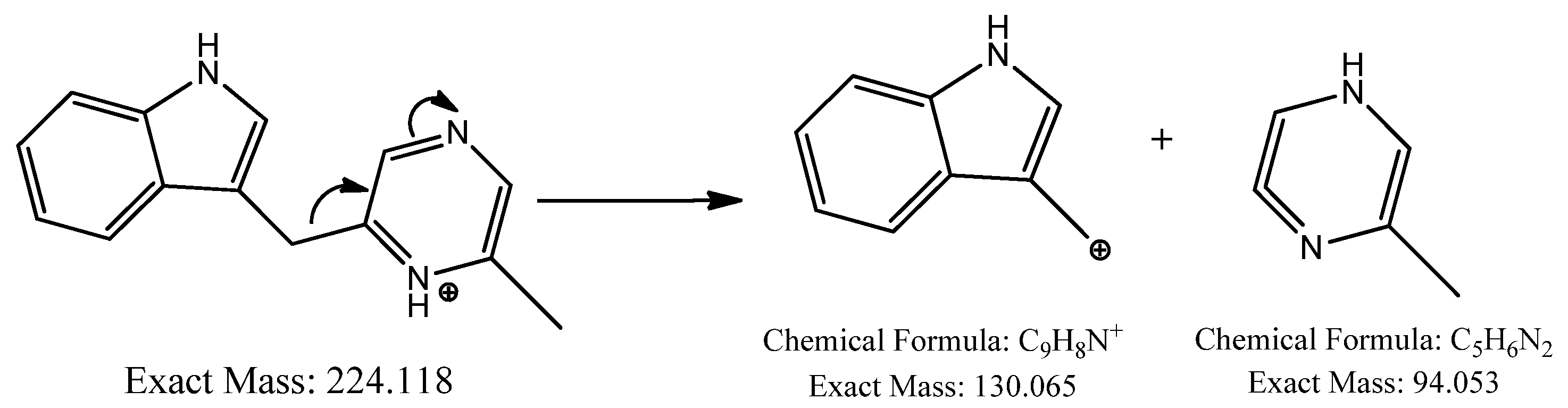
Another precursor ion was detected at a retention time of 12.09 min with m/z 224.118 [M+H]+. Its MS2 profile (Figure 2b) displayed a distinct peak at m/z 130.065 [M+H−94]+ due to the C9H8N+ ion attributed to the loss of a neutral C5H6N2 unit, as shown in Figure 4. Thus, this compound was putatively identified as 3-[(6-methylpyrazin-2-yl) methyl]-1H-indole, isolated and identified previously as a new alkaloid from marine Serinicoccus profundi sp. by Yang et al. [50]. To the best of our knowledge, this compound was observed for the first time in Streptomyces sp.
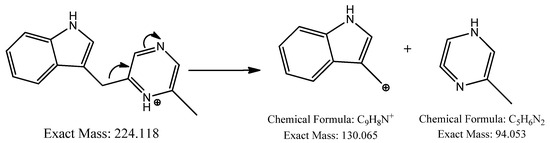
Figure 4.
Observed fragmentation pattern of 3-[(6-methylpyrazin-2-yl) methyl]-1H-indole in (+)-ESI mode.
Furthermore, a molecular ion was observed at m/z 237.185 [M+H]+ in a retention time of 10.27 min, with its MS2 spectrum shown in Figure 5a.
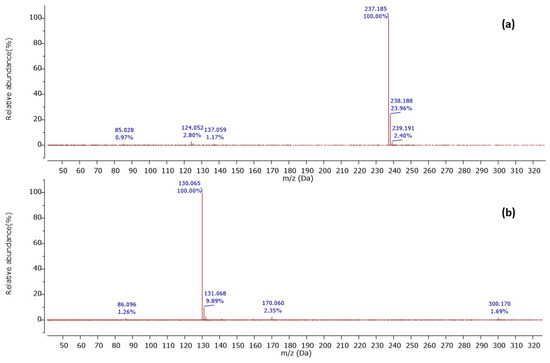
Figure 5.
Observed MS/MS profiles of the precursor protonated molecules at m/z 237.185 (a) and m/z 300.170 (b).
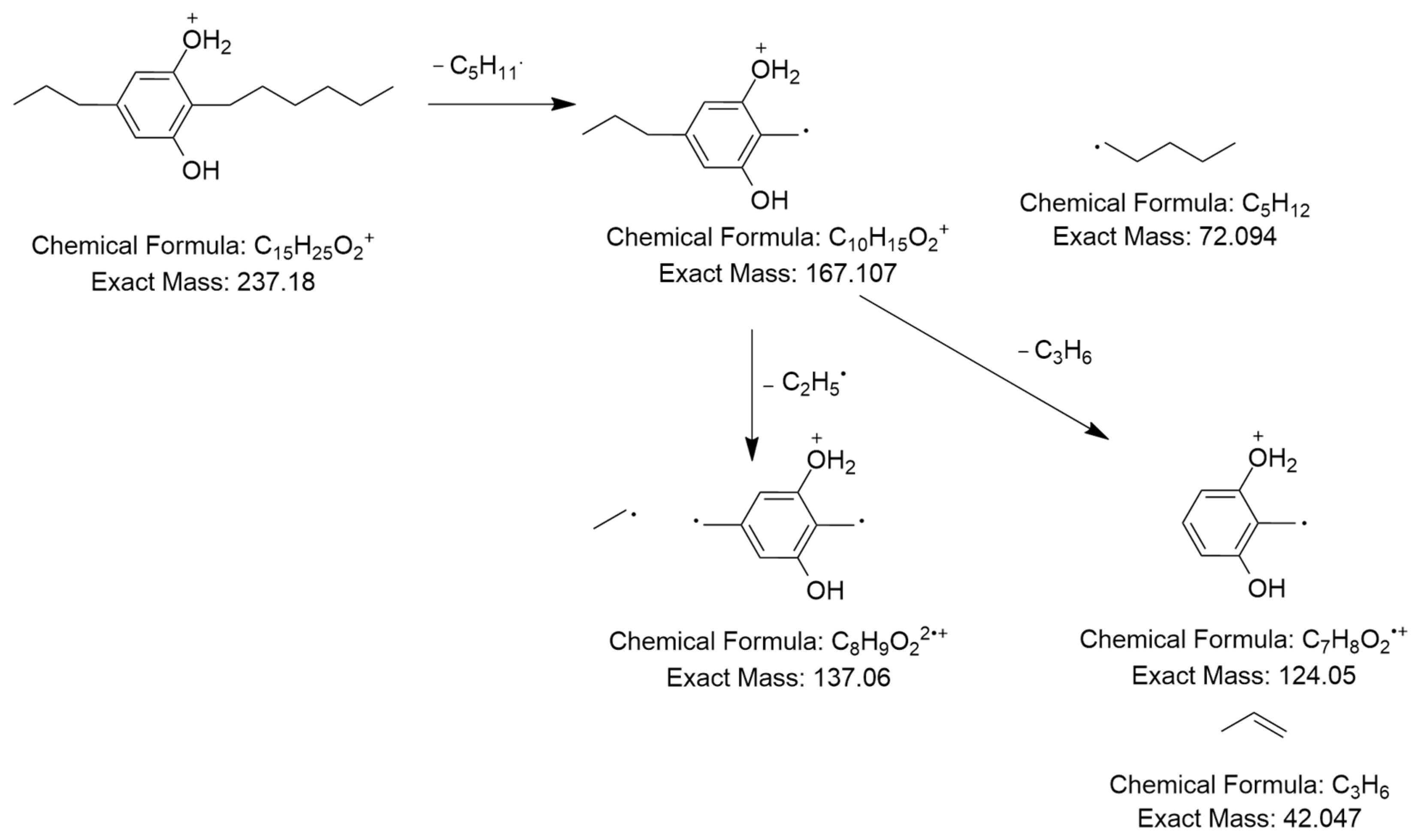
The MS2 spectrum displayed a molecular ion peak [C15H25O2]+ as a base peak which may be due to incomplete fragmentation. Further, fragment ions were detected at m/z 124 [C7H10O2]•+ because of [M+H−C5H11−C3H6]•+ and m/z 137 owing to [M+H−C5H11−C2H5]2•+, as shown in Figure 6. Thus, this compound was tentatively identified as 2-n-hexyl-5-n-propylresorcinol. This alkyl resorcinol, previously reported in Pseudomonas chlororaphis PCL1606, has now been identified in Streptomyces sp. for the first time [52].
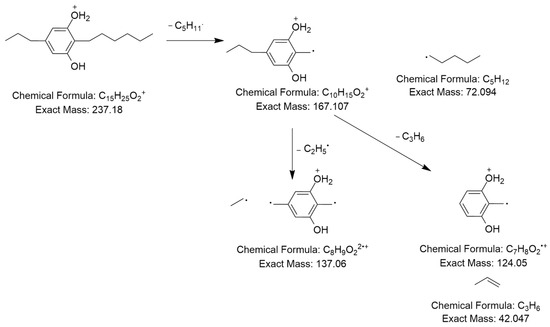
Figure 6.
Observed fragmentation pattern of 2-n-hexyl-5-n-propylresorcinol in (+)-ESI mode.
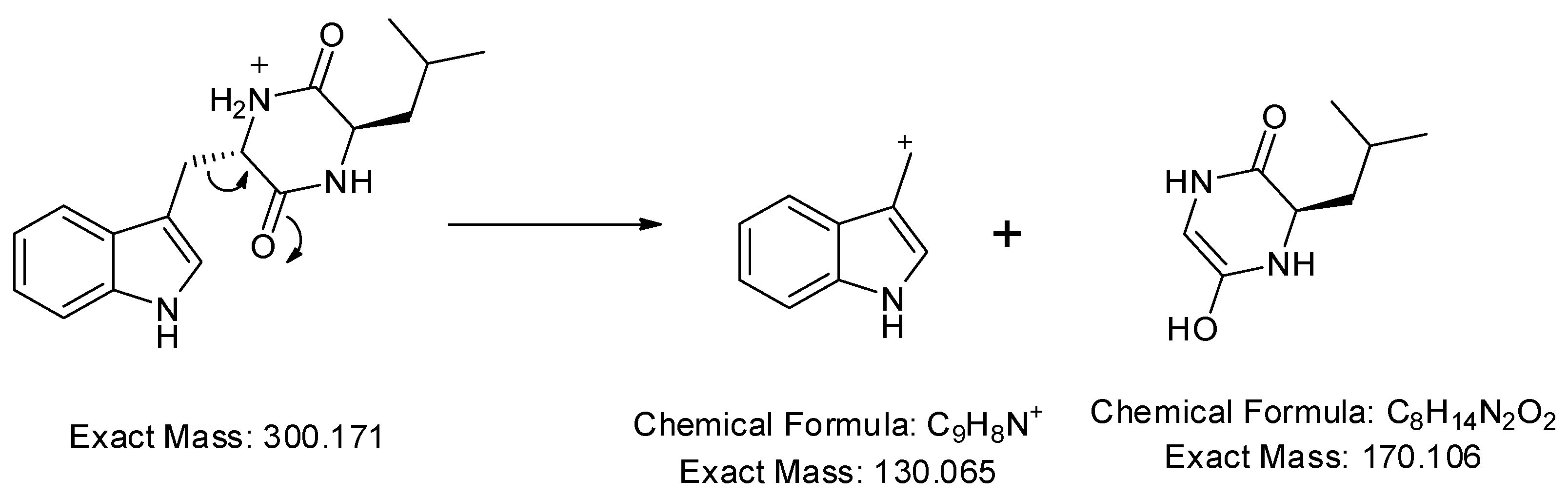
In addition, a protonated ion was detected at m/z 300.171 [M+H]+ in a retention time of 10.26 min. Its MS2 profile (Figure 5b) revealed a base peak with m/z 130 [M+H−170]+ due to the C9H9N+ ion formed by the loss of the C8H14N2O2 unit as shown in Figure 7. Thus, this compound was tentatively identified as cyclo-(d-Leu-l-Trp). Although this compound was previously reported in Penicillium brevicompactum [43], this is the first time it was observed in Streptomyces sp.
