Retinoic Acid Signaling in Vertebrate Hindbrain Segmentation: Evolution and Diversification
Abstract
:1. Introduction
2. RA Signaling and Vertebrate Hindbrain Segmentation
2.1. RA Signaling and Its Roles in Development
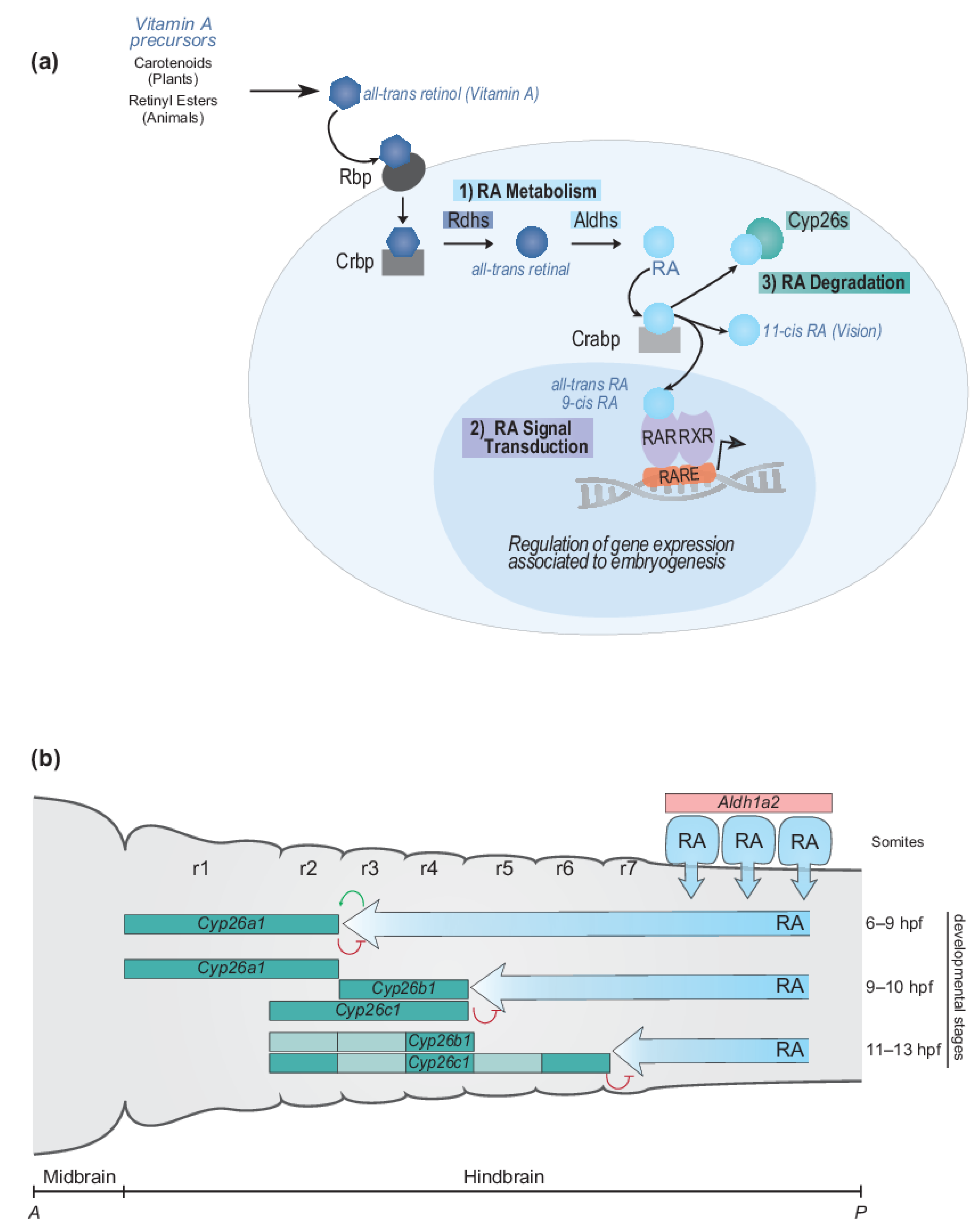
2.1.1. RA Metabolism, Signal Transduction and Degradation
2.1.2. Dynamic Regulation of Endogenous RA Levels during Hindbrain Development
2.1.3. Temporal Dynamics of Cyp26 Gene Expression in the Developing Hindbrain
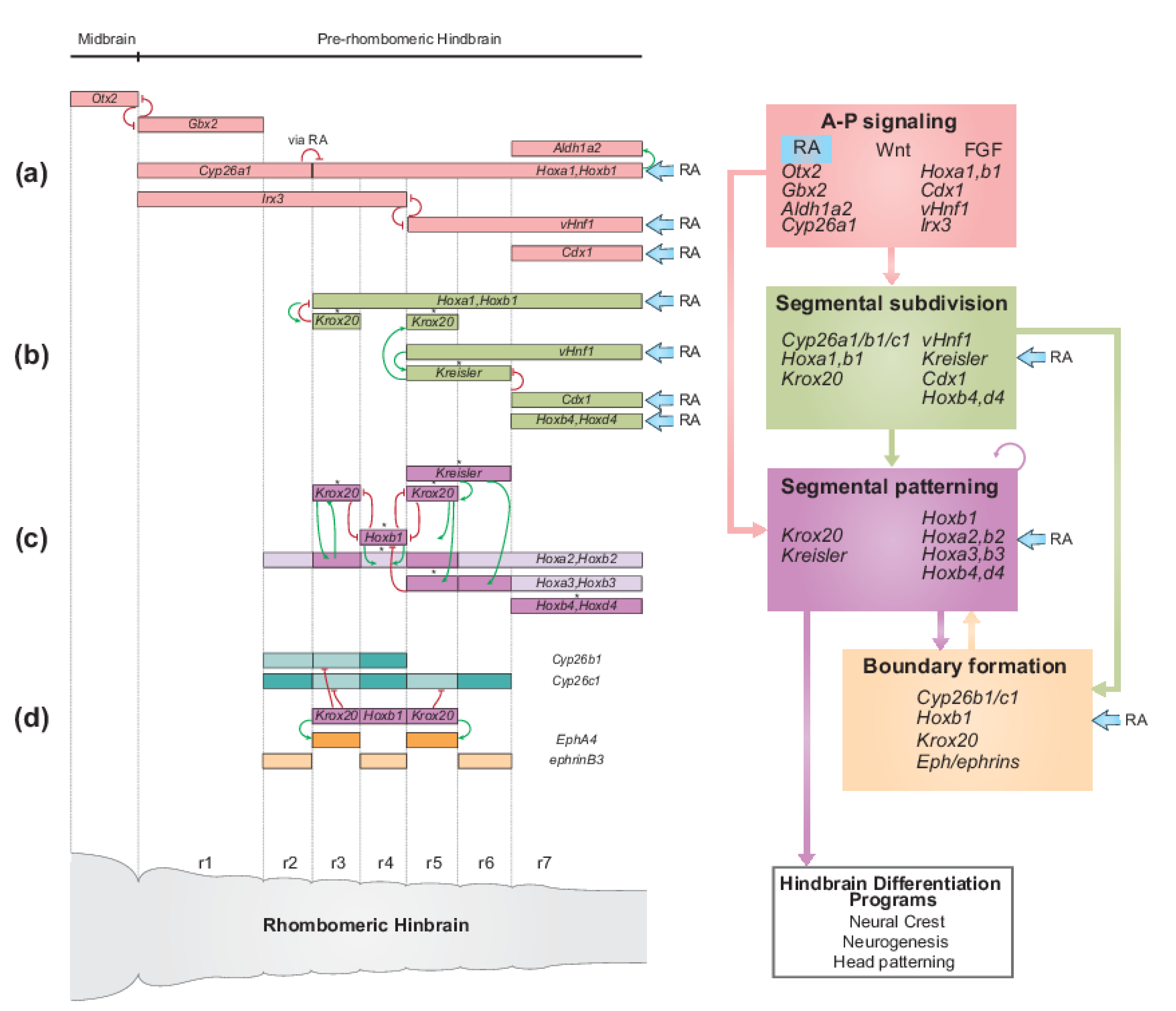
2.2. The Role of RA Signaling in the GRN for Hindbrain Segmentation
2.2.1. RA Plays a Pivotal Role in Multiple Aspects of the Jawed Vertebrate Hindbrain GRN
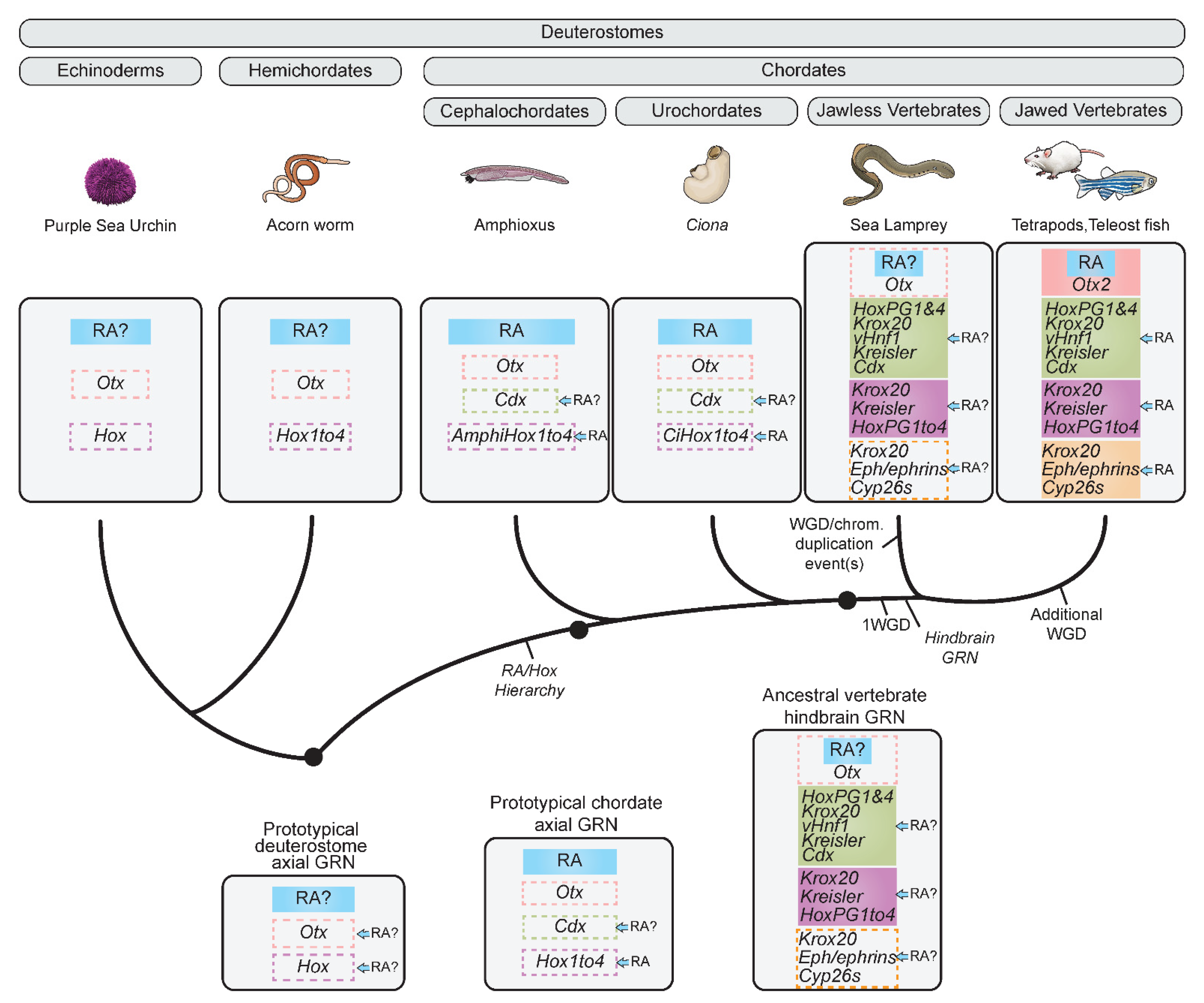
2.2.2. Retracing the Evolution of the Hindbrain GRN Using Jawless Vertebrates
2.3. Origins and Evolution of the Role of RA in the Hindbrain GRN
A Prototypical Axial GRN Integrating RA Can Be Rooted to the Base of Chordates
2.4. Evolution of the RA Signaling Pathway in Chordates
2.4.1. The Metabolic Pathway
2.4.2. RARs and the RA Signal Transduction Pathway
2.4.3. RA Degradation by Cyp26s
2.5. Origins and Evolution of RA Signaling beyond Chordates—Insights from Non-Chordate Deuterostomes
2.5.1. Nervous System Patterning in Hemichordates and Echinoderms
2.5.2. Hemichordates and Echinoderms Models and the Evolution of the RA Machinery
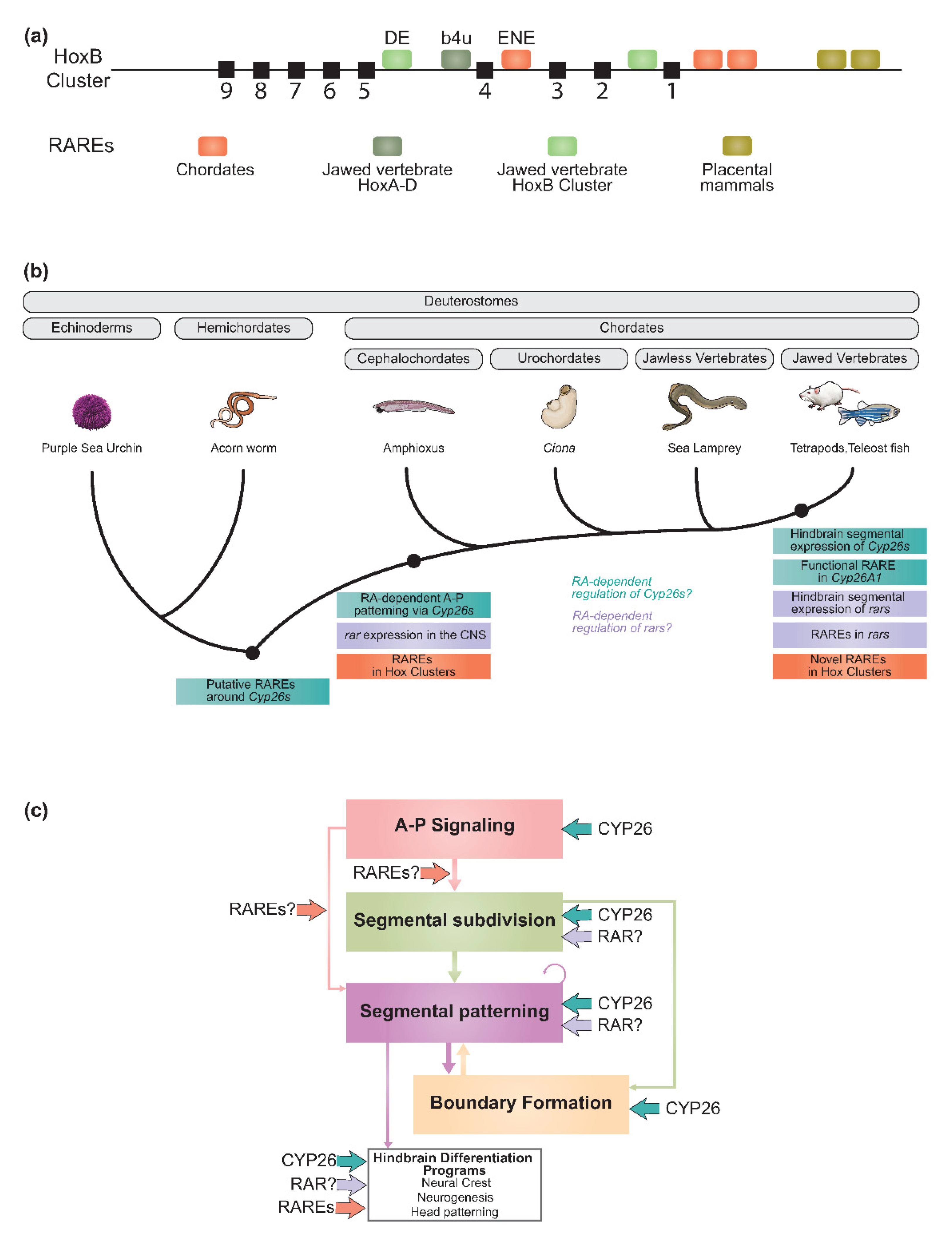
2.6. Regulatory Diversification of the RA Machinery and Evolution of the Vertebrate Hindbrain GRN
2.6.1. Evolving Roles of RAREs
2.6.2. Regulatory Evolution of the Cyp26, Rar, and Crabp Gene Families
Cyp26 Genes
Rar Genes
Crabp Genes
2.7. Lamprey as a Model for Understanding the Origin of the Hindbrain RA/Segmentation Hierarchy in Vertebrates
3. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Carroll, S.B. Homeotic genes and the evolution of arthropods and chordates. Nature 1995, 376, 479–485. [Google Scholar] [CrossRef] [PubMed]
- Martin, A.; Serano, J.M.; Jarvis, E.; Bruce, H.S.; Wang, J.; Ray, S.; Barker, C.A.; O’Connell, L.C.; Patel, N.H. CRISPR/Cas9 Mutagenesis Reveals Versatile Roles of Hox Genes in Crustacean Limb Specification and Evolution. Curr. Biol. 2016, 26, 14–26. [Google Scholar] [CrossRef] [Green Version]
- McGinnis, W.; Krumlauf, R. Homeobox genes and axial patterning. Cell 1992, 68, 283–302. [Google Scholar] [CrossRef]
- Mallo, M.; Wellik, D.M.; Deschamps, J. Hox genes and regional patterning of the vertebrate body plan. Dev. Biol. 2010, 344, 7–15. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Diederich, R.J.; Pattatucci, A.M.; Kaufman, T.C. Developmental and evolutionary implications of labial, Deformed and engrailed expression in the Drosophila head. Development 1991, 113, 273–281. [Google Scholar] [CrossRef] [PubMed]
- Lutz, B.; Lu, H.C.; Eichele, G.; Miller, D.; Kaufman, T.C. Rescue of Drosophila labial null mutant by the chicken ortholog Hoxb-1 demonstrates that the function of Hox genes is phylogenetically conserved. Genes Dev. 1996, 10, 176–184. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Powers, T.P.; Hogan, J.; Ke, Z.; Dymbrowski, K.; Wang, X.; Collins, F.H.; Kaufman, T.C. Characterization of the Hox cluster from the mosquito Anopheles gambiae (Diptera: Culicidae). Evol. Dev. 2000, 2, 311–325. [Google Scholar] [CrossRef]
- Hughes, C.L.; Kaufman, T.C. Exploring the myriapod body plan: Expression patterns of the ten Hox genes in a centipede. Development 2002, 129, 1225–1238. [Google Scholar] [CrossRef]
- Singh, N.P.; De Kumar, B.; Paulson, A.; Parrish, M.E.; Zhang, Y.; Florens, L.; Conaway, J.W.; Si, K.; Krumlauf, R. A six-amino-acid motif is a major determinant in functional evolution of HOX1 proteins. Genes Dev. 2020, 34, 1680–1696. [Google Scholar] [CrossRef]
- Ferrier, D.E.; Holland, P.W. Ancient origin of the Hox gene cluster. Nat. Rev. Genet. 2001, 2, 33–38. [Google Scholar] [CrossRef]
- Duboule, D.; Dolle, P. The structural and functional organization of the murine HOX gene family resembles that of Drosophila homeotic genes. EMBO J. 1989, 8, 1497–1505. [Google Scholar] [CrossRef]
- Graham, A.; Papalopulu, N.; Krumlauf, R. The murine and Drosophila homeobox gene complexes have common features of organization and expression. Cell 1989, 57, 367–378. [Google Scholar] [CrossRef]
- Lewis, E.B. A gene complex controlling segmentation in Drosophila. Nature 1978, 276, 565–570. [Google Scholar] [CrossRef] [PubMed]
- Scott, M.P.; Weiner, A.J. Structural relationships among genes that control development: Sequence homology between the Antennapedia, Ultrabithorax and fushi tarazu loci of Drosophila. Proc. Natl. Acad. Sci. USA 1984, 81, 4115–4119. [Google Scholar] [CrossRef] [Green Version]
- Scott, M.; Weiner, A.; Hazelrigg, T.; Polisky, B.; Pirotta, V.; Scalenghe, F.; Kaufman, M. The molecular organization of the Antennapedia locus of Drosophila. Cell 1983, 35, 763–776. [Google Scholar] [CrossRef]
- McGinnis, W.; Garber, R.L.; Wirz, J.; Kuroiwa, A.; Gehring, W. A homologous protein-coding sequence in Drosophila homeotic genes and its conservation in other metazoans. Cell 1984, 37, 403–408. [Google Scholar] [CrossRef]
- McGinnis, W.; Levine, M.S.; Hafen, E.; Kuroiwa, A.; Gehring, W. A conserved DNA sequence in homeotic genes of the Drosophila Antennapedia and bithorax complexes. Nature 1984, 308, 428–433. [Google Scholar] [CrossRef]
- Harding, K.; Wedeen, C.; McGinnis, W.; Levine, M. Spatially regulated expression of homeotic genes in Drosophila. Science 1985, 229, 1236–1242. [Google Scholar] [CrossRef] [PubMed]
- Dekker, E.J.; Pannese, M.; Houtzager, E.; Boncinelli, E.; Durston, A. Colinearity in the Xenopus laevis Hox-2 complex. Mech. Dev. 1992, 40, 3–12. [Google Scholar] [CrossRef]
- Kmita, M.; Duboule, D. Organizing axes in time and space; 25 years of colinear tinkering. Science 2003, 301, 331–333. [Google Scholar] [CrossRef] [Green Version]
- Deschamps, J.; Duboule, D. Embryonic timing, axial stem cells, chromatin dynamics, and the Hox clock. Genes Dev. 2017, 31, 1406–1416. [Google Scholar] [CrossRef] [Green Version]
- Duboule, D.; Morata, G. Colinearity and functional hierarchy among genes of the homeotic complexes. Trends Genet. 1994, 10, 358–364. [Google Scholar] [CrossRef]