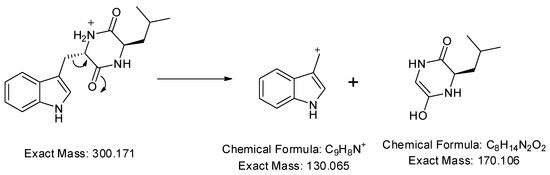
Figure 7.
Observed fragmentation pattern of cyclo-(d-Leu-l-Trp) in (+)-ESI mode.
2.5. GNPS-Based Molecular Networking
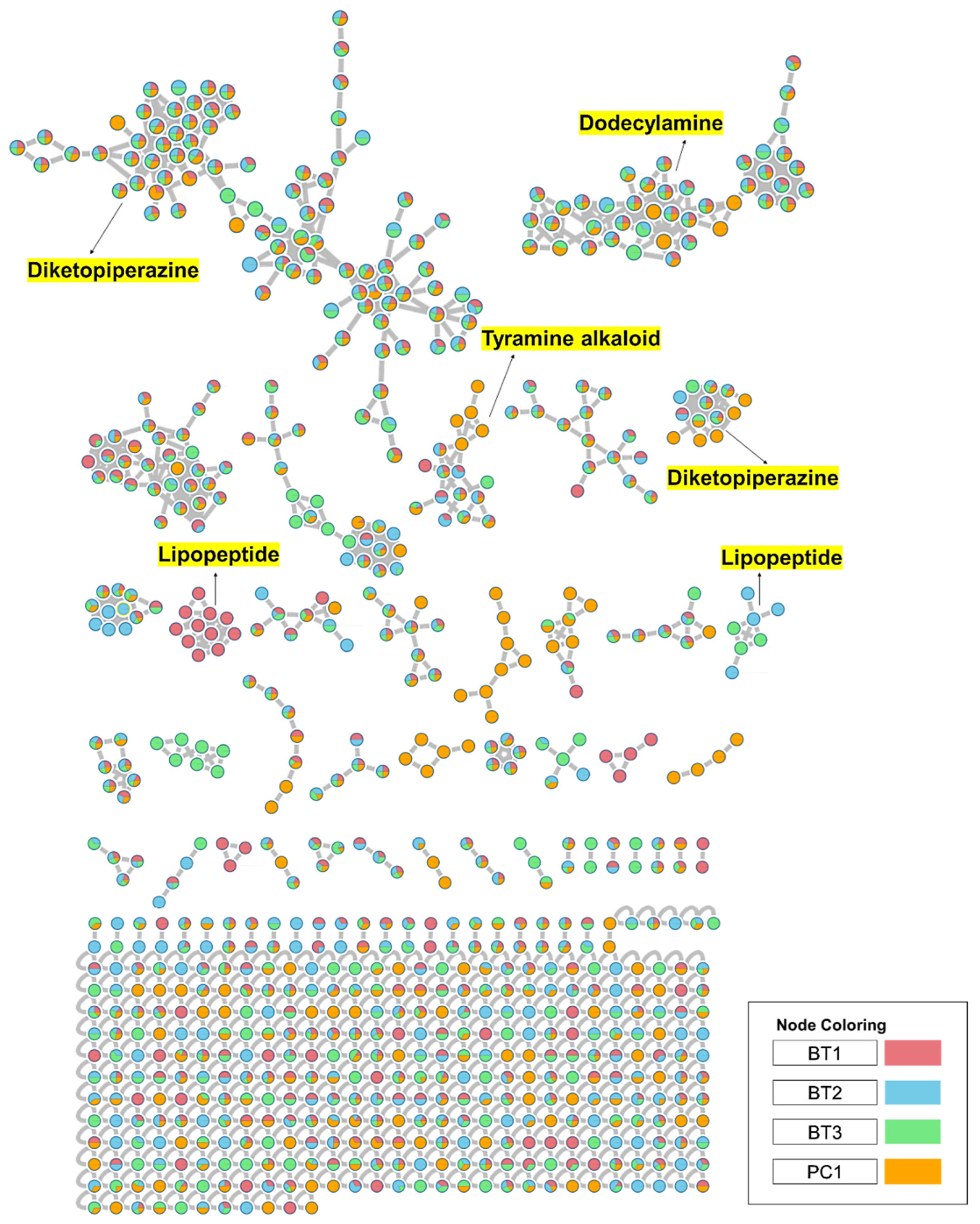
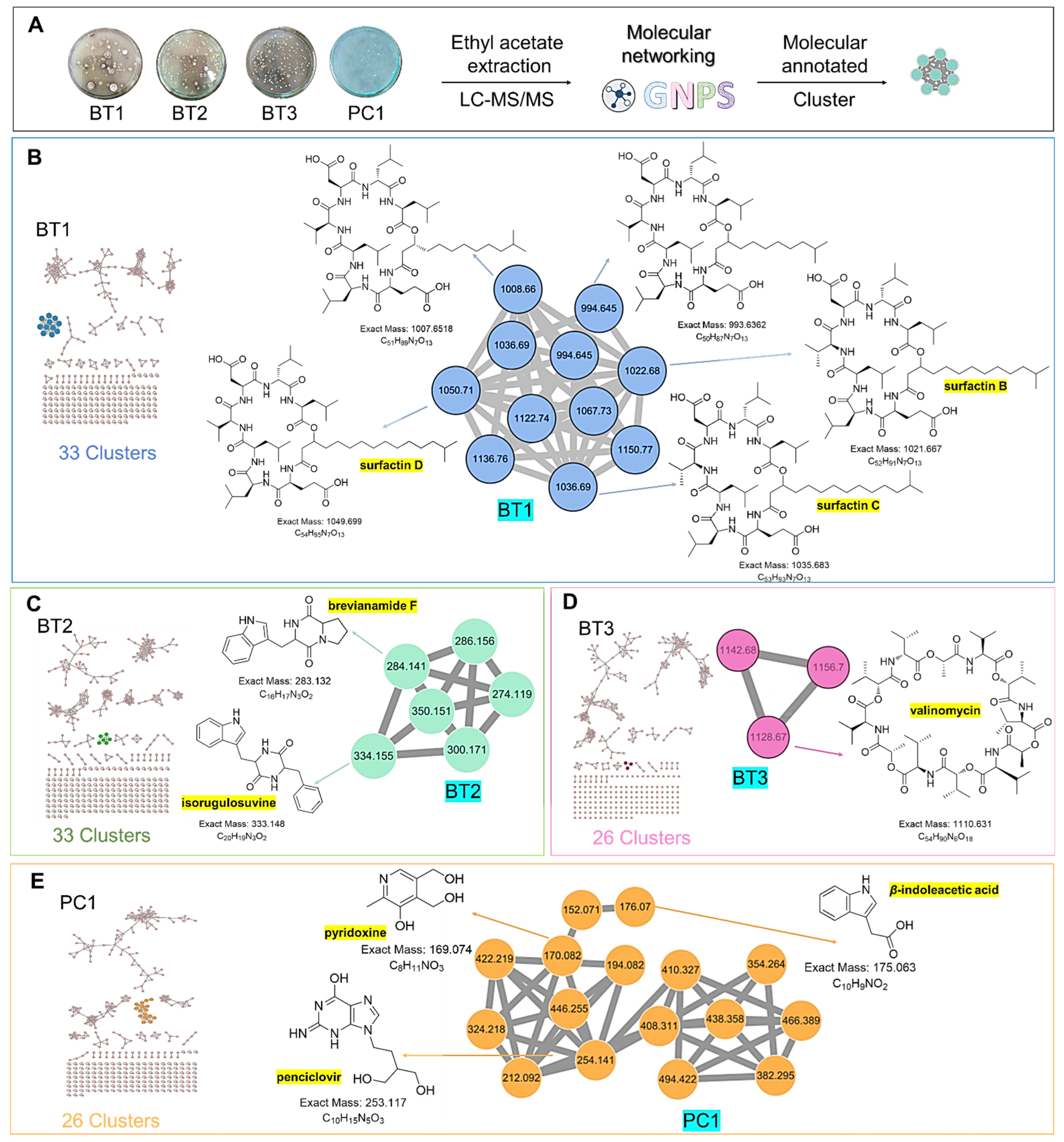
To comprehensively investigate the detailed metabolite profile of four isolates, namely Streptomyces sp. BT1, BT2, BT3, and PC1, we conducted MS2 and GNPS metabolic profiling as illustrated in Figure 8. A total of 378 molecular ions were observed to have MS2 spectra of four samples represented by nodes that were connected by 788 edges in the molecular network. Out of these 378 ions, 62 ion pairs formed two-node clusters, while the remaining 335 molecular ions were self-looped. We successfully identified and dereplicated 25 known compounds through the GNPS library (Table 3), and you can find more detailed information on the GNPS website [58]. The GNPS approach revealed that the extracts from the four isolates consist of various compounds, including diketopiperazines, lipopeptides, dodecylamines, and tyramine alkaloids.
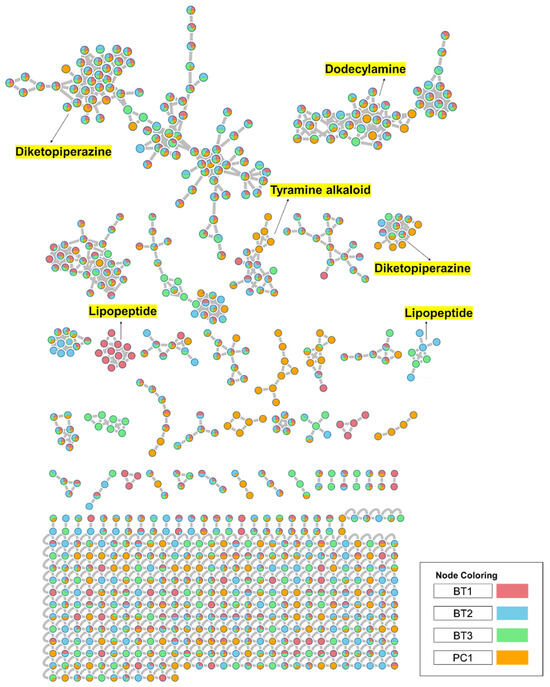
Figure 8.
Full molecular networking was created using MS/MS data in positive mode from extracts of Streptomyces species BT1, BT2, BT3, and PC1. Nodes are labeled with parent mass. The networking is displayed as a pie chart with pink, blue, green, and orange indicating the chemical composition distribution of Streptomyces species BT1, BT2, BT3, and Streptomyces sp.PC1 extracts, respectively.

Table 3.
The list of annotated compounds using GNPS in Streptomyces species BT1, BT2, BT3, and PC1.
The extracts of the four samples (Streptomyces sp. BT1, BT2, BT3, and PC1) underwent metabolic profiling through MS2 data with positive ion mode and built a network using GNPS (Figure 9A). In Figure 9B, the ions with m/z 1022.680, 1036.690, and 1050.710 corresponded to surfactins B, C, and D, respectively. Intriguingly, the presence of an ion with m/z 1008.660, 16 Da lower than m/z 1022.680, suggests the removal of a methylene group from surfactin B, with a similar pattern observed for the ion at m/z 994.645 relative to m/z 1008.660, indicating structural similarities and related metabolic pathways. Notably, it was revealed that the BT1 isolate extract contained lipopeptides, as indicated by the base peak ion MS chromatograms. Additionally, Figure 9C revealed that the BT2 extract contained a significant amount of tyramine alkaloid, as suggested by the presence of annotated compounds isorugulosuvine and brevianamide F. Furthermore, the cluster associated with BT3 displayed an ammonium-adduct ion peak [M+NH4]+ at m/z 1128.670, corresponding to valinomycin (Figure 9D). In contrast to BT1, BT2, and BT3, PC1 was found to lack large molecules of lipopeptides. Instead, it contained smaller nitrogen-containing compounds, including pyridoxine, penciclovir, and β-indoleacetic acid (Figure 9E). These findings collectively highlight the diverse and distinct chemical profiles of the four isolates, shedding light on their unique metabolic pathways and chemical compositions.
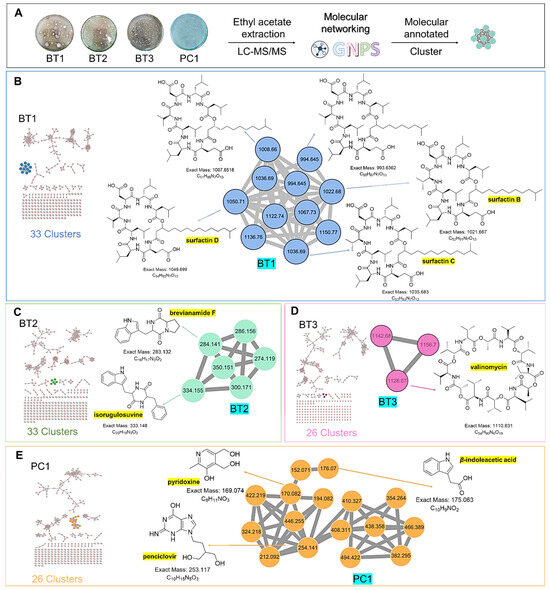
Figure 9.
(A) Molecular networking analysis and identification of compounds in four samples. (B) Zoomed-in molecular networking of annotated compounds from BT1. (C) Zoomed-in molecular networking of annotated compounds from BT2. (D) Zoomed-in molecular networking of annotated compounds from BT3. (E) Zoomed-in molecular networking of annotated compounds from PC1.
3. Discussion
The improper usage and overprescription of pharmaceuticals have led to MDR pathogens, which have become a serious global issue [79]. Structural modification of a drug molecule is one adaptive strategy employed by bacteria to develop resistance to drugs. Thus, an alternative strategy to solve the problem of drug resistance could be the discovery of secondary metabolites that have the same therapeutic benefits and therefore can aid in the drug discovery program.
Soil ecosystem serves as key sources for Streptomyces isolation, exhibiting a broad range of metabolites production, with novel compounds emerging in response to varying nutritional or environmental factors [80]. Precise identification of bacterial isolates at the species level is essential, providing valuable insights into the microorganism, potential bioactive chemicals, and its distinctiveness [81]. Morphology also proves to be an essential factor in differentiating Streptomyces from other spore-forming actinomycetes, and in characterizing distinct species within the Streptomyces genus. Further, sequencing of 16S rRNA was performed for accurate identification of these strains suggesting that isolates are Streptomyces species including Streptomyces sp. BT1, Streptomyces sp. BT2, Streptomyces sp. BT3, and Streptomyces sp. PC1.
The total ion chromatogram of ethyl acetate extracts of Streptomyces sp. BT1, BT2, BT3, and PC1, shown in Figure 1 illustrated an identical chromatogram, signifying the elution of the same type of metabolites in the particular retention time. Despite originating from distinct ecological niches, we observed quite similar metabolic profiles across the species studied. This intriguing finding may be attributed to the uniformity in cultivation conditions experienced by all species. Specifically, each species was grown under an identical culture media, maintaining consistent temperature, and environmental conditions throughout the cultivation process. This standardized approach ensured that external factors did not introduce variability, allowing us to confidently attribute the observed similarities in metabolomic profiles to intrinsic biological factors shared among the studied species. Changes in the metabolites have been reported in Streptomyces species when altering the growth media. For instance, the use of four different media in the culture resulted in the generation of many bioactive compounds, specifically, three new macrolides were discovered when using a YMG agar medium, five additional polyketides from sterilized Waksman synthetic medium, and three newly discovered naphthomycins from oatmeal medium [82,83,84].
In our study, 37 metabolites were identified in Streptomyces sp. BT1, BT2, BT3, and PC1 using LC-HRMS/MS analysis in which many compounds belong to the diketopiperazine (DKP) class. DKP is a unique class of organic compounds resembling piperazine with two amide linkages, produced by microorganisms involved in fermentation processes, particularly crucial in the food and beverage industry. It has gained a massive interest nowadays due to its variety of biological activities. The majority of indole DKPs showed notable bioactivities, including cytotoxic, antibacterial, anti-inflammatory, and antiviral activities; as a result, these substances may serve as the basis for novel drug development [85]. Almost all the metabolites identified in this study demonstrate a potent antimicrobial effect as per the existing literature, leading the bacterial strain towards a good antimicrobial activity. Cyclo-(d-Pro-d-Phe) showed antimicrobial activity against phytopathogenic Rhodococcus fascians [86]. Similarly, cyclo-(l-Pro-l-OMet) demonstrated antimicrobial activity against Escherichia coli, Pseudomonas aeruginosa, and Staphylococcus aureus showing a 100 µg/mL MIC value along with antifungal activity with a 50 µg/mL MIC value [87]. Due to the potent antimicrobial activity and synergistic impact in the development of enterococci, which are resistant to vancomycin, with MIC between 0.25 and 0.5 µg/mL, cyclo-(l-Leu-l-Pro) has a wide therapeutic application [88]. The ability to demonstrate antimicrobial activity against Vibrio anguillarum with a MIC value of 2.68 × 10−7 µg/mL makes cyclo-(l-Phenylalanyl-trans-4-hydroxy-l-Proline) a successful candidate that might show therapeutic applications [89]. Proline-rich antimicrobial peptides, or PrAMPs, have a significant amount of antibacterial activity and minimal cytotoxicity, making them prospective agents against infections that are resistant to several drugs [90]. Likewise, cyclo-(d-Pro-l-Tyr) is reported to demonstrate a potential antimicrobial effect against various plant pathogenic bacteria [32]. Cyclo-(Pro-Val) exhibited an antimicrobial effect against Vibrio anguillarum with a MIC value of 7.14 × 10−7 µg/mL [89], but this metabolite did not stop the proliferation of cancer cells. Another metabolite N-acetyltyramine detected in this work highlights a great potential to be used as a successful candidate for further study against drug-resistant bacteria as it shows a MIC value of 30 mg/mL [35]. A cyclic dipeptide called maculosin, which was isolated from Pseudomonas rhizosphaerae, has been shown to have antibacterial properties against a range of marine bacteria, including Bacillus cereus, Ruegeria sp., and Pseudoalteromonas piscida [91]. Strong antibacterial action (26–37 µg/mL) was demonstrated by a glycoside of maculosin that was isolated from marine Streptomyces sp. ZZ446, against methicillin-resistant Staphylococcus aureus, Escherichia coli, and Candida albicans [20]. Hence, most of the diketopiperazines identified in our study might be responsible for the observed antimicrobial potential.
Earlier research indicates that brevianamide F could potentially be employed in the treatment of cardiovascular dysfunction, and bacterial infection [92,93]. Apart from functioning as a glycopeptide-like antibiotic, cyclo-(Phenylalanyl-Prolyl) has demonstrated various significant roles, including inhibiting membrane permeability and decelerating DNA synthesis [94]. Due to its potent antioxidant capabilities and non-toxic nature, maculosin could be a promising candidate for diverse applications in cosmetics and therapeutics [20]. Prior research revealed that cyclo-(Gly-Leu) interacts with dopamine receptors, suggesting a potential involvement of central dopamine receptors in the pathophysiology of hypertension [95]. However, cyclo-(d-Ala-l-Pro) isolated from the fungus Colpoma sp. was reported to show poor antimicrobial activity [96]. It was reported that at an optimal concentration of 10 μg/mL, cyclo-(Tyr-Leu) was able to enhance the mycelial growth of H. marmoreus [97]. It was reported that N-phenethylacetamide hindered the TGF-β/Smad pathway, restraining the metastasis of A549 cells by impacting TGF-β-induced epithelial-mesenchymal transition (EMT) [98]. Further, cyclo-(l-Ala-l-Leu) develops disease resistance contrary to Pseudomonas syringae attack, but it does not directly stop the growth of fungi [99].
As per the existing literature, surfactin C13 demonstrated cytotoxic potential against various cancer cell lines [100]. One of the most common uses of phthalic anhydride is the production of phthalate esters, which can be used as plasticizers [101]. Phytoceramide is reported to promote hydration and enhance the healing process of damaged human stratum corneum in human skin and can be used in the cosmetic industry for creating skin barrier moisturizers [102]. Moreover, phytoceramides were found to be cytotoxic to the MES-SA, MCF-7, and HK-2 cell lines in the previous study of Monanchora clathrata, and have further proven to be used in the prevention of neurodegeneration in both in vivo and in vitro [103]. The ability of cyclo-(Pro-Gly) to decrease motor neuronal death demonstrates the neuroprotective function after brain injury and this compound also shows anxiolytic activity [104,105]. Similarly, coronafacoyl-l-isoleucine is a biosynthetic intermediate of coronatine, which seems to amplify the degree of illness symptoms brought on by pathogenic microorganisms during host infection [106,107]. N-lauryl diethanolamine is a plastic antistatic agent with high proton affinity, thus detected in positive ionization mode and reported as an interference substance that leaches from plastic microtubes during sample pretreatment [51]. Neomarinone was reported to show in vitro cytotoxicity, with an IC50 value of 8 µg/mL against HCT-116 colon cancer cells [21]. Cyclo-(d-Pro-d-Phe) has been reported to show antifungal, quorum sensing, and antimicrobial activities, and it does not show any toxicity effect up to 200 µg/mL to human cell lines [108]. Cyclo-(l-Val-l-Leu) metabolite shows a 50% inhibition rate against PANC-1 and Hela S3 cancer cells [109]. Dibutyl phthalate demonstrated a toxic effect in humans causing headaches and vertigo and could affect the throat and nose severely. Furthermore, this compound has long-term negative effects on the developing fetus and testicles [110,111]. However, despite all the negative effects, it does show antimicrobial and antifungal activities [112]. Dibutyl phthalate isolated from Streptomyces albidoflavus exhibited potent antimicrobial activity against both Gram-positive and Gram-negative bacteria [22]. The ability of pyridoxine to maintain the proper ratio of potassium and sodium in the body followed by enhancement in the production of red blood cells aids in the prevention of homocysteine synthesis and develops immunity against cancer [113].
In general, compounds of class β-carboline are known to have biological activities including antiviral, antibiotic, anticancer, and antimalarial properties [114,115]. Similarly, alkyl resorcinol, an important structural group of amphiphilic phenolic lipids, is reported to show a variety of biological activities, such as cytotoxic, genotoxic, antioxidant, and signaling capabilities [116]. 2-n-hexyl-5-n-propylresorcinol (HPR) identified in our study is reported to demonstrate antimicrobial properties against both fungi and bacteria [117]. HPR is a tiny chemical produced by several bacteria from the cell that exhibits some antibacterial action in the surroundings [52]. In our research, we identified cyclo-(d-Leu-l-Trp) in Streptomyces sp. for the first time, and researchers have reported that it could enhance the root growth of seedlings [43]. However, 3-((6-methylpyrazin-2-yl)methyl)-1H-indole was reported to show a low cytotoxicity effect against human liver cell lines and demonstrated poor antibacterial activity [50]. Hence, previously reported activities of compounds have shown the factor responsible for those antimicrobial activities, and further studies on these annotated compounds can lead to the discovery of novel antimicrobials.