- Lowe, C.J.; Wu, M.; Salic, A.; Evans, L.; Lander, E.; Stange-Thomann, N.; Gruber, C.E.; Gerhart, J.; Kirschner, M. Anteroposterior patterning in hemichordates and the origins of the chordate nervous system. Cell 2003, 113, 853–865. [Google Scholar] [CrossRef] [Green Version]
- Serano, J.M.; Martin, A.; Liubicich, D.M.; Jarvis, E.; Bruce, H.S.; La, K.; Browne, W.E.; Grimwood, J.; Patel, N.H. Comprehensive analysis of Hox gene expression in the amphipod crustacean Parhyale hawaiensis. Dev. Biol. 2016, 409, 297–309. [Google Scholar] [CrossRef] [PubMed]
- Bachiller, D.; Macias, A.; Duboule, D.; Morata, G. Conservation of a functional hierarchy between mammalian and insect Hox/HOM genes. EMBO J. 1994, 13, 1930–1941. [Google Scholar] [CrossRef] [PubMed]
- Gaunt, S.J.; Strachan, L. Temporal colinearity in expression of anterior Hox genes in developing chick embryos. Dev. Dyn. 1996, 207, 270–280. [Google Scholar] [CrossRef]
- Duboule, D. Temporal colinearity and phylotypic progression: A basis for the stability of a vertebrate Bauplan and the evolution of morphologies through heterochrony. Development 1994, 135–142. [Google Scholar] [CrossRef]
- Duboule, D. Vertebrate hox gene regulation: Clustering and/or colinearity? Curr. Opin. Genet. Dev. 1998, 8, 514–518. [Google Scholar] [CrossRef]
- Gaunt, S.J.; Sharpe, P.T.; Duboule, D. Spatially restricted domains of homeo-gene transcripts in mouse embryos: Relation to a segmented body plan. Development 1988, 104, 169–181. [Google Scholar] [CrossRef]
- Krumlauf, R. Hox genes in vertebrate development. Cell 1994, 78, 191–201. [Google Scholar] [CrossRef]
- Lowe, C.J.; Clarke, D.N.; Medeiros, D.M.; Rokhsar, D.S.; Gerhart, J. The deuterostome context of chordate origins. Nature 2015, 520, 456–465. [Google Scholar] [CrossRef]
- Kessel, M.; Gruss, P. Homeotic transformations of murine vertebrae and concomitant alteration of Hox codes induced by retinoic acid. Cell 1991, 67, 89–104. [Google Scholar] [CrossRef] [Green Version]
- Parker, H.J.; Bronner, M.E.; Krumlauf, R. An atlas of anterior hox gene expression in the embryonic sea lamprey head: Hox-code evolution in vertebrates. Dev. Biol. 2019, 453, 19–33. [Google Scholar] [CrossRef] [Green Version]
- Parker, H.J.; Krumlauf, R. Segmental arithmetic: Summing up the Hox gene regulatory network for hindbrain development in chordates. Wiley Interdiscip. Rev. Dev. Biol. 2017, 6, e286. [Google Scholar] [CrossRef]
- Wilkinson, D.G.; Bhatt, S.; Cook, M.; Boncinelli, E.; Krumlauf, R. Segmental expression of Hox-2 homoeobox-containing genes in the developing mouse hindbrain. Nature 1989, 341, 405–409. [Google Scholar] [CrossRef]
- Hunt, P.; Gulisano, M.; Cook, M.; Sham, M.H.; Faiella, A.; Wilkinson, D.; Boncinelli, E.; Krumlauf, R. A distinct Hox code for the branchial region of the vertebrate head. Nature 1991, 353, 861–864. [Google Scholar] [CrossRef]
- Prince, V.E.; Moens, C.B.; Kimmel, C.B.; Ho, R.K. Zebrafish hox genes: Expression in the hindbrain region of wild-type and mutants of the segmentation gene, valentino. Development 1998, 125, 393–406. [Google Scholar] [CrossRef] [PubMed]
- Lumsden, A. Segmentation and compartition in the early avian hindbrain. Mech. Dev. 2004, 121, 1081–1088. [Google Scholar] [CrossRef] [PubMed]
- Murphy, P.; Davidson, D.R.; Hill, R.E. Segment-specific expression of a homeobox-containing gene in the mouse hindbrain. Nature 1989, 341, 156–159. [Google Scholar] [CrossRef]
- Murphy, P.; Hill, R.E. Expression of the mouse labial-like homeobox-containing genes, Hox 2.9 and Hox 1.6, during segmentation of the hindbrain. Development 1991, 111, 61–74. [Google Scholar] [CrossRef] [PubMed]
- Kessel, M.; Balling, R.; Gruss, P. Variations of cervical vertebrae after expression of a Hox-1.1 transgene in mice. Cell 1990, 61, 301–308. [Google Scholar] [CrossRef]
- Alexander, T.; Nolte, C.; Krumlauf, R. Hox genes and segmentation of the hindbrain and axial skeleton. Annu. Rev. Cell Dev. Biol. 2009, 25, 431–456. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Krumlauf, R.; Wilkinson, D.G. Segmentation and patterning of the vertebrate hindbrain. Development 2021, 148, dev186460. [Google Scholar] [CrossRef]
- Balling, R.; Mutter, G.; Gruss, P.; Kessel, M. Craniofacial abnormalities induced by ectopic expression of the homeobox gene Hox-1.1 in transgenic mice. Cell 1989, 58, 337–347. [Google Scholar] [CrossRef] [Green Version]
- Zhang, M.; Kim, H.J.; Marshall, H.; Gendron-Maguire, M.; Lucas, D.A.; Baron, A.; Gudas, L.J.; Gridley, T.; Krumlauf, R.; Grippo, J.F. Ectopic Hoxa-1 induces rhombomere transformation in mouse hindbrain. Development 1994, 120, 2431–2442. [Google Scholar] [CrossRef]
- Alexandre, D.; Clarke, J.D.; Oxtoby, E.; Yan, Y.L.; Jowett, T.; Holder, N. Ectopic expression of Hoxa-1 in the zebrafish alters the fate of the mandibular arch neural crest and phenocopies a retinoic acid-induced phenotype. Development 1996, 122, 735–746. [Google Scholar] [CrossRef]
- Di Bonito, M.; Narita, Y.; Avallone, B.; Sequino, L.; Mancuso, M.; Andolfi, G.; Franze, A.M.; Puelles, L.; Rijli, F.M.; Studer, M. Assembly of the auditory circuitry by a Hox genetic network in the mouse brainstem. PLoS Genet. 2013, 9, e1003249. [Google Scholar] [CrossRef] [Green Version]
- Geisen, M.J.; Di Meglio, T.; Pasqualetti, M.; Ducret, S.; Brunet, J.F.; Chedotal, A.; Rijli, F.M. Hox paralog group 2 genes control the migration of mouse pontine neurons through slit-robo signaling. PLoS Biol. 2008, 6, e142. [Google Scholar] [CrossRef] [Green Version]
- Chatonnet, F.; Wrobel, L.J.; Mezieres, V.; Pasqualetti, M.; Ducret, S.; Taillebourg, E.; Charnay, P.; Rijli, F.M.; Champagnat, J. Distinct roles of Hoxa2 and Krox20 in the development of rhythmic neural networks controlling inspiratory depth, respiratory frequency, and jaw opening. Neural Dev. 2007, 2, 19. [Google Scholar] [CrossRef]
- Oury, F.; Murakami, Y.; Renaud, J.S.; Pasqualetti, M.; Charnay, P.; Ren, S.Y.; Rijli, F.M. Hoxa2- and rhombomere-dependent development of the mouse facial somatosensory map. Science 2006, 313, 1408–1413. [Google Scholar] [CrossRef] [Green Version]