Furthermore, GNPS-based molecular networking was employed for the molecular annotation of metabolites in addition to manual interpretation of LC-MS/MS data. GNPS is a web-based mass spectrometry platform accessible to the public and offers several tools for analyzing MS/MS data. This platform enables the annotation of compounds either by using spectral library search or molecular networking-based grouping of compounds into families or clusters of molecules [92]. Different compounds were annotated from GNPS including different classes’ majority of diketopiperazines, lipopeptides, dodecyl amine, and tyramine alkaloids (Figure 8). In the manual interpretation of MS/MS data, we used several natural product-based databases. Whereas, GNPS uses its library and some other reference libraries for spectral hitting, and approximately 1.8% to 2% of metabolites are annotated in untargeted metabolomics experiments due to a lack of sufficient chemical space coverage [118,119]. Moreover, ions with very low intensity lying near noise level are generally neglected in manual interpretation but GNPS workflow utilizes all ions including ions with low intensity for spectral hitting. Thus, these factors may be responsible for the variation in the number and metabolites detected in manual interpretation and GNPS spectral library hitting. 3-epi-xestoaminol C, a stereoisomer of xestoaminol C, was identified via molecular networking. This compound was reported to demonstrate IC50 values of 19.4 μM against M. tuberculosis H37Ra, 8.8 μM against HL-60 cells, and 18.0 μM against HEK cells, as reported in the literature [59]. These results emphasize the varied and distinct metabolite profiles of four isolates, providing insights into their metabolic pathways, chemical compositions, and bioactivities.
4. Materials and Methods
4.1. Isolation and Characterization of Streptomyces Species
Soil samples were collected from various ecosystems ranging up to 2743 m, altitudes in Nepal. The soils were taken from a depth of 10–15 cm beneath the earth’s surface representing diverse sampling habitats including agriculture fields, forests, and hilly regions (Table S1). The collected soil samples were kept in a sterilized zip bag and stored at 4 °C in the laboratory.
Isolation of Streptomyces species was carried out using the spread plate technique developed by William and Davies, amended with antibiotics supplements [120]. One gram of each soil sample was dissolved in 9 mL of distilled and autoclaved water and thoroughly mixed by using a vertex shaker. A three-fold serial dilution was carried out to lower the bacterial population. Then, 100 µL of each serially diluted soil suspension was spread over the ISP4 medium with the help of a sterile glass spreader. 20 mg/L nalidixic acid and 50 mg/L cycloheximide were also added to the ISP4 medium to inhibit Gram-negative bacteria and fungus species, respectively. Finally, the plates were incubated at 28 °C for 7 days [121].
4.2. Genomic DNA Extraction and 16S rRNA Gene Sequencing
The genomic DNA of Streptomyces species was isolated using the phenol-chloroform method as described in the standard protocol of molecular biology [122]. For taxonomy identification, 16S rRNA gene was amplified using oligonucleotides 27F: 5′-AGAGTTTGATCCTGGCTCAG-3′, and 1492R: 5′-GGTTACCTTGTTACGACTT-3′. The Polymerase Chain Reaction (PCR) was carried out in a 50 μL reaction mixture using 5xTaq-PCR Premix (GenoTech Corporation, Daejeon, Republic of Korea) containing Taq-polymerase within 30 cycles. A cycle was programmed with denaturation at 98 °C (10 s), an annealing at 54 °C (10 s), and an extension at 72 °C (2 min). The PCR products were isolated from low-melting agarose gel and were purified by using the QIAquick Gel Extraction Kit (Qiagen, Germantown, MD, USA). The purified PCR products were sequenced using the same primers (27F and 1492F) through the Sangar dideoxy method by GenoTech Co., Daejeon, Republic of Korea. Then, the BLAST tool of the National Center for Biotechnology Information (NCBI) was used to compare the 16S rRNA gene sequences of our isolates with those in the GenBank database [123]. Then, 12 sequences that were highly similar to our amplified 16S rRNA genes were subjected to multiple sequence alignment with our sequences, followed by the generation of a phylogenetic tree using the neighbor-joining method with the MEGA Software (version 11.0.13) (https://www.megasoftware.net/) [17].
4.3. Fermentation and Extraction of Metabolites
The seed culture of Streptomyces species was carried out in Tryptic Soy Broth (TSB) medium (Tryptone 17.0 g, Soytone 3.0 g, Glucose 2.5 g, Sodium Chloride 5.0 g, Dipotassium Phosphate 2.5 g, pH 7.3 ± 0.2 at 28 °C; volume 1 L water). After sufficient growth of Streptomyces species, 1 mL (1%) of bacterial suspension was transferred into 100 mL of freshly prepared TSB medium for fermentation (for production of secondary metabolites). The incubation was conducted at 28 °C for 5–7 days at 180 rpm in a shaking incubator until bacterial growth reached the stationary phase. Secondary metabolites were harvested by mixing an equal volume of ethyl acetate with culture broth. The clear supernatant was transferred into a clean and dry beaker and evaporated in a water bath at 37 °C for 2–3 days to obtain the crude bacterial extracts. Then, the dried extract was placed at 4 °C until use [124].
4.4. Antimicrobial Assays
The primary screening of the isolates was carried out in Mueller Hinton Agar (MHA) medium by a perpendicular streaking method. The agar-well diffusion method was used for the secondary screening of crude extracts [125]. In this method, the standard culture of test organisms was swabbed over the MHA medium with the help of sterile cotton buds. For this, Gram-positive pathogen Staphylococcus aureus ATCC 43300, and various Gram-negative pathogens, Shigella sonnei ATCC 25931, Salmonella typhi ATCC 14028, Klebsiella pneumoniae ATCC 700603, and Escherichia coli ATCC 25922 were tested. These tested pathogens were incubated in Mueller Hinton Broth (MHB) medium at 37 °C for 24 h. Then, their turbidity was adjusted to that of standard 0.5 McFarland (1.5 × 108 CFU/mL) for further use. Then, the wells were made with the help of sterile cork borers of 6 mm in diameter. Finally, the wells were filled with the positive control (1 mg/mL neomycin), negative control (50% DMSO), and a working solution of extract dissolved in 50% DMSO. At last, the plates were incubated at 37 °C for 24 h and observed for the clear zone of inhibition.
The MIC and MBC of the crude extract were determined by the broth microdilution method according to the Clinical Laboratory Standard Institute (CLSI) [126]. A series of two-fold dilutions of extract were prepared directly in sterile 96-well microdilution plates containing MHB to obtain a range of concentrations. The bacterial inoculum was added at a final concentration of 1.5 × 108 CFU/mL by diluting 1:100 after matching the turbidity of the 0.5 McFarland turbidity culture in MHB. Finally, 30 μL of bacteria were added to each well except for the negative control. The MBC was determined by streaking the good contents onto nutrient agar plates, followed by incubation for over 18 h at 37 °C [127].
4.5. Metabolic Comparison
Ethyl acetate (EA) extracts of Streptomyces species were then subjected to liquid chromatography-high resolution tandem mass spectrometric analysis (LC–HRMS/MS) by employing an Agilent G6545B quadrupole time-of-flight (Q-TOF) mass spectrometer (Agilent Technologies, Santa Clara, CA, USA) equipped with a heated electrospray ion source at Sungkyunkwan University, Suwon, Republic of Korea. For MS/MS analysis, four samples (BT1, BT2, BT3, and PC1) were prepared by dissolving EA extracts in HPLC-grade solvent (methanol) at a concentration of 1 mg/mL. A volume of 150 μL from each sample was transferred to HPLC autosampler vials. Chromatographic separation was achieved using an Acquity® (Indio, CA, USA) UPLC BEH reverse-phase C18 column (150 mm × 2.1 mm, 1.7 μM). The mobile phases, acidified with 0.1% formic acid, consisted of H2O (A) and acetonitrile (B). The composition of the organic solvent was used as follows: 5% from 0.00 to 2.00 min, 20% at 5.00 min, 100% at 20.00 min, and then returning to 5% from 23.00 to 25.00 min. The injection volume for each sample was maintained at 3 μL, and a constant flow rate of 0.5 mL/min was maintained. The MS/MS data acquisition was performed using the electrospray ionization (ESI) technique in positive ion mode with a m/z range of 50–1200 Da, collision energies set at 15 V and 40 V, and a full width at half maximum (FWHM) of 3000.
The raw data (.d format) were converted into .mzXML format [128] and further annotated using CSI: FingerID, which is a graphical interface incorporated in SIRIUS software (version 5.8.0) [129]. The calculated mass, absolute error, RDBE, and molecular formulae were generated by MestReNova software (version 12.0.0) (accessed on 10–30 December 2023) and were compared with the formula generated by SIRIUS. Furthermore, the annotated compounds were validated via the literature survey using SciFinder (https://scifinder-n.cas.org/, accessed on 12–14 January 2024), and natural products-based databases such as PubChem (https://pubchem.ncbi.nlm.nih.gov/, accessed on 25–30 December 2023), ChemSpider (https://www.chemspider.com/, accessed on 25–30 December 2023), Natural Products Atlas (https://www.npatlas.org/, accessed on 25–30 December 2023), LOTUS (https://lotus.naturalproducts.net/, accessed on 25–30 December 2023), and libraries search using SIRIUS software (version 5.8.0). SIRIUS score, generated by software, serves as a parameter for gauging the confidence of molecular annotation, with a higher score indicating greater confidence in the annotation.
4.6. GNPS-Based Molecular Networking Analysis
EA extracts of four Streptomyces species were prepared for MS/MS analysis by dissolving them in HPLC-grade MeOH (1 mg/mL), with 150 μL of each sample transferred to an HPLC autosampler vial. Metabolomic profiling was conducted using an Agilent G6545B quadrupole time-of-flight (Q-TOF) mass spectrometer (Agilent Technologies, Santa Clara, CA, USA) equipped with a heated electrospray ion source (HESI). Chromatographic separation was achieved using an Acquity® UPLC BEH reverse-phase column C18 (150 mm × 2.1 mm, 1.7 μM). The mobile phase consisted of 0.1% formic acid in H2O (A), and acetonitrile (B) in varying proportions: 5% (B) from 0 to 2 min, 5–20% (B) from 2 to 5 min, 20–100% (B) from 5 to 20 min, 100% (B) from 20 to 23 min, and 100–5% (B) from 23 to 25 min. Each sample was injected at a volume of 3 μL, with a flow rate of 0.3 mL/min maintained. MS/MS analysis was conducted using electrospray ionization (ESI) in positive ion mode. Spectral hits were performed using a modified version with an m/z range of 50–1700, with collision energies set at 15 V and 40 V, capillary voltage (2.5 kV), and a full width at half maximum (FWHM) of 3000 [129]. The raw data ‘.d format’ files were first converted to ‘.mzXML’ format using open-source MSConvert software (https://proteowizard.sourceforge.io/download.html) (Version: 3.0). To upload the files, the recommended FTP client WinSCP was utilized, and they were transferred to the GNPS platform. Visualizing the MS/MS data followed established GNPS-based procedures (accessed on 17 January 2023). Molecular networks generated in GNPS were further exported to Cytoscape (version 3.9.1.) in ‘.graphml’ format to enable customized visualization and additional analysis.
5. Conclusions
Streptomyces genus can produce a wide range of bioactive secondary metabolites that have good efficacy against several MDR pathogens. In this study, Streptomyces species were isolated from the soils collected from various habitats in Nepal and characterized by using 16S rRNA gene sequencing. Further, the ethyl acetate extracts of Streptomyces sp. were subjected to LC-HRMS/MS analysis and GNPS-based molecular networking. We found a similar metabolite profile in the mentioned species. Thirty-seven different secondary metabolites encompassing a range of compounds, including polypeptides, bacterial alkaloids, amino compounds, and diketopiperazines were annotated in our study. These metabolites, specifically diketopiperazines are reported to demonstrate an effective antimicrobial activity based on existing literature. In addition, to the best of our knowledge, four metabolites, namely cyclo-(Ile-Ser), 2-n-hexyl-5-n-propylresorcinol, 3-(6-methylpyrazin-2-yl) methyl)-1H-indole and cyclo-(d-Leu-l-Trp) were reported for the first time in Streptomyces species. In addition, these findings contribute new additional value by revealing new sources for the isolation of these metabolites, which can aid in the drug discovery process. Furthermore, despite being isolated from different ecological niches, we observed similar metabolome profiles in the studied species. These results could be supported by the fact that all the species were cultivated under identical culture media, temperature, and conditions. Therefore, further study can be conducted on the Streptomyces species by varying fermentation media and culture conditions to unveil new metabolites. Moreover, the newly annotated metabolites in this study can further be validated through NMR, followed by their biological evaluation, which could facilitate the drug discovery process.
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/ijms25084193/s1.
Author Contributions
Conceptualization, J.K.S., N.P. and K.H.K.; methodology, B.B.T., R.B., P.C. and P.B.P.; software, B.B.T., C.H., S.J. and R.B.; formal analysis, R.B., C.H. and R.T.M.; writing—original draft preparation, B.B.T., R.B., P.C., S.J. and N.P.; writing—review and editing, J.K.S., K.H.K. and P.B.P.; supervision, J.K.S., N.P. and K.H.K.; funding acquisition, J.K.S., K.H.K. and N.P. All authors have read and agreed to the published version of the manuscript.
Funding
This research was supported by the National Research Foundation (NRF) of Korea (Grant Number RS-2023-00259697) through the Ministry of Science and ICT. Similarly, it was supported by the NRF (MSIT; Grant numbers 2019R1A5A2027340 and 2021R1A2C2007937). This project was supported by the University Grants Commission, Nepal (Award No. CRIG-78/79-S&T-01).
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
Data available on request from the corresponding author.
Acknowledgments
We are thankful to Bibek Raj Bhattarai and Sajan Shakya for their support.
Conflicts of Interest
The authors declare no conflicts of interest.
References
- Toner, E.; Adalja, A.; Gronvall, G.K.; Cicero, A.; Inglesby, T.V. Antimicrobial Resistance Is a Global Health Emergency. Health Secur. 2015, 13, 153–155. [Google Scholar] [CrossRef] [PubMed]
- Clish, C.B. Metabolomics: An Emerging but Powerful Tool for Precision Medicine. Cold Spring Harb. Mol. Case Stud. 2015, 1, a000588. [Google Scholar] [CrossRef] [PubMed]
- Zhou, B.; Xiao, J.F.; Tuli, L.; Ressom, H.W. LC-MS-Based Metabolomics. Mol. Biosyst. 2012, 8, 470. [Google Scholar] [CrossRef] [PubMed]
- Quinn, R.A.; Nothias, L.-F.; Vining, O.; Meehan, M.; Esquenazi, E.; Dorrestein, P.C. Molecular Networking as a Drug Discovery, Drug Metabolism, and Precision Medicine Strategy. Trends Pharmacol. Sci. 2017, 38, 143–154. [Google Scholar] [CrossRef] [PubMed]
- Sanglier, J.J.; Haag, H.; Huck, T.A.; Fehr, T. Novel Bioactive Compounds from Actinomycetes: A Short Review (1988–1992). Res. Microbiol. 1993, 144, 633–642. [Google Scholar] [CrossRef] [PubMed]
- Pacios-Michelena, S.; Aguilar González, C.N.; Alvarez-Perez, O.B.; Rodriguez-Herrera, R.; Chávez-González, M.; Arredondo Valdés, R.; Ascacio Valdés, J.A.; Govea Salas, M.; Ilyina, A. Application of Streptomyces Antimicrobial Compounds for the Control of Phytopathogens. Front. Sustain. Food Syst. 2021, 5, 696518. [Google Scholar] [CrossRef]
- Gopalakrishnan, S.; Srinivas, V.; Prasanna, S.L. Chapter 5—Streptomyces. In Beneficial Microbes in Agro-Ecology; Amaresan, N., Senthil Kumar, M., Annapurna, K., Kumar, K., Sankaranarayanan, A., Eds.; Academic Press: Cambridge, MA, USA, 2020; pp. 55–71. ISBN 978-0-12-823414-3. [Google Scholar]
- Arasu, M.V.; Duraipandiyan, V.; Agastian, P.; Ignacimuthu, S. Antimicrobial Activity of Streptomyces Spp. ERI-26 Recovered from Western Ghats of Tamil Nadu. J. Mycol. Méd. 2008, 18, 147–153. [Google Scholar] [CrossRef]
- Yang, L.; Li, X.; Wu, P.; Xue, J.; Xu, L.; Li, H.; Wei, X. Streptovertimycins A–H, New Fasamycin-Type Antibiotics Produced by a Soil-Derived Streptomyces morookaense Strain. J. Antibiot. 2020, 73, 283–289. [Google Scholar] [CrossRef]
- Fang, Y.; Wang, J.; Tang, Y.; Guo, Z.; Bai, J.; Wu, L.; Su, J.; Cen, S.; Yu, L.; Zhang, D. Geninthiocins E and F, Two New Cyclic Thiopeptides with Antiviral Activities from Soil-Derived Streptomyces Sp. CPCC 200267 Using OSMAC Strategy. J. Antibiot. 2023, 76, 101–104. [Google Scholar] [CrossRef]
- Liu, M.; Wan, Z.; Yang, S.; Liu, F.; Yang, X.; Wu, Z.; Zhang, Y.; Wang, K.; Fang, W. Two New Dipimprinine Alkaloids from Soil-Derived Streptomyces Sp. 44414B. J. Antibiot. 2021, 74, 474–476. [Google Scholar] [CrossRef]
- Maiti, P.K.; Das, S.; Sahoo, P.; Mandal, S. Streptomyces Sp SM01 Isolated from Indian Soil Produces a Novel Antibiotic Picolinamycin Effective against Multi Drug Resistant Bacterial Strains. Sci. Rep. 2020, 10, 10092. [Google Scholar] [CrossRef] [PubMed]
- Hu, L.; Chen, X.; Han, L.; Zhao, L.; Miao, C.; Huang, X.; Chen, Y.; Li, P.; Li, Y. Two New Phenazine Metabolites with Antimicrobial Activities from Soil-Derived Streptomyces Species. J. Antibiot. 2019, 72, 574–577. [Google Scholar] [CrossRef] [PubMed]
- Jiang, Z.; Guo, L.; Chen, C.; Liu, S.; Zhang, L.; Dai, S.; He, Q.; You, X.; Hu, X.; Tuo, L.; et al. Xiakemycin A, a Novel Pyranonaphthoquinone Antibiotic, Produced by the Streptomyces Sp. CC8-201 from the Soil of a Karst Cave. J. Antibiot. 2015, 68, 771–774. [Google Scholar] [CrossRef] [PubMed]
- Ganesan, P.; Reegan, A.D.; David, R.H.A.; Gandhi, M.R.; Paulraj, M.G.; Al-Dhabi, N.A.; Ignacimuthu, S. Antimicrobial Activity of Some Actinomycetes from Western Ghats of Tamil Nadu, India. Alex. J. Med. 2017, 53, 101–110. [Google Scholar] [CrossRef]
- Goodfellow, M.; Kämpfer, P.; Busse, H.-J.; Trujillo, M.E.; Suzuki, K.; Ludwig, W.; Whitman, W.B. (Eds.) Bergey’s Manual® of Systematic Bacteriology; Springer: New York, NY, USA, 2012; ISBN 978-0-387-95043-3. [Google Scholar]
- Tamura, K.; Stecher, G.; Kumar, S. MEGA11: Molecular Evolutionary Genetics Analysis Version 11. Mol. Biol. Evol. 2021, 38, 3022–3027. [Google Scholar] [CrossRef]
- Singh, V.K.; Mishra, A.; Jha, B. 3-Benzyl-Hexahydro-Pyrrolo[1,2-a]Pyrazine-1,4-Dione Extracted from Exiguobacterium Indicum Showed Anti-Biofilm Activity against Pseudomonas Aeruginosa by Attenuating Quorum Sensing. Front. Microbiol. 2019, 10, 1269. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Dobretsov, S.; Xu, Y.; Xiao, X.; Hung, O.S.; Qian, P. Antifouling Diketopiperazines Produced by a Deep-Sea Bacterium, Streptomyces fungicidicus. Biofouling 2006, 22, 187–194. [Google Scholar] [CrossRef]
- Paudel, B.; Maharjan, R.; Rajbhandari, P.; Aryal, N.; Aziz, S.; Bhattarai, K.; Baral, B.; Malla, R.; Bhattarai, H.D. Maculosin, a Non-Toxic Antioxidant Compound Isolated from Streptomyces Sp. KTM18. Pharm. Biol. 2021, 59, 931–934. [Google Scholar] [CrossRef]
- Hardt, I.H.; Jensen, P.R.; Fenical, W. Neomarinone, and New Cytotoxic Marinone Derivatives, Produced by a Marine Filamentous Bacterium (Actinomycetales). Tetrahedron Lett. 2000, 41, 2073–2076. [Google Scholar] [CrossRef]
- Roy, R.N.; Laskar, S.; Sen, S.K. Dibutyl Phthalate, the Bioactive Compound Produced by Streptomyces Albidoflavus 321.2. Microbiol. Res. 2006, 161, 121–126. [Google Scholar] [CrossRef]
- Dempsey, W.B. Synthesis of Pyridoxine by a Pyridoxal Auxotroph of Escherichia coli. J. Bacteriol. 1966, 92, 333–337. [Google Scholar] [CrossRef] [PubMed]
- Yang, X.-Q.; Yang, Y.-B.; Zhou, H.; He, G.-W.; Zhao, L.-X.; Xu, L.-H.; Ding, Z.-T. New Megastigmane Glycoside and Alkaloids from Streptomyces Sp. YIM 63342. Nat. Prod. Res. 2013, 27, 1191–1196. [Google Scholar] [CrossRef] [PubMed]
- Hoefler, B.C.; Gorzelnik, K.V.; Yang, J.Y.; Hendricks, N.; Dorrestein, P.C.; Straight, P.D. Enzymatic Resistance to the Lipopeptide Surfactin as Identified through Imaging Mass Spectrometry of Bacterial Competition. Proc. Natl. Acad. Sci. USA 2012, 109, 13082–13087. [Google Scholar] [CrossRef] [PubMed]
- Dashti, Y.; Grkovic, T.; Abdelmohsen, U.; Hentschel, U.; Quinn, R. Production of Induced Secondary Metabolites by a Co-Culture of Sponge-Associated Actinomycetes, Actinokineospora Sp. EG49 and Nocardiopsis Sp. RV163. Mar. Drugs 2014, 12, 3046–3059. [Google Scholar] [CrossRef] [PubMed]
- Zhou, H.; Yang, Y.; Peng, T.; Li, W.; Zhao, L.; Xu, L.; Ding, Z. Metabolites of Streptomyces Sp., an Endophytic Actinomycete from Alpinia oxyphylla. Nat. Prod. Res. 2014, 28, 265–267. [Google Scholar] [CrossRef] [PubMed]
- Sriragavi, G.; Sangeetha, M.; Santhakumar, M.; Lokesh, E.; Nithyalakshmi, M.; Saleel, C.A.; Balagurunathan, R. Exploring Antibacterial Properties of Bioactive Compounds Isolated from Streptomyces Sp. in Bamboo Rhizosphere Soil. ACS Omega 2023, 8, 36333–36343. [Google Scholar] [CrossRef] [PubMed]