- Studer, M.; Lumsden, A.; Ariza-McNaughton, L.; Bradley, A.; Krumlauf, R. Altered segmental identity and abnormal migration of motor neurons in mice lacking Hoxb-1. Nature 1996, 384, 630–634. [Google Scholar] [CrossRef] [PubMed]
- Gavalas, A.; Ruhrberg, C.; Livet, J.; Henderson, C.E.; Krumlauf, R. Neuronal defects in the hindbrain of Hoxa1, Hoxb1 and Hoxb2 mutants reflect regulatory interactions among these Hox genes. Development 2003, 130, 5663–5679. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gavalas, A.; Studer, M.; Lumsden, A.; Rijli, F.M.; Krumlauf, R.; Chambon, P. Hoxa1 and Hoxb1 synergize in patterning the hindbrain, cranial nerves and second pharyngeal arch. Development 1998, 125, 1123–1136. [Google Scholar] [CrossRef]
- Davenne, M.; Maconochie, M.K.; Neun, R.; Pattyn, A.; Chambon, P.; Krumlauf, R.; Rijli, F.M. Hoxa2 and Hoxb2 control dorsoventral patterns of neuronal development in the rostral hindbrain. Neuron 1999, 22, 677–691. [Google Scholar] [CrossRef] [Green Version]
- Gaufo, G.O.; Wu, S.; Capecchi, M.R. Contribution of Hox genes to the diversity of the hindbrain sensory system. Development 2004, 131, 1259–1266. [Google Scholar] [CrossRef] [Green Version]
- Gaufo, G.O.; Thomas, K.R.; Capecchi, M.R. Hox3 genes coordinate mechanisms of genetic suppression and activation in the generation of branchial and somatic motoneurons. Development 2003, 130, 5191–5201. [Google Scholar] [CrossRef] [Green Version]
- Arenkiel, B.R.; Tvrdik, P.; Gaufo, G.O.; Capecchi, M.R. Hoxb1 functions in both motoneurons and in tissues of the periphery to establish and maintain the proper neuronal circuitry. Genes Dev. 2004, 18, 1539–1552. [Google Scholar] [CrossRef] [Green Version]
- Briscoe, J.; Wilkinson, D.G. Establishing neuronal circuitry: Hox genes make the connection. Genes Dev. 2004, 18, 1643–1648. [Google Scholar] [CrossRef] [Green Version]
- Ramirez-Solis, R.; Zheng, H.; Whiting, J.; Krumlauf, R.; Bradley, A. Hoxb-4 (Hox-2.6) mutant mice show homeotic transformation of a cervical vertebra and defects in the closure of the sternal rudiments. Cell 1993, 73, 279–294. [Google Scholar] [CrossRef]
- Condie, B.G.; Capecchi, M.R. Mice homozygous for a targeted disruption of Hoxd-3(Hox-4.1) exhibit anterior transformations of the first and second cervical vertebrae, the atlas and axis. Development 1993, 119, 579–595. [Google Scholar] [CrossRef]
- Kostic, D.; Capecchi, M.R. Targeted disruptions of the murine Hoxa-4 and Hoxa-6 genes result in homeotic transformations of components of the vertebral column. Mech. Dev. 1994, 46, 231–247. [Google Scholar] [CrossRef]
- Guerreiro, I.; Nunes, A.; Woltering, J.M.; Casaca, A.; Novoa, A.; Vinagre, T.; Hunter, M.E.; Duboule, D.; Mallo, M. Role of a polymorphism in a Hox/Pax-responsive enhancer in the evolution of the vertebrate spine. Proc. Natl. Acad. Sci. USA 2013, 110, 10682–10686. [Google Scholar] [CrossRef] [Green Version]
- Jungbluth, S.; Bell, E.; Lumsden, A. Specification of distinct motor neuron identities by the singular activities of individual Hox genes. Development 1999, 126, 2751–2758. [Google Scholar] [CrossRef]
- Meyer, A.; Schartl, M. Gene and genome duplications in vertebrates: The one-to-four (-to-eight in fish) rule and the evolution of novel gene functions. Curr. Opin. Cell Biol. 1999, 11, 699–704. [Google Scholar] [CrossRef] [Green Version]
- Taylor, J.S.; Braasch, I.; Frickey, T.; Meyer, A.; Van de Peer, Y. Genome duplication, a trait shared by 22,000 species of ray-finned fish. Genome Res. 2003, 13, 382–390. [Google Scholar] [CrossRef] [Green Version]
- Vandepoele, K.; De Vos, W.; Taylor, J.S.; Meyer, A.; Van de Peer, Y. Major events in the genome evolution of vertebrates: Paranome age and size differ considerably between ray-finned fishes and land vertebrates. Proc. Natl. Acad. Sci. USA 2004, 101, 1638–1643. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Siegel, N.; Hoegg, S.; Salzburger, W.; Braasch, I.; Meyer, A. Comparative genomics of ParaHox clusters of teleost fishes: Gene cluster breakup and the retention of gene sets following whole genome duplications. BMC Genom. 2007, 8, 312. [Google Scholar] [CrossRef] [Green Version]
- Kuraku, S.; Meyer, A.; Kuratani, S. Timing of genome duplications relative to the origin of the vertebrates: Did cyclostomes diverge before or after? Mol. Biol. Evol. 2009, 26, 47–59. [Google Scholar] [CrossRef] [Green Version]
- Holland, P.W.; Takahashi, T. The evolution of homeobox genes: Implications for the study of brain development. Brain Res. Bull. 2005, 66, 484–490. [Google Scholar] [CrossRef]
- Holland, P.W. Evolution of homeobox genes. Wiley Interdiscip. Rev. Dev. Biol. 2013, 2, 31–45. [Google Scholar] [CrossRef]
- Smith, J.J.; Keinath, M.C. The sea lamprey meiotic map improves resolution of ancient vertebrate genome duplications. Genome Res. 2015, 25, 1081–1090. [Google Scholar] [CrossRef] [Green Version]
- Smith, J.J.; Timoshevskaya, N.; Ye, C.; Holt, C.; Keinath, M.C.; Parker, H.J.; Cook, M.E.; Hess, J.E.; Narum, S.R.; Lamanna, F.; et al. The sea lamprey germline genome provides insights into programmed genome rearrangement and vertebrate evolution. Nat. Genet. 2018, 50, 270–277. [Google Scholar] [CrossRef]
- Duboule, D. The rise and fall of Hox gene clusters. Development 2007, 134, 2549–2560. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kuraku, S.; Meyer, A. The evolution and maintenance of Hox gene clusters in vertebrates and the teleost-specific genome duplication. Int. J. Dev. Biol. 2009, 53, 765–773. [Google Scholar] [CrossRef] [Green Version]
- Pascual-Anaya, J.; Sato, I.; Sugahara, F.; Higuchi, S.; Paps, J.; Ren, Y.; Takagi, W.; Ruiz-Villalba, A.; Ota, K.G.; Wang, W.; et al. Hagfish and lamprey Hox genes reveal conservation of temporal colinearity in vertebrates. Nat. Ecol. Evol. 2018, 2, 859–866. [Google Scholar] [CrossRef] [PubMed]