- Stankeviciute, G.; Tang, P.; Ashley, B.; Chamberlain, J.D.; Hansen, M.E.B.; Coleman, A.; D’Emilia, R.; Fu, L.; Mohan, E.C.; Nguyen, H.; et al. Convergent Evolution of Bacterial Ceramide Synthesis. Nat. Chem. Biol. 2022, 18, 305–312. [Google Scholar] [CrossRef] [PubMed]
- Ser, H.-L.; Palanisamy, U.D.; Yin, W.-F.; Abd Malek, S.N.; Chan, K.-G.; Goh, B.-H.; Lee, L.-H. Presence of Antioxidative Agent, Pyrrolo[1,2-a]Pyrazine-1,4-Dione, Hexahydro- in Newly Isolated Streptomyces mangrovisoli Sp. Nov. Front. Microbiol. 2015, 6. [Google Scholar] [CrossRef] [PubMed]
- Zin, N.M.; Baba, M.S.; Zainal-Abidin, A.H.; Latip, J.; Mazlan, N.W.; Edrada-Ebel, R. Gancidin W, a Potential Low-Toxicity Antimalarial Agent Isolated from an Endophytic Streptomyces SUK10. Drug Des. Dev. Ther. 2017, 11, 351–363. [Google Scholar] [CrossRef]
- Wattana-Amorn, P.; Charoenwongsa, W.; Williams, C.; Crump, M.P.; Apichaisataienchote, B. Antibacterial Activity of Cyclo (L-Pro-L-Tyr) and Cyclo (D-Pro-L-Tyr) from Streptomyces Sp. Strain 22-4 against Phytopathogenic Bacteria. Nat. Prod. Res. 2016, 30, 1980–1983. [Google Scholar] [CrossRef]
- Fyans, J.K.; Altowairish, M.S.; Li, Y.; Bignell, D.R.D. Characterization of the Coronatine-Like Phytotoxins Produced by the Common Scab Pathogen Streptomyces Scabies. Mol. Plant-Microbe Interactions 2015, 28, 443–454. [Google Scholar] [CrossRef] [PubMed]
- Pettit, G.R.; Du, J.; Pettit, R.K.; Richert, L.A.; Hogan, F.; Mukku, V.J.R.V.; Hoard, M.S. Antineoplastic Agents. 554. The Manitoba Bacterium Streptomyces Sp. J. Nat. Prod. 2006, 69, 804–806. [Google Scholar] [CrossRef] [PubMed]
- Driche, E.H.; Badji, B.; Bijani, C.; Belghit, S.; Pont, F.; Mathieu, F.; Zitouni, A. A New Saharan Strain of Streptomyces Sp. GSB-11 Produces Maculosin and N-Acetyltyramine Active against Multidrug-Resistant Pathogenic Bacteria. Curr. Microbiol. 2022, 79, 298. [Google Scholar] [CrossRef] [PubMed]
- Cho, J.Y.; Kang, J.Y.; Hong, Y.K.; Baek, H.H.; Shin, H.W.; Kim, M.S. Isolation and Structural Determination of the Antifouling Diketopiperazines from Marine-Derived Streptomyces Praecox 291-11. Biosci. Biotechnol. Biochem. 2012, 76, 1116–1121. [Google Scholar] [CrossRef] [PubMed]
- Wei, H.-X.; Fang, X.-W.; Xie, X.-S.; Zhang, S.-P.; Jiang, Y.; Wu, S.-H. Secondary Metabolites of a Soil-Derived Streptomyces Kunmingensis. Chem. Nat. Compd. 2017, 53, 794–796. [Google Scholar] [CrossRef]
- Bhandari, S.; Bhattarai, B.R.; Adhikari, A.; Aryal, B.; Shrestha, A.; Aryal, N.; Lamichhane, U.; Thapa, R.; Thapa, B.B.; Yadav, R.P.; et al. Characterization of Streptomyces Species and Validation of Antimicrobial Activity of Their Metabolites through Molecular Docking. Processes 2022, 10, 2149. [Google Scholar] [CrossRef]
- Macherla, V.R.; Liu, J.; Bellows, C.; Teisan, S.; Nicholson, B.; Lam, K.S.; Potts, B.C.M. Glaciapyrroles A, B, and C, Pyrrolosesquiterpenes from a Streptomyces Sp. Isolated from an Alaskan Marine Sediment. J. Nat. Prod. 2005, 68, 780–783. [Google Scholar] [CrossRef]
- Elleuch, L.; Shaaban, M.; Smaoui, S.; Mellouli, L.; Karray-Rebai, I.; Fourati-Ben Fguira, L.; Shaaban, K.A.; Laatsch, H. Bioactive Secondary Metabolites from a New Terrestrial Streptomyces Sp. TN262. Appl. Biochem. Biotechnol. 2010, 162, 579–593. [Google Scholar] [CrossRef] [PubMed]
- Ding, W.-J.; Zhang, S.-Q.; Wang, J.-H.; Lin, Y.-X.; Liang, Q.-X.; Zhao, W.-J.; Li, C.-Y. A New Di-O-Prenylated Flavone from an Actinomycete Streptomyces Sp. MA-12. J. Asian Nat. Prod. Res. 2013, 15, 209–214. [Google Scholar] [CrossRef]
- Al-Dhabi, N.A.; Esmail, G.A.; Duraipandiyan, V.; Arasu, M.V. Chemical Profiling of Streptomyces Sp. Al-Dhabi-2 Recovered from an Extreme Environment in Saudi Arabia as a Novel Drug Source for Medical and Industrial Applications. Saudi J. Biol. Sci. 2019, 26, 758–766. [Google Scholar] [CrossRef]
- Kimura, Y.; Sawada, A.; Kuramata, M.; Kusano, M.; Fujioka, S.; Kawano, T.; Shimada, A. Brevicompanine C, Cyclo-(d-Ile-l-Trp), and Cyclo-(d-Leu-l-Trp), Plant Growth Regulators from Penicillium Brevi-Compactum. J. Nat. Prod. 2005, 68, 237–239. [Google Scholar] [CrossRef]
- Srivastava, V.; Singla, R.K.; Dubey, A.K. Inhibition of Biofilm and Virulence Factors of Candida albicans by Partially Purified Secondary Metabolites of Streptomyces chrestomyceticus Strain ADP4. Curr. Top. Med. Chem. 2018, 18, 925–945. [Google Scholar] [CrossRef] [PubMed]
- Zhou, H.; Yang, Y.; Yang, X.; Li, W.; Xiong, Z.; Zhao, L.; Xu, L.; Ding, Z. A New Cyclic Tetrapeptide from an Endophytic Streptomyces Sp. YIM67005. Nat. Prod. Res. 2014, 28, 318–323. [Google Scholar] [CrossRef] [PubMed]
- Kornsakulkarn, J.; Saepua, S.; Veeranondha, S.; Rachtawee, P.; Isaka, M.; Thongpanchang, C. Carboline Alkaloids and Isocoumarins from the Wasp Pathogenic Fungus Ophiocordyceps sphecocephala BCC 2661. Phytochem. Lett. 2018, 27, 134–138. [Google Scholar] [CrossRef]
- Tan, L.T.-H.; Chan, K.-G.; Pusparajah, P.; Yin, W.-F.; Khan, T.M.; Lee, L.-H.; Goh, B.-H. Mangrove Derived Streptomyces Sp. MUM265 as a Potential Source of Antioxidant and Anticolon-Cancer Agents. BMC Microbiol. 2019, 19, 38. [Google Scholar] [CrossRef] [PubMed]
- Feng, K.-J.; Cheng, M.-J.; Yang, S.-S.; Wu, M.-D.; Hsieh, S.-Y.; Chan, H.-Y.; Su, Y.-S.; Chou, Y.-T.; Chang, H.-S. Chemical Constituents of the Endophytic Fungus Ophiocordyceps sobolifera. Chem. Nat. Compd. 2019, 55, 309–312. [Google Scholar] [CrossRef]
- Wei, J.; Zhang, X.-Y.; Deng, S.; Cao, L.; Xue, Q.-H.; Gao, J.-M. α-Glucosidase Inhibitors and Phytotoxins from Streptomyces xanthophaeus. Nat. Prod. Res. 2017, 31, 2062–2066. [Google Scholar] [CrossRef] [PubMed]
- Yang, X.-W.; Zhang, G.-Y.; Ying, J.-X.; Yang, B.; Zhou, X.-F.; Steinmetz, A.; Liu, Y.-H.; Wang, N. Isolation, Characterization, and Bioactivity Evaluation of 3-((6-Methylpyrazin-2-Yl)Methyl)-1H-Indole, a New Alkaloid from a Deep-Sea-Derived Actinomycete Serinicoccus Profundi Sp. Nov. Mar. Drugs 2013, 11, 33–39. [Google Scholar] [CrossRef] [PubMed]
- Chai, Y.; Chen, H.; Gao, G.; Liu, X.; Lu, C. Identification of New Interferences Leached from Plastic Microcentrifuge Tubes in Electrospray Ionization Mass Spectrometry. Rapid Commun. Mass. Spectrom. 2019, 33, 969–977. [Google Scholar] [CrossRef]
- Calderón, C.E.; Tienda, S.; Heredia-Ponce, Z.; Arrebola, E.; Cárcamo-Oyarce, G.; Eberl, L.; Cazorla, F.M. The Compound 2-Hexyl, 5-Propyl Resorcinol Has a Key Role in Biofilm Formation by the Biocontrol Rhizobacterium Pseudomonas chlororaphis PCL1606. Front. Microbiol. 2019, 10, 396. [Google Scholar] [CrossRef]
- Álvarez-Álvarez, R.; Martínez-Burgo, Y.; Rodríguez-García, A.; Liras, P. Discovering the Potential of S. Clavuligerus for Bioactive Compound Production: Cross-Talk between the Chromosome and the pSCL4 Megaplasmid. BMC Genom. 2017, 18, 907. [Google Scholar] [CrossRef] [PubMed]
- Alshaibani, M.M.; Zin, N.M.; Jalil, J.; Sidik, N.M.; Ahmad, S.J.; Kamal, N.; Edrada-Ebel, R. Isolation, Purification, and Characterization of Five Active Diketopiperazine Derivatives from Endophytic Streptomyces SUK 25 with Antimicrobial and Cytotoxic Activities. J. Microbiol. Biotechnol. 2017, 27, 1249–1256. [Google Scholar] [CrossRef] [PubMed]
- Abdelkader, M.S.A.; Philippon, T.; Asenjo, J.A.; Bull, A.T.; Goodfellow, M.; Ebel, R.; Jaspars, M.; Rateb, M.E. Asenjonamides A–C, Antibacterial Metabolites Isolated from Streptomyces asenjonii Strain KNN 42.f from an Extreme-Hyper Arid Atacama Desert Soil. J. Antibiot. 2018, 71, 425–431. [Google Scholar] [CrossRef] [PubMed]
- Bown, L.; Li, Y.; Berrué, F.; Verhoeven, J.T.P.; Dufour, S.C.; Bignell, D.R.D. Coronafacoyl Phytotoxin Biosynthesis and Evolution in the Common Scab Pathogen Streptomyces scabiei. Appl. Env. Microbiol. 2017, 83, e01169-e17. [Google Scholar] [CrossRef] [PubMed]
- Wang, T.; Jiang, Y.; Ma, K.; Li, Y.; Huang, R.; Xie, X.; Wu, S. Two New Butenolides Produced by an Actinomycete Streptomyces Sp. Chem. Biodivers. 2014, 11, 929–933. [Google Scholar] [CrossRef] [PubMed]
- GNPS—Analyze, Connect, and Network with Your Mass Spectrometry Data. Available online: https://gnps.ucsd.edu/ProteoSAFe/static/gnps-splash.jsp (accessed on 3 December 2023).