- Mehta, T.K.; Ravi, V.; Yamasaki, S.; Lee, A.P.; Lian, M.M.; Tay, B.H.; Tohari, S.; Yanai, S.; Tay, A.; Brenner, S.; et al. Evidence for at least six Hox clusters in the Japanese lamprey (Lethenteron japonicum). Proc. Natl. Acad. Sci. USA 2013, 110, 16044–16049. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Holland, P.W.H.; Garcia-Fernandez, J. Hox genes and chordate evolution. Dev. Biol. 1996, 173, 382–395. [Google Scholar] [CrossRef] [Green Version]
- Shimeld, S.M.; Holland, P.W. Vertebrate innovations. Proc. Natl. Acad. Sci. USA 2000, 97, 4449–4452. [Google Scholar] [CrossRef] [Green Version]
- Duboule, D.; Deschamps, J. Colinearity loops out. Dev. Cell 2004, 6, 738–740. [Google Scholar] [CrossRef] [PubMed]
- Durston, A.J.; Jansen, H.J.; Wacker, S.A. Review: Time-space translation regulates trunk axial patterning in the early vertebrate embryo. Genomics 2010, 95, 250–255. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Narendra, V.; Rocha, P.P.; An, D.; Raviram, R.; Skok, J.A.; Mazzoni, E.O.; Reinberg, D. CTCF establishes discrete functional chromatin domains at the Hox clusters during differentiation. Science 2015, 347, 1017–1021. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Durston, A.J. Vertebrate hox temporal collinearity: Does it exist and what is it’s function? Cell Cycle 2019, 18, 523–530. [Google Scholar] [CrossRef] [Green Version]
- Kmita, M.; van Der Hoeven, F.; Zakany, J.; Krumlauf, R.; Duboule, D. Mechanisms of Hox gene colinearity: Transposition of the anterior Hoxb1 gene into the posterior HoxD complex. Genes Dev. 2000, 14, 198–211. [Google Scholar] [PubMed]
- Ahn, Y.; Mullan, H.E.; Krumlauf, R. Long-range regulation by shared retinoic acid response elements modulates dynamic expression of posterior Hoxb genes in CNS development. Dev. Biol. 2014, 388, 134–144. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nolte, C.; Jinks, T.; Wang, X.; Martinez Pastor, M.T.; Krumlauf, R. Shadow enhancers flanking the HoxB cluster direct dynamic Hox expression in early heart and endoderm development. Dev. Biol. 2013, 383, 158–173. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kondo, T.; Duboule, D. Breaking colinearity in the mouse HoxD complex. Cell 1999, 97, 407–417. [Google Scholar] [CrossRef]
- Darras, S.; Fritzenwanker, J.H.; Uhlinger, K.R.; Farrelly, E.; Pani, A.M.; Hurley, I.A.; Norris, R.P.; Osovitz, M.; Terasaki, M.; Wu, M.; et al. Anteroposterior axis patterning by early canonical Wnt signaling during hemichordate development. PLoS Biol. 2018, 16, e2003698. [Google Scholar] [CrossRef] [Green Version]
- Neijts, R.; Amin, S.; van Rooijen, C.; Tan, S.; Creyghton, M.P.; de Laat, W.; Deschamps, J. Polarized regulatory landscape and Wnt responsiveness underlie Hox activation in embryos. Genes Dev. 2016, 30, 1937–1942. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pownall, M.; Tucker, A.; Slack, J.; Isaacs, H. eFGF, Xcad3 and Hox genes form a molecular pathway that establishes the anteroposterior axis in Xenopus. Development 1996, 122, 3881–3892. [Google Scholar] [CrossRef]
- Pownall, M.E.; Isaacs, H.V.; Slack, J.M. Two phases of Hox gene regulation during early Xenopus development. Curr. Biol. 1998, 8, 673–676. [Google Scholar] [CrossRef] [Green Version]
- Bel-Vialar, S.; Itasaki, N.; Krumlauf, R. Initiating Hox gene expression: In the early chick neural tube differential sensitivity to FGF and RA signaling subdivides the HoxB genes in two distinct groups. Development 2002, 129, 5103–5115. [Google Scholar] [CrossRef]
- In der Rieden, P.M.; Vilaspasa, F.L.; Durston, A.J. Xwnt8 directly initiates expression of labial Hox genes. Dev. Dyn. 2010, 239, 126–139. [Google Scholar] [CrossRef]
- Frank, D.; Sela-Donenfeld, D. Hindbrain induction and patterning during early vertebrate development. Cell Mol. Life Sci. 2019, 76, 941–960. [Google Scholar] [CrossRef]
- Deschamps, J.; van Nes, J. Developmental regulation of the Hox genes during axial morphogenesis in the mouse. Development 2005, 132, 2931–2942. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Young, T.; Rowland, J.E.; van de Ven, C.; Bialecka, M.; Novoa, A.; Carapuco, M.; van Nes, J.; de Graaff, W.; Duluc, I.; Freund, J.N.; et al. Cdx and Hox genes differentially regulate posterior axial growth in mammalian embryos. Dev. Cell 2009, 17, 516–526. [Google Scholar] [CrossRef] [Green Version]
- Diez del Corral, R.; Olivera-Martinez, I.; Goriely, A.; Gale, E.; Maden, M.; Storey, K. Opposing FGF and retinoid pathways control ventral neural pattern, neuronal differentiation, and segmentation during body axis extension. Neuron 2003, 40, 65–79. [Google Scholar] [CrossRef] [Green Version]
- Diez del Corral, R.; Storey, K.G. Opposing FGF and retinoid pathways: A signalling switch that controls differentiation and patterning onset in the extending vertebrate body axis. BioEssays 2004, 26, 857–869. [Google Scholar] [CrossRef] [PubMed]
- Ye, Z.; Kimelman, D. Hox13 genes are required for mesoderm formation and axis elongation during early zebrafish development. Development 2020, 147, dev185298. [Google Scholar] [CrossRef]
- Bel-Vialar, S.; Core, N.; Terranova, R.; Goudot, V.; Boned, A.; Djabali, M. Altered retinoic acid sensitivity and temporal expression of Hox genes in polycomb-M33-deficient mice. Dev. Biol. 2000, 224, 238–249. [Google Scholar] [CrossRef] [Green Version]
- Parker, H.J.; Krumlauf, R. A Hox gene regulatory network for hindbrain segmentation. Curr. Top. Dev. Biol. 2020, 139, 169–203. [Google Scholar] [CrossRef]
- White, R.J.; Schilling, T.F. How degrading: Cyp26s in hindbrain development. Dev. Dyn. 2008, 237, 2775–2790. [Google Scholar] [CrossRef] [Green Version]
- Sosnik, J.; Zheng, L.; Rackauckas, C.V.; Digman, M.; Gratton, E.; Nie, Q.; Schilling, T.F. Noise modulation in retinoic acid signaling sharpens segmental boundaries of gene expression in the embryonic zebrafish hindbrain. Elife 2016, 5, e14034. [Google Scholar] [CrossRef]
- Sirbu, I.O.; Gresh, L.; Barra, J.; Duester, G. Shifting boundaries of retinoic acid activity control hindbrain segmental gene expression. Development 2005, 132, 2611–2622. [Google Scholar] [CrossRef] [Green Version]
- Begemann, G.; Meyer, A. Hindbrain patterning revisited: Timing and effects of retinoic acid signalling. BioEssays 2001, 23, 981–986. [Google Scholar] [CrossRef]
- Durston, A.J.; Timmermans, J.P.; Hage, W.J.; Hendriks, H.F.; de Vries, N.J.; Heideveld, M.; Nieuwkoop, P.D. Retinoic acid causes an anteroposterior transformation in the developing central nervous system. Nature 1989, 340, 140–144. [Google Scholar] [CrossRef] [PubMed]
- Godsave, S.F.; Koster, C.H.; Getahun, A.; Mathu, M.; Hooiveld, M.; van der Wees, J.; Hendriks, J.; Durston, A.J. Graded retinoid responses inthe developing hindbrain. Dev. Dyn. 1998, 213, 39–49. [Google Scholar] [CrossRef]
- van der Wees, J.; Schilthuis, J.G.; Koster, C.H.; Diesveld-Schipper, H.; Folkers, G.E.; van der Saag, P.T.; Dawson, M.I.; Shudo, K.; van der Burg, B.; Durston, A.J. Inhibition of retinoic acid receptor-mediated signalling alters positional identity in the developing hindbrain. Development 1998, 125, 545–556. [Google Scholar] [CrossRef] [PubMed]
- Gale, E.; Prince, V.; Lumsden, A.; Clarke, J.; Holder, N.; Maden, M. Late effects of retinoic acid on neural crest and aspects of rhombomere. Development 1996, 122, 783–793. [Google Scholar] [CrossRef] [PubMed]
- Maden, M.; Hunt, P.; Eriksson, U.; Kuroiwa, A.; Krumlauf, R.; Summerbell, D. Retinoic acid-binding protein, rhombomeres and the neural crest. Development 1991, 111, 35–43. [Google Scholar] [CrossRef]
- Maden, M.; Holder, N. Retinoic acid and development of the central nervous system. Bioessays 1992, 14, 431–438. [Google Scholar] [CrossRef]
- Maden, M.; Gale, E.; Kostetskii, I.; Zile, M. Vitamin A deficient quail embryos have half a hindbrain and other neural defects. Curr. Biol. 1996, 6, 417–426. [Google Scholar] [CrossRef] [Green Version]
- Gale, E.; Zile, M.; Maden, M. Hindbrain respecification in the retinoid-deficient quail. Mech. Dev. 1999, 89, 43–54. [Google Scholar] [CrossRef]
- Maden, M. Retinoid signalling in the development of the central nervous system. Nat. Rev. Neurosci. 2002, 3, 843–853. [Google Scholar] [CrossRef] [PubMed]
- Marshall, H.; Nonchev, S.; Sham, M.H.; Muchamore, I.; Lumsden, A.; Krumlauf, R. Retinoic acid alters hindbrain Hox code and induces transformation of rhombomeres 2/3 into a 4/5 identity. Nature 1992, 360, 737–741. [Google Scholar] [CrossRef] [PubMed]
- Studer, M.; Popperl, H.; Marshall, H.; Kuroiwa, A.; Krumlauf, R. Role of a conserved retinoic acid response element in rhombomere restriction of Hoxb-1. Science 1994, 265, 1728–1732. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Marshall, H.; Morrison, A.; Studer, M.; Popperl, H.; Krumlauf, R. Retinoids and Hox genes. FASEB J. 1996, 10, 969–978. [Google Scholar] [CrossRef] [Green Version]
- Gould, A.; Itasaki, N.; Krumlauf, R. Initiation of rhombomeric Hoxb4 expression requires induction by somites and a retinoid pathway. Neuron 1998, 21, 39–51. [Google Scholar] [CrossRef] [Green Version]
- Sundin, O.; Eichele, G. An early marker of axial pattern in the chick embryo and its respecification by retinoic acid. Development 1992, 114, 841–852. [Google Scholar] [CrossRef]
- Dupe, V.; Lumsden, A. Hindbrain patterning involves graded responses to retinoic acid signalling. Development 2001, 128, 2199–2208. [Google Scholar] [CrossRef]
- Lloret-Vilaspasa, F.; Jansen, H.J.; de Roos, K.; Chandraratna, R.A.; Zile, M.H.; Stern, C.D.; Durston, A.J. Retinoid signalling is required for information transfer from mesoderm to neuroectoderm during gastrulation. Int. J. Dev. Biol. 2010, 54, 599–608. [Google Scholar] [CrossRef] [Green Version]
- Dekker, E.J.; Pannese, M.; Houtzager, E.; Timmermans, A.; Boncinelli, E.; Durston, A. Xenopus Hox-2 genes are expressed sequentially after the onset of gastrulation and are differentially inducible by retinoic acid. Dev. Suppl. 1992, 195–202. [Google Scholar] [CrossRef]
- Durston, A.J. What are the roles of retinoids, other morphogens, and Hox genes in setting up the vertebrate body axis? Genesis 2019, 57, e23296. [Google Scholar] [CrossRef] [PubMed]
- Papalopulu, N.; Clarke, J.D.; Bradley, L.; Wilkinson, D.; Krumlauf, R.; Holder, N. Retinoic acid causes abnormal development and segmental patterning of the anterior hindbrain in Xenopus embryos. Development 1991, 113, 1145–1158. [Google Scholar] [CrossRef] [PubMed]
- Skromne, I.; Thorsen, D.; Hale, M.; Prince, V.E.; Ho, R.K. Repression of the hindbrain developmental program by Cdx factors is required for the specification of the vertebrate spinal cord. Development 2007, 134, 2147–2158. [Google Scholar] [CrossRef] [Green Version]
- Simeone, A.; Acampora, D.; Arcioni, L.; Andrews, P.W.; Boncinelli, E.; Mavilio, F. Sequential activation of HOX2 homeobox genes by retinoic acid in human embryonal carcinoma cells. Nature 1990, 346, 763–766. [Google Scholar] [CrossRef] [PubMed]
- Simeone, A.; Acampora, D.; Nigro, V.; Faiella, A.; D’Esposito, M.; Stornaiuolo, A.; Mavilio, F.; Boncinelli, E. Differential regulation by retinoic acid of the homeobox genes of the four HOX loci in human embryonal carcinoma cells. Mech. Dev. 1991, 33, 215–227. [Google Scholar] [CrossRef]
- Papalopulu, N.; Lovell-Badge, R.; Krumlauf, R. The expression of murine Hox-2 genes is dependent on the differentiation pathway and displays a collinear sensitivity to retinoic acid in F9 cells and Xenopus embryos. Nucleic Acids Res. 1991, 19, 5497–5506. [Google Scholar] [CrossRef] [PubMed]
- De Kumar, B.; Parrish, M.E.; Slaughter, B.D.; Unruh, J.R.; Gogol, M.; Seidel, C.; Paulson, A.; Li, H.; Gaudenz, K.; Peak, A.; et al. Analysis of dynamic changes in retinoid-induced transcription and epigenetic profiles of murine Hox clusters in ES cells. Genome Res. 2015, 25, 1229–1243. [Google Scholar] [CrossRef] [Green Version]
- Mazzoni, E.O.; Mahony, S.; Peljto, M.; Patel, T.; Thornton, S.R.; McCuine, S.; Reeder, C.; Boyer, L.A.; Young, R.A.; Gifford, D.K.; et al. Saltatory remodeling of Hox chromatin in response to rostrocaudal patterning signals. Nat. Neurosci. 2013, 16, 1191–1198. [Google Scholar] [CrossRef] [Green Version]
- Pani, A.M.; Mullarkey, E.E.; Aronowicz, J.; Assimacopoulos, S.; Grove, E.A.; Lowe, C.J. Ancient deuterostome origins of vertebrate brain signalling centres. Nature 2012, 483, 289–294. [Google Scholar] [CrossRef] [Green Version]