- Dasyam, N.; Munkacsi, A.B.; Fadzilah, N.H.; Senanayake, D.S.; O’Toole, R.F.; Keyzers, R.A. Identification and Bioactivity of 3-Epi-Xestoaminol C Isolated from the New Zealand Brown Alga Xiphophora chondrophylla. J. Nat. Prod. 2014, 77, 1519–1523. [Google Scholar] [CrossRef] [PubMed]
- Induja, D.K.; Jesmina, A.R.S.; Joseph, M.M.; Shamjith, S.; Ingaladal, N.; Maiti, K.K.; Kumar, B.S.D.; Lankalapalli, R.S. Isolation of Two New Stereochemical Variants of Streptophenazine by Cocultivation of Streptomyces NIIST-D31, Streptomyces NIIST-D47, and Streptomyces NIIST-D63 Strains in 3C2 Combinations. J. Antibiot. 2023, 76, 567–578. [Google Scholar] [CrossRef]
- Mondal, H.; Thomas, J. Isolation and Characterization of a Novel Actinomycete Isolated from Marine Sediments and Its Antibacterial Activity against Fish Pathogens. Antibiotics 2022, 11, 1546. [Google Scholar] [CrossRef] [PubMed]
- Nakano, C.; Kudo, F.; Eguchi, T.; Ohnishi, Y. Genome Mining Reveals Two Novel Bacterial Sesquiterpene Cyclases: (−)-Germacradien-4-ol and (−)-Epi-α-Bisabolol Synthases from Streptomyces citricolor. ChemBioChem 2011, 12, 2271–2275. [Google Scholar] [CrossRef]
- Murao, S.; Hayashi, H.; Tarui, N. Anthranilic Acid, as a Fruiting Body Inducing Substance in Favolus arcularius, from a Strain TA 7 of Actinomycetes. Agric. Biol. Chem. 1984, 48, 1669–1671. [Google Scholar] [CrossRef]
- Xie, C.-L.; Niu, S.; Xia, J.-M.; Peng, K.; Zhang, G.-Y.; Yang, X.-W. Saccharopolytide A, a New Cyclic Tetrapeptide with Rare 4-Hydroxy-Proline Moieties from the Deep-Sea Derived Actinomycete Saccharopolyspora cebuensis MCCC 1A09850. Nat. Prod. Res. 2018, 32, 1627–1631. [Google Scholar] [CrossRef] [PubMed]
- Chen, C.; Ye, Y.; Wang, R.; Zhang, Y.; Wu, C.; Debnath, S.C.; Ma, Z.; Wang, J.; Wu, M. Streptomyces nigra Sp. Nov. Is a Novel Actinobacterium Isolated from Mangrove Soil and Exerts a Potent Antitumor Activity in Vitro. Front. Microbiol. 2018, 9, 1587. [Google Scholar] [CrossRef] [PubMed]
- Gao, Y.; Yu, L.; Peng, C.; Li, Z.; Guo, Y. Diketopiperazines from Two Strains of South China Sea Sponge-Associated Microorganisms. Biochem. Syst. Ecol. 2010, 38, 931–934. [Google Scholar] [CrossRef]
- Bhattarai, B.R.; Khadayat, K.; Aryal, N.; Aryal, B.; Lamichhane, U.; Bhattarai, K.; Rana, N.; Regmi, B.P.; Adhikari, A.; Thapa, S.; et al. Untargeted Metabolomics of Streptomyces Species Isolated from Soils of Nepal. Processes 2022, 10, 1173. [Google Scholar] [CrossRef]
- Guo, Z.K.; Wang, R.; Chen, F.X.; Liu, T.M.; Yang, M.Q. Bioactive Aromatic Metabolites from the Sea Urchin-Derived Actinomycete Streptomyces spectabilis Strain HDa1. Phytochem. Lett. 2018, 25, 132–135. [Google Scholar] [CrossRef]
- Liu, X.-Y.; Yang, S.-Z.; Mu, B.-Z. Isolation and Characterization of a C12-Lipopeptide Produced by Bacillus subtilis HSO 121. J. Pept. Sci. 2008, 14, 864–875. [Google Scholar] [CrossRef]
- Barale, S.S.; Ghane, S.G.; Sonawane, K.D. Purification and Characterization of Antibacterial Surfactin Isoforms Produced by Bacillus velezensis SK. AMB Expr. 2022, 12, 7. [Google Scholar] [CrossRef]
- Atikana, A.; Sukmarini, L.; Warsito, M.F.; Untari, F.; Murniasih, T.; Rahmawati, S.I.; Qodria, L.; Siwi, O.R.; Ratnakomala, S.; Prasetyoputri, A.; et al. Bioactivity Profiles of Actinobacterium Strain BTA 1-131 (InaCC A1205) Isolated from Indonesian Sponge Melophlus sarassinorum. Indones. J. Pharm. 2023, 34, 280–290. [Google Scholar] [CrossRef]
- Zhang, F.; Li, B.; Wen, Y.; Liu, Y.; Liu, R.; Liu, J.; Liu, S.; Jiang, Y. An Integrated Strategy for the Comprehensive Profiling of the Chemical Constituents of Aspongopus chinensis Using UPLC-QTOF-MS Combined with Molecular Networking. Pharm. Biol. 2022, 60, 1349–1364. [Google Scholar] [CrossRef]
- Gos, F.M.W.R.; Savi, D.C.; Shaaban, K.A.; Thorson, J.S.; Aluizio, R.; Possiede, Y.M.; Rohr, J.; Glienke, C. Antibacterial Activity of Endophytic Actinomycetes Isolated from the Medicinal Plant Vochysia Divergens (Pantanal, Brazil). Front. Microbiol. 2017, 8, 1642. [Google Scholar] [CrossRef]
- Clark, A.M.; Hufford, C.D. Use of Microorganisms for the Study of Drug Metabolism: An Update. Med. Res. Rev. 1991, 11, 473–501. [Google Scholar] [CrossRef]
- Hordern, J.; Johnson, R.H.; McLennan, B.D. N6-(Δ2-isopentenyl)Adenosine: Hydrolysis by a Nucleosidase Isolated from Lactobacillus acidophilus Cells. Can. J. Microbiol. 1975, 21, 633–638. [Google Scholar] [CrossRef]
- Korkmaz, Ç.A.; Hameş-Kocabaş, E.E.; Uzel, A.; Bedir, E. Tryptamine Derived Amides with Thiazole Ring System from Thermoactinomyces Strain TA66-2. Magn. Reson. Chem. 2008, 46, 80–83. [Google Scholar] [CrossRef] [PubMed]
- Roshchina, V.V. New Trends and Perspectives in the Evolution of Neurotransmitters in Microbial, Plant, and Animal Cells. In Microbial Endocrinology: Interkingdom Signaling in Infectious Disease and Health; Lyte, M., Ed.; Advances in Experimental Medicine and Biology Series; Springer International Publishing: Cham, Switzerland, 2016; Volume 874, pp. 25–77. ISBN 978-3-319-20214-3. [Google Scholar]
- Chen, Y.; Zhou, D.; Qi, D.; Gao, Z.; Xie, J.; Luo, Y. Growth Promotion and Disease Suppression Ability of a Streptomyces Sp. CB-75 from Banana Rhizosphere Soil. Front. Microbiol. 2018, 8, 2704. [Google Scholar] [CrossRef]
- Zhai, X.; Wu, G.; Tao, X.; Yang, S.; Lv, L.; Zhu, Y.; Dong, D.; Xiang, H. Success Stories of Natural Product-Derived Compounds from Plants as Multidrug Resistance Modulators in Microorganisms. RSC Adv. 2023, 13, 7798–7817. [Google Scholar] [CrossRef] [PubMed]
- Abdella, B.; Abdella, M.; ElSharif, H.A.; ElAhwany, A.M.D.; El-Sersy, N.A.; Ghozlan, H.A.; Sabry, S.A. Identification of Potent Anti-Candida Metabolites Produced by the Soft Coral Associated Streptomyces Sp. HC14 Using Chemoinformatics. Sci. Rep. 2023, 13, 12564. [Google Scholar] [CrossRef] [PubMed]
- Adegboye, M.F.; Babalola, O.O. Phylogenetic Characterization of Culturable Antibiotic Producing Streptomyces from Rhizospheric Soils. Mol. Biol. 2013, S1, 001. [Google Scholar] [CrossRef]
- Yang, Y.-H.; Fu, X.-L.; Li, L.-Q.; Zeng, Y.; Li, C.-Y.; He, Y.-N.; Zhao, P.-J. Naphthomycins L–N, Ansamycin Antibiotics from Streptomyces Sp. CS. J. Nat. Prod. 2012, 75, 1409–1413. [Google Scholar] [CrossRef]
- Li, J.; Lu, C.; Shen, Y. Novel Polyketides Isolated from Streptomyces Sp. Helv. Chim. Acta 2008, 91, 741–745. [Google Scholar] [CrossRef]
- Lu, C.; Shen, Y. A New Macrolide Antibiotic with Antitumor Activity Produced by Streptomyces Sp. CS, a Commensal Microbe of Maytenus hookeri. J. Antibiot. 2003, 56, 415–418. [Google Scholar] [CrossRef]
- Kingston, D.G.I. Comprehensive Natural Products Chemistry, Volumes 1−9 Edited by Sir Derek Barton (Texas A&M University) and Koji Nakanishi (Columbia University); Executive Editor Otto Meth-Cohn. Elsevier Science, Inc., New York, NY. 1999. 19.5 × 28 Cm. $3744.00. ISBN 0-08-042709-X. J. Nat. Prod. 2000, 63, 286–287. [Google Scholar] [CrossRef]
- Cimmino, A.; Bejarano, A.; Masi, M.; Puopolo, G.; Evidente, A. Isolation of 2,5-Diketopiperazines from Lysobacter capsici AZ78 with Activity against Rhodococcus fascians. Nat. Prod. Res. 2021, 35, 4969–4977. [Google Scholar] [CrossRef] [PubMed]
- Abdelhameed, R.F.A.; Elhady, S.S.; Noor, A.O.; Almasri, D.M.; Bagalagel, A.A.; Maatooq, G.T.; Khedr, A.I.M.; Yamada, K. Production of a New Cyclic Depsipeptide by the Culture Broth of Staphylococcus Sp. Isolated from Corallina officinalis L. Metabolites 2019, 9, 273. [Google Scholar] [CrossRef] [PubMed]
- Rhee, K.-H. Cyclic Dipeptides Exhibit Synergistic, Broad Spectrum Antimicrobial Effects and Have Anti-Mutagenic Properties. Int. J. Antimicrob. Agents 2004, 24, 423–427. [Google Scholar] [CrossRef] [PubMed]
- Fdhila, F.; Vázquez, V.; Sánchez, J.L.; Riguera, R. dd -Diketopiperazines: Antibiotics Active against Vibrio a Nguillarum Isolated from Marine Bacteria Associated with Cultures of Pecten maximus. J. Nat. Prod. 2003, 66, 1299–1301. [Google Scholar] [CrossRef] [PubMed]
- Mardirossian, M.; Sola, R.; Beckert, B.; Collis, D.W.P.; Di Stasi, A.; Armas, F.; Hilpert, K.; Wilson, D.N.; Scocchi, M. Proline-Rich Peptides with Improved Antimicrobial Activity against E. coli, K. pneumoniae, and A. baumannii. ChemMedChem 2019, 14, 2025–2033. [Google Scholar] [CrossRef] [PubMed]
- Qi, S.-H.; Xu, Y.; Gao, J.; Qian, P.-Y.; Zhang, S. Antibacterial and Antilarval Compounds from Marine bacterium Pseudomonas rhizosphaerae. Ann. Microbiol. 2009, 59, 229–233. [Google Scholar] [CrossRef]
- Tangerina, M.M.P.; Furtado, L.C.; Leite, V.M.B.; Bauermeister, A.; Velasco-Alzate, K.; Jimenez, P.C.; Garrido, L.M.; Padilla, G.; Lopes, N.P.; Costa-Lotufo, L.V.; et al. Metabolomic Study of Marine Streptomyces Sp.: Secondary Metabolites and the Production of Potential Anticancer Compounds. PLoS ONE 2020, 15, e0244385. [Google Scholar] [CrossRef]