- Gerhart, J.; Lowe, C.; Kirschner, M. Hemichordates and the origin of chordates. Curr. Opin. Genet. Dev. 2005, 15, 461–467. [Google Scholar] [CrossRef] [Green Version]
- Kiecker, C.; Lumsden, A. Compartments and their boundaries in vertebrate brain development. Nat. Rev. Neurosci. 2005, 6, 553–564. [Google Scholar] [CrossRef]
- Gilland, E.; Baker, R. Conservation of neuroepithelial and mesodermal segments in the embryonic vertebrate head. Acta Anat. 1993, 148, 110–123. [Google Scholar] [PubMed]
- Gilland, E.; Baker, R. Evolutionary patterns of cranial nerve efferent nuclei in vertebrates. Brain Behav. Evol. 2005, 66, 234–254. [Google Scholar] [CrossRef]
- Pasqualetti, M.; Diaz, C.; Renaud, J.S.; Rijli, F.M.; Glover, J.C. Fate-mapping the mammalian hindbrain: Segmental origins of vestibular projection neurons assessed using rhombomere-specific Hoxa2 enhancer elements in the mouse embryo. J. Neurosci. 2007, 27, 9670–9681. [Google Scholar] [CrossRef] [PubMed]
- Samad, O.A.; Geisen, M.J.; Caronia, G.; Varlet, I.; Zappavigna, V.; Ericson, J.; Goridis, C.; Rijli, F.M. Integration of anteroposterior and dorsoventral regulation of Phox2b transcription in cranial motoneuron progenitors by homeodomain proteins. Development 2004, 131, 4071–4083. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lumsden, A.; Keynes, R. Segmental patterns of neuronal development in the chick hindbrain. Nature 1989, 337, 424–428. [Google Scholar] [CrossRef]
- Minoux, M.; Rijli, F.M. Molecular mechanisms of cranial neural crest cell migration and patterning in craniofacial development. Development 2010, 137, 2605–2621. [Google Scholar] [CrossRef] [Green Version]
- Le Douarin, N. The Neural Crest; Cambridge University Press: Cambridge, UK, 1983. [Google Scholar]
- Le Douarin, N.; Kalcheim, C. The Neural Crest, 2nd ed.; Cambridge University Press: Cambridge, UK; New York, NY, USA, 1999; 445p. [Google Scholar]
- Keynes, R.; Lumsden, A. Segmentation and the origins of regional diversity in the vertebrate central nervous system. Neuron 1990, 4, 1–9. [Google Scholar] [CrossRef]
- Guthrie, S.; Lumsden, A. Formation and regeneration of rhombomere boundaries in the developing chick hindbrain. Development 1991, 112, 221–229. [Google Scholar] [CrossRef]
- Lumsden, A.; Krumlauf, R. Patterning the vertebrate neuraxis. Science 1996, 274, 1109–1115. [Google Scholar] [CrossRef]
- Moens, C.B.; Prince, V.E. Constructing the hindbrain: Insights from the zebrafish. Dev. Dyn. 2002, 224. [Google Scholar] [CrossRef] [PubMed]
- Moens, C.B.; Kimmel, C.B. Hindbrain patterning in the zebrafish embryo. Soc. Neurosci. Abstr. 1995, 21, 118.118. [Google Scholar]
- Moens, C.B.; Cordes, S.P.; Giorgianni, M.W.; Barsh, G.S.; Kimmel, C.B. Equivalence in the genetic control of hindbrain segmentation in fish and mouse. Development 1998, 125, 381–391. [Google Scholar] [CrossRef]
- Waskiewicz, A.J.; Rikhof, H.A.; Moens, C.B. Eliminating zebrafish pbx proteins reveals a hindbrain ground state. Dev. Cell 2002, 3, 723–733. [Google Scholar] [CrossRef] [Green Version]
- Kolm, P.; Apekin, V.; Sive, H. Xenopus hindbrain patterning requires retinoid signaling. Dev. Biol. 1997, 192, 1–16. [Google Scholar] [CrossRef]
- Godsave, S.; Dekker, E.J.; Holling, T.; Pannese, M.; Boncinelli, E.; Durston, A. Expression patterns of Hoxb genes in the Xenopus embryo suggest roles in anteroposterior specification of the hindbrain and in dorsoventral patterning of the mesoderm. Dev. Biol. 1994, 166, 465–476. [Google Scholar] [CrossRef]
- Wilkinson, D.G.; Bhatt, S.; Chavrier, P.; Bravo, R.; Charnay, P. Segment-specific expression of a zinc-finger gene in the developing nervous system of the mouse. Nature 1989, 337, 461–464. [Google Scholar] [CrossRef] [PubMed]
- Xu, Q.; Mellitzer, G.; Robinson, V.; Wilkinson, D.G. In vivo cell sorting in complementary segmental domains mediated by Eph receptors and ephrins. Nature 1999, 399, 267–271. [Google Scholar] [CrossRef]
- Addison, M.; Xu, Q.; Cayuso, J.; Wilkinson, D.G. Cell Identity Switching Regulated by Retinoic Acid Signaling Maintains Homogeneous Segments in the Hindbrain. Dev. Cell 2018, 45, 606–620.e3. [Google Scholar] [CrossRef] [Green Version]
- Morrison, A.; Chaudhuri, C.; Ariza-McNaughton, L.; Muchamore, I.; Kuroiwa, A.; Krumlauf, R. Comparative analysis of chicken Hoxb-4 regulation in transgenic mice. Mech. Dev. 1995, 53, 47–59. [Google Scholar] [CrossRef]
- Gould, A.; Morrison, A.; Sproat, G.; White, R.A.; Krumlauf, R. Positive cross-regulation and enhancer sharing: Two mechanisms for specifying overlapping Hox expression patterns. Genes Dev. 1997, 11, 900–913. [Google Scholar] [CrossRef] [Green Version]
- Manzanares, M.; Nardelli, J.; Gilardi-Hebenstreit, P.; Marshall, H.; Giudicelli, F.; Martinez-Pastor, M.T.; Krumlauf, R.; Charnay, P. Krox20 and kreisler co-operate in the transcriptional control of segmental expression of Hoxb3 in the developing hindbrain. EMBO J. 2002, 21, 365–376. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Manzanares, M.; Bel-Vialer, S.; Ariza-McNaughton, L.; Ferretti, E.; Marshall, H.; Maconochie, M.K.; Blasi, F.; Krumlauf, R. Independent regulation of initiation and maintenance phases of Hoxa3 expression in the vertebrate hindbrain involves auto and cross-regulatory mechanisms. Development 2001, 128, 3595–3607. [Google Scholar] [CrossRef] [PubMed]
- Nonchev, S.; Maconochie, M.; Vesque, C.; Aparicio, S.; Ariza-McNaughton, L.; Manzanares, M.; Maruthainar, K.; Kuroiwa, A.; Brenner, S.; Charnay, P.; et al. The conserved role of Krox-20 in directing Hox gene expression during vertebrate hindbrain segmentation. Proc. Natl. Acad. Sci. USA 1996, 93, 9339–9345. [Google Scholar] [CrossRef] [Green Version]
- Nonchev, S.; Vesque, C.; Maconochie, M.; Seitanidou, T.; Ariza-McNaughton, L.; Frain, M.; Marshall, H.; Sham, M.H.; Krumlauf, R.; Charnay, P. Segmental expression of Hoxa-2 in the hindbrain is directly regulated by Krox-20. Development 1996, 122, 543–554. [Google Scholar] [CrossRef]
- Vesque, C.; Maconochie, M.; Nonchev, S.; Ariza-McNaughton, L.; Kuroiwa, A.; Charnay, P.; Krumlauf, R. Hoxb-2 transcriptional activation in rhombomeres 3 and 5 requires an evolutionarily conserved cis-acting element in addition to the Krox-20 binding site. EMBO J. 1996, 15, 5383–5396. [Google Scholar] [CrossRef]
- Sham, M.-H.; Hunt, P.; Nonchev, S.; Papalopulu, N.; Graham, A.; Boncinelli, E.; Krumlauf, R. Analysis of the murine Hox-2.7 gene: Conserved alternative transcripts with differential distributions in the nervous system and the potential for shared regulatory regions. EMBO J. 1992, 11, 1825–1836. [Google Scholar] [CrossRef]
- Morrison, A.; Moroni, M.C.; Ariza-McNaughton, L.; Krumlauf, R.; Mavilio, F. In vitro and transgenic analysis of a human HOXD4 retinoid-responsive enhancer. Development 1996, 122, 1895–1907. [Google Scholar] [CrossRef]
- Popperl, H.; Bienz, M.; Studer, M.; Chan, S.K.; Aparicio, S.; Brenner, S.; Mann, R.S.; Krumlauf, R. Segmental expression of Hoxb-1 is controlled by a highly conserved autoregulatory loop dependent upon exd/pbx. Cell 1995, 81, 1031–1042. [Google Scholar] [CrossRef] [Green Version]
- Marshall, H.; Studer, M.; Popperl, H.; Aparicio, S.; Kuroiwa, A.; Brenner, S.; Krumlauf, R. A conserved retinoic acid response element required for early expression of the homeobox gene Hoxb-1. Nature 1994, 370, 567–571. [Google Scholar] [CrossRef]
- Parker, H.J.; Bronner, M.E.; Krumlauf, R. A Hox regulatory network of hindbrain segmentation is conserved to the base of vertebrates. Nature 2014, 514, 490–493. [Google Scholar] [CrossRef] [Green Version]
- McNulty, C.L.; Peres, J.N.; Bardine, N.; van den Akker, W.M.; Durston, A.J. Knockdown of the complete Hox paralogous group 1 leads to dramatic hindbrain and neural crest defects. Development 2005, 132, 2861–2871. [Google Scholar] [CrossRef] [Green Version]
- Tvrdik, P.; Capecchi, M.R. Reversal of hox1 gene subfunctionalization in the mouse. Dev. Cell 2006, 11, 239–250. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rijli, F.; Gavalas, A.; Chambon, P. Segmentation and specification in the branchial region of the head: The role of Hox selector genes. Int. J. Dev. Biol. 1998, 42, 393–401. [Google Scholar] [PubMed]
- Dupe, V.; Davenne, M.; Brocard, J.; Dolle, P.; Mark, M.; Dierich, A.; Chambon, P.; Rijli, F.M. In vivo functional analysis of the Hoxa-1 3’ retinoic acid response element (3′RARE). Development 1997, 124, 399–410. [Google Scholar] [CrossRef] [PubMed]
- Rijli, F.M.; Mark, M.; Lakkaraju, S.; Dierich, A.; Dollé, P.; Chambon, P. A homeotic transformation is generated in the rostral branchial region of the head by disruption of Hoxa-2, which acts as a selector gene. Cell 1993, 75, 1333–1349. [Google Scholar] [CrossRef]
- Parker, H.J.; Pushel, I.; Krumlauf, R. Coupling the roles of Hox genes to regulatory networks patterning cranial neural crest. Dev. Biol. 2018, 444 (Suppl. 1), S67–S78. [Google Scholar] [CrossRef] [PubMed]
- Baltzinger, M.; Ori, M.; Pasqualetti, M.; Nardi, I.; Rijli, F.M. Hoxa2 knockdown in Xenopus results in hyoid to mandibular homeosis. Dev. Dyn. 2005, 234, 858–867. [Google Scholar] [CrossRef] [PubMed]
- Santagati, F.; Minoux, M.; Ren, S.Y.; Rijli, F.M. Temporal requirement of Hoxa2 in cranial neural crest skeletal morphogenesis. Development 2005, 132, 4927–4936. [Google Scholar] [CrossRef] [Green Version]
- Kulesa, P.M.; Fraser, S.E. In ovo time-lapse analysis of chick hindbrain neural crest cell migration shows cell interactions during migration to the branchial arches. Development 2000, 127, 1161–1172. [Google Scholar] [CrossRef] [PubMed]
- Trainor, P.A.; Sobieszczuk, D.; Wilkinson, D.; Krumlauf, R. Signalling between the hindbrain and paraxial tissues dictates neural crest migration pathways. Development 2002, 129, 433–442. [Google Scholar] [CrossRef] [PubMed]
- Graham, A.; Heyman, I.; Lumsden, A. Even-numbered rhombomeres control the apoptotic elimination of neural crest cells from odd-numbered rhombomeres in the chick hindbrain. Development 1993, 119, 233–245. [Google Scholar] [CrossRef]
- Sechrist, J.; Serbedzija, G.N.; Scherson, T.; Fraser, S.E.; Bronner-Fraser, M. Segmental migration of the hindbrain neural crest does not arise from its segmental generation. Development 1993, 118, 691–703. [Google Scholar] [CrossRef] [PubMed]
- Maconochie, M.; Nonchev, S.; Morrison, A.; Krumlauf, R. Paralogous Hox genes: Function and regulation. Annu. Rev. Genet. 1996, 30, 529–556. [Google Scholar] [CrossRef]
- Schilling, T.F.; Nie, Q.; Lander, A.D. Dynamics and precision in retinoic acid morphogen gradients. Curr. Opin. Genet. Dev. 2012, 22, 562–569. [Google Scholar] [CrossRef] [Green Version]
- Parker, H.J.; Bronner, M.E.; Krumlauf, R. The vertebrate Hox gene regulatory network for hindbrain segmentation: Evolution and diversification: Coupling of a Hox gene regulatory network to hindbrain segmentation is an ancient trait originating at the base of vertebrates. BioEssays 2016, 38, 526–538. [Google Scholar] [CrossRef]
- Sauka-Spengler, T.; Meulemans, D.; Jones, M.; Bronner-Fraser, M. Ancient evolutionary origin of the neural crest gene regulatory network. Dev. Cell 2007, 13, 405–420. [Google Scholar] [CrossRef] [Green Version]
- Parker, H.J.; De Kumar, B.; Green, S.A.; Prummel, K.D.; Hess, C.; Kaufman, C.K.; Mosimann, C.; Wiedemann, L.M.; Bronner, M.E.; Krumlauf, R. A Hox-TALE regulatory circuit for neural crest patterning is conserved across vertebrates. Nat. Commun. 2019, 10, 1189. [Google Scholar] [CrossRef] [Green Version]
- Buckingham, M.; Meilhac, S.; Zaffran, S. Building the mammalian heart from two sources of myocardial cells. Nat. Rev. Genet. 2005, 6, 826–835. [Google Scholar] [CrossRef]
- Vincent, S.D.; Buckingham, M.E. How to make a heart: The origin and regulation of cardiac progenitor cells. Curr. Top. Dev. Biol. 2010, 90, 1–41. [Google Scholar] [CrossRef]
- Uehara, M.; Yashiro, K.; Takaoka, K.; Yamamoto, M.; Hamada, H. Removal of maternal retinoic acid by embryonic CYP26 is required for correct Nodal expression during early embryonic patterning. Genes Dev. 2009, 23, 1689–1698. [Google Scholar] [CrossRef] [Green Version]
- Kudoh, T.; Wilson, S.W.; Dawid, I.B. Distinct roles for Fgf, Wnt and retinoic acid in posteriorizing the neural ectoderm. Development 2002, 129, 4335–4346. [Google Scholar] [CrossRef] [PubMed]