- Mehetre, G.T.; Vinodh, J.S.; Burkul, B.B.; Desai, D.; Santhakumari, B.; Dharne, M.S.; Dastager, S.G. Bioactivities and Molecular Networking-Based Elucidation of Metabolites of Potent Actinobacterial Strains Isolated from the Unkeshwar Geothermal Springs in India. RSC Adv. 2019, 9, 9850–9859. [Google Scholar] [CrossRef]
- Husain, D.R.; Wardhani, R. Antibacterial Activity of Endosymbiotic Bacterial Compound from Pheretima Sp. Earthworms Inhibit the Growth of Salmonella Typhi and Staphylococcus aureus: In Vitro and in Silico Approach. Iran. J. Microbiol. 2021, 13, 537–543. [Google Scholar] [CrossRef]
- Bhargava, H.N. Effect of Cyclo(Leucyl-Glycine) on [3H]Spiroperidol Binding in the Corpus Striatum and Hypothalamus of Spontaneously Hypertensive Rats. Eur. J. Pharmacol. 1984, 100, 109–112. [Google Scholar] [CrossRef] [PubMed]
- Lee, S.; Tamayo-Castillo, G.; Pang, C.; Clardy, J.; Cao, S.; Kim, K.H. Diketopiperazines from Costa Rican Endolichenic Fungus Colpoma Sp. CR1465A. Bioorg. Med. Chem. Lett. 2016, 26, 2438–2441. [Google Scholar] [CrossRef] [PubMed]
- Cao, J.; Xie, J.; Yu, M.; Xu, T.; Zhang, H.; Chen, L.; Sun, S. The Promoting Mechanism of the Sterile Fermentation Filtrate of Serratia odorifera on Hypsizygus marmoreus by Means of Metabolomics Analysis. Biomolecules 2023, 13, 1804. [Google Scholar] [CrossRef] [PubMed]
- Lee, M.J.; Kim, G.J.; Shin, M.-S.; Moon, J.; Kim, S.; Nam, J.-W.; Kang, K.S.; Choi, H. Chemical Investigation of Diketopiperazines and N-Phenethylacetamide Isolated from Aquimarina Sp. MC085 and Their Effect on TGF-β-Induced Epithelial–Mesenchymal Transition. Appl. Sci. 2021, 11, 8866. [Google Scholar] [CrossRef]
- Noh, S.W.; Seo, R.; Park, J.-K.; Manir, M.M.; Park, K.; Sang, M.K.; Moon, S.-S.; Jung, H.W. Cyclic Dipeptides from Bacillus vallismortis BS07 Require Key Components of Plant Immunity to Induce Disease Resistance in Arabidopsis against Pseudomonas Infection. Plant Pathol. J. 2017, 33, 402–409. [Google Scholar] [CrossRef]
- Liu, X.; Tao, X.; Zou, A.; Yang, S.; Zhang, L.; Mu, B. Effect of Themicrobial Lipopeptide on Tumor Cell Lines: Apoptosis Induced by Disturbing the Fatty Acid Composition of Cell Membrane. Protein Cell 2010, 1, 584–594. [Google Scholar] [CrossRef] [PubMed]
- Lorz, P.M.; Towae, F.K.; Enke, W.; Jäckh, R.; Bhargava, N.; Hillesheim, W. Phthalic Acid and Derivatives. In Ullmann’s Encyclopedia of Industrial Chemistry; Wiley: Hoboken, NJ, USA, 2007; ISBN 978-3-527-30385-4. [Google Scholar]
- Oh, M.J.; Cho, Y.H.; Cha, S.Y.; Lee, E.O.; Kim, J.W.; Kim, S.K.; Park, C.S. Novel Phytoceramides Containing Fatty Acids of Diverse Chain Lengths Are Better than a Single C18-Ceramide N-Stearoyl Phytosphingosine to Improve the Physiological Properties of Human Stratum Corneum. Clin. Cosmet. Investig. Dermatol. 2017, 10, 363–371. [Google Scholar] [CrossRef] [PubMed]
- Santalova, E.A.; Kuzmich, A.S.; Chingizova, E.A.; Menchinskaya, E.S.; Pislyagin, E.A.; Dmitrenok, P.S. Phytoceramides from the Marine Sponge Monanchora Clathrata: Structural Analysis and Cytoprotective Effects. Biomolecules 2023, 13, 677. [Google Scholar] [CrossRef] [PubMed]
- Bellezza, I.; Peirce, M.J.; Minelli, A. Cyclic Peptides in Neurological Disorders: The Case of Cyclo(His-Pro). In Quorum Sensing; Elsevier: Amsterdam, The Netherlands, 2019; pp. 257–286. ISBN 978-0-12-814905-8. [Google Scholar]
- Guan, J. Insulin-Like Growth Factor-1 and Its Derivatives: Potential Pharmaceutical Application for Ischemic Brain Injury. Recent Patents CNS Drug Discov. 2008, 3, 112–127. [Google Scholar] [CrossRef]
- Mitchell, R.E.; Young, H. N-Coronafacoyl-L-Isoleucine and N-Coronafacoyl-L-Alloisoleucine, Potential Biosynthetic Intermediates of the Phytotoxin Coronatine. Phytochemistry 1985, 24, 2716–2717. [Google Scholar] [CrossRef]
- Bignell, D.R.D.; Cheng, Z.; Bown, L. The Coronafacoyl Phytotoxins: Structure, Biosynthesis, Regulation and Biological Activities. Antonie van Leeuwenhoek 2018, 111, 649–666. [Google Scholar] [CrossRef] [PubMed]
- Nishanth Kumar, S.; Nath, V.S.; Pratap Chandran, R.; Nambisan, B. Cyclic Dipeptides from Rhabditid Entomopathogenic Nematode-Associated Bacillus cereus Have Antimicrobial Activities. World J. Microbiol. Biotechnol. 2014, 30, 439–449. [Google Scholar] [CrossRef] [PubMed]
- Lin, W.-X.; Xie, C.-L.; Zhou, M.; Xia, M.-L.; Zhou, T.-T.; Chen, H.-F.; Yang, X.-W.; Yang, Q. Chemical Constituents from the Deep Sea-Derived Streptomyces xiamenensis MCCC 1A01570 and Their Effects on RXRα Transcriptional Regulation. Nat. Prod. Res. 2020, 34, 1461–1464. [Google Scholar] [CrossRef] [PubMed]
- Yuan, L.; Liu, J.; Huang, Y.; Shen, G.; Pang, S.; Wang, C.; Li, Y.; Mu, X. Integrated Toxicity Assessment of DEHP and DBP toward Aquatic Ecosystem Based on Multiple Trophic Model Assays. Env. Sci. Pollut. Res. 2022, 29, 87402–87412. [Google Scholar] [CrossRef] [PubMed]
- Wu, L.; Shi, F.; Zhang, Y.; Xu, X.; Xie, Z.; Hua, S.; Xia, S.; Jiang, J. Maternal Exposure to Dibutyl Phthalate (DBP) Impairs Angiogenesis and AR Signalling Pathway through Suppression of TGFB1I1 in Hypospadias Offspring. Ecotoxicol. Environ. Saf. 2024, 270, 115941. [Google Scholar] [CrossRef] [PubMed]
- Khatiwora, E.; Adsul, V.B.; Kulkarni, M.; Deshpande, N.R.; Kashalkar, R.V. Antibacterial Activity of Dibutyl Phthalate: A Secondary Metabolite Isolated from Ipomoea Carnea Stem. J. Pharm. Res. 2012, 5, 150–152. [Google Scholar]
- T3DB: Pyridoxine. Available online: http://www.t3db.ca/toxins/T3D2701 (accessed on 11 January 2024).
- Patel, K.; Gadewar, M.; Tripathi, R.; Prasad, S.; Patel, D.K. A Review on Medicinal Importance, Pharmacological Activity and Bioanalytical Aspects of Beta-Carboline Alkaloid “Harmine”. Asian Pac. J. Trop. Biomed. 2012, 2, 660–664. [Google Scholar] [CrossRef] [PubMed]
- Cao, R.; Peng, W.; Wang, Z.; Xu, A. β-Carboline Alkaloids: Biochemical and Pharmacological Functions. Curr. Med. Chem. 2007, 14, 479–500. [Google Scholar] [CrossRef] [PubMed]
- Stasiuk, M.; Kozubek, A. Biological Activity of Phenolic Lipids. Cell. Mol. Life Sci. 2010, 67, 841–860. [Google Scholar] [CrossRef]
- Kanda, N.; Ishizaki, N.; Inoue, N.; Oshima, M.; Handa, A.; Kitahara, T. DB-2073, a New Alkylresorcinol Antibiotic. I. Taxonomy, Isolation and Characterization. J. Antibiot. 1975, 28, 935–942. [Google Scholar] [CrossRef][Green Version]
- Da Silva, R.R.; Dorrestein, P.C.; Quinn, R.A. Illuminating the Dark Matter in Metabolomics. Proc. Natl. Acad. Sci. USA 2015, 112, 12549–12550. [Google Scholar] [CrossRef] [PubMed]
- Wang, M.; Carver, J.J.; Phelan, V.V.; Sanchez, L.M.; Garg, N.; Peng, Y.; Nguyen, D.D.; Watrous, J.; Kapono, C.A.; Luzzatto-Knaan, T.; et al. Sharing and Community Curation of Mass Spectrometry Data with Global Natural Products Social Molecular Networking. Nat. Biotechnol. 2016, 34, 828–837. [Google Scholar] [CrossRef] [PubMed]
- Williams, S.T.; Davies, F.L. Use of Antibiotics for Selective Isolation and Enumeration of Actinomycetes in Soil. J. Gen. Microbiol. 1965, 38, 251–261. [Google Scholar] [CrossRef] [PubMed]
- Kharel, M.K.; Shepherd, M.D.; Nybo, S.E.; Smith, M.L.; Bosserman, M.A.; Rohr, J. Isolation of Streptomyces Species from Soil. Curr. Protoc. Microbiol. 2010, 19, 10E.4.1–10E.4.5. [Google Scholar] [CrossRef]
- Green, M.R.; Sambrook, J. Molecular Cloning: A Laboratory Manual, 4th ed.; Cold Spring Harbor Laboratory Press: Cold Spring Harbor, NY, USA, 2012; ISBN 978-1-936113-41-5. [Google Scholar]
- Johnson, M.; Zaretskaya, I.; Raytselis, Y.; Merezhuk, Y.; McGinnis, S.; Madden, T.L. NCBI BLAST: A Better Web Interface. Nucleic Acids Res. 2008, 36, W5–W9. [Google Scholar] [CrossRef] [PubMed]
- Rajan, B.M.; Kannabiran, K. Extraction and Identification of Antibacterial Secondary Metabolites from Marine Streptomyces Sp. VITBRK2. Int. J. Mol. Cell Med. 2014, 3, 130–137. [Google Scholar] [PubMed]
- Valgas, C.; Souza, S.M.; Smânia, E.F.A.; Smânia, A., Jr. Screening Methods to Determine Antibacterial Activity of Natural Products. Braz. J. Microbiol. 2007, 38, 369–380. [Google Scholar] [CrossRef]
- Wiegand, I.; Hilpert, K.; Hancock, R.E.W. Agar and Broth Dilution Methods to Determine the Minimal Inhibitory Concentration (MIC) of Antimicrobial Substances. Nat. Protoc. 2008, 3, 163–175. [Google Scholar] [CrossRef] [PubMed]
- Chater, K.F.; Biró, S.; Lee, K.J.; Palmer, T.; Schrempf, H. The Complex Extracellular Biology of Streptomyces. FEMS Microbiol. Rev. 2010, 34, 171–198. [Google Scholar] [CrossRef]
- Chambers, M.C.; Maclean, B.; Burke, R.; Amodei, D.; Ruderman, D.L.; Neumann, S.; Gatto, L.; Fischer, B.; Pratt, B.; Egertson, J.; et al. A Cross-Platform Toolkit for Mass Spectrometry and Proteomics. Nat. Biotechnol. 2012, 30, 918–920. [Google Scholar] [CrossRef]
- Dührkop, K.; Shen, H.; Meusel, M.; Rousu, J.; Böcker, S. Searching Molecular Structure Databases with Tandem Mass Spectra Using CSI:FingerID. Proc. Natl. Acad. Sci. USA 2015, 112, 12580–12585. [Google Scholar] [CrossRef] [PubMed]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).