- Kam, R.K.; Deng, Y.; Chen, Y.; Zhao, H. Retinoic acid synthesis and functions in early embryonic development. Cell Biosci. 2012, 2, 11. [Google Scholar] [CrossRef] [Green Version]
- Rhinn, M.; Dolle, P. Retinoic acid signalling during development. Development 2012, 139, 843–858. [Google Scholar] [CrossRef] [Green Version]
- Niederreither, K.; Dolle, P. Retinoic acid in development: Towards an integrated view. Nat. Rev. Genet. 2008, 9, 541–553. [Google Scholar] [CrossRef] [PubMed]
- Evans, R.M.; Mangelsdorf, D.J. Nuclear Receptors, RXR, and the Big Bang. Cell 2014, 157, 255–266. [Google Scholar] [CrossRef] [Green Version]
- Mangelsdorf, D.J.; Thummel, C.; Beato, M.; Herrlich, P.; Schutz, G.; Umesono, K.; Blumberg, B.; Kastner, P.; Mark, M.; Chambon, P.; et al. The nuclear receptor superfamily: The second decade. Cell 1995, 83, 835–839. [Google Scholar] [CrossRef] [Green Version]
- Xu, Q.; Jitkaew, S.; Choksi, S.; Kadigamuwa, C.; Qu, J.; Choe, M.; Jang, J.; Liu, C.; Liu, Z.G. The cytoplasmic nuclear receptor RARgamma controls RIP1 initiated cell death when cIAP activity is inhibited. Nat. Commun. 2017, 8, 425. [Google Scholar] [CrossRef] [Green Version]
- Spiegler, E.; Kim, Y.K.; Wassef, L.; Shete, V.; Quadro, L. Maternal-fetal transfer and metabolism of vitamin A and its precursor beta-carotene in the developing tissues. Biochim. Biophys. Acta 2012, 1821, 88–98. [Google Scholar] [CrossRef] [Green Version]
- Lampert, J.M.; Holzschuh, J.; Hessel, S.; Driever, W.; Vogt, K.; von Lintig, J. Provitamin A conversion to retinal via the beta,beta-carotene-15,15′-oxygenase (bcox) is essential for pattern formation and differentiation during zebrafish embryogenesis. Development 2003, 130, 2173–2186. [Google Scholar] [CrossRef] [Green Version]
- Mahony, S.; Mazzoni, E.O.; McCuine, S.; Young, R.A.; Wichterle, H.; Gifford, D.K. Ligand-dependent dynamics of retinoic acid receptor binding during early neurogenesis. Genome Biol. 2011, 12, R2. [Google Scholar] [CrossRef] [Green Version]
- Zechel, C. Synthetic retinoids dissociate coactivator binding from corepressor release. J. Recept. Signal. Transduct. Res. 2002, 22, 31–61. [Google Scholar] [CrossRef]
- Chen, J.D.; Evans, R.M. A transcriptional co-repressor that interacts with nuclear hormone receptors. Nature 1995, 377, 454–457. [Google Scholar] [CrossRef]
- Hernandez, R.E.; Putzke, A.P.; Myers, J.P.; Margaretha, L.; Moens, C.B. Cyp26 enzymes generate the retinoic acid response pattern necessary for hindbrain development. Development 2007, 134, 177–187. [Google Scholar] [CrossRef] [Green Version]
- Sakai, Y.; Meno, C.; Fujii, H.; Nishino, J.; Shiratori, H.; Saijoh, Y.; Rossant, J.; Hamada, H. The retinoic acid-inactivating enzyme CYP26 is essential for establishing an uneven distribution of retinoic acid along the anterio-posterior axis within the mouse embryo. Genes Dev. 2001, 15, 213–225. [Google Scholar] [CrossRef] [Green Version]
- Zhao, D.; McCaffery, P.; Ivins, K.J.; Neve, R.L.; Hogan, P.; Chin, W.W.; Drager, U.C. Molecular identification of a major retinoic-acid-synthesizing enzyme, a retinaldehyde-specific dehydrogenase. Eur. J. Biochem. 1996, 240, 15–22. [Google Scholar] [CrossRef]
- Niederreither, K.; McCaffery, P.; Drager, U.C.; Chambon, P.; Dolle, P. Restricted expression and retinoic acid-induced downregulation of the retinaldehyde dehydrogenase type 2 (RALDH-2) gene during mouse development. Mech. Dev. 1997, 62, 67–78. [Google Scholar] [CrossRef]
- Begemann, G.; Schilling, T.F.; Rauch, G.J.; Geisler, R.; Ingham, P.W. The zebrafish neckless mutation reveals a requirement for raldh2 in mesodermal signals that pattern the hindbrain. Development 2001, 128, 3081–3094. [Google Scholar] [CrossRef]
- Berggren, K.; McCaffery, P.; Drager, U.; Forehand, C.J. Differential distribution of retinoic acid synthesis in the chicken embryo as determined by immunolocalization of the retinoic acid synthetic enzyme, RALDH-2. Dev. Biol. 1999, 210, 288–304. [Google Scholar] [CrossRef] [Green Version]
- Lin, C.; Garrett, A.S.; De Kumar, B.; Smith, E.R.; Gogol, M.; Seidel, C.; Krumlauf, R.; Shilatifard, A. Dynamic transcriptional events in embryonic stem cells mediated by the super elongation complex (SEC). Genes Dev. 2011, 25, 1486–1498. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chan, S.K.; Ryoo, H.D.; Gould, A.; Krumlauf, R.; Mann, R.S. Switching the in vivo specificity of a minimal HOX-responsive element. Development 1997, 124, 2007–2014. [Google Scholar] [CrossRef]
- Shimozono, S.; Iimura, T.; Kitaguchi, T.; Higashijima, S.; Miyawaki, A. Visualization of an endogenous retinoic acid gradient across embryonic development. Nature 2013, 496, 363–366. [Google Scholar] [CrossRef]
- Schilling, T.F.; Sosnik, J.; Nie, Q. Visualizing retinoic acid morphogen gradients. Methods Cell Biol. 2016, 133, 139–163. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- White, R.J.; Nie, Q.; Lander, A.D.; Schilling, T.F. Complex regulation of cyp26a1 creates a robust retinoic acid gradient in the zebrafish embryo. PLoS Biol. 2007, 5, e304. [Google Scholar] [CrossRef]
- Rhinn, M.; Lun, K.; Luz, M.; Werner, M.; Brand, M. Positioning of the midbrain-hindbrain boundary organizer through global posteriorization of the neuroectoderm mediated by Wnt8 signaling. Development 2005, 132, 1261–1272. [Google Scholar] [CrossRef] [Green Version]
- Kiecker, C.; Lumsden, A. The role of organizers in patterning the nervous system. Annu. Rev. Neurosci. 2012, 35, 347–367. [Google Scholar] [CrossRef] [Green Version]
- Joyner, A.; Herrup, K.; Auerbach, A.; Davis, C.; Rossant, J. Subtle cerebellar phenotype in mice homozygous for a targeted deletion of the En-2 homeobox. Science 1991, 251, 1239–1243. [Google Scholar] [CrossRef]
- McMahon, A.P.; Joyner, A.L.; Bradley, A.; McMahon, J.A. The midbrain-hindbrain phenotype of Wnt-1-/Wnt-1- mice results from stepwise deletion of engrailed-expressing cells by 9.5 days postcoitum. Cell 1992, 69, 581–595. [Google Scholar] [CrossRef]
- Millen, K.J.; Wurst, W.; Herrup, K.; Joyner, A.L. Abnormal embryonic cerebellar development and patterning of postnatal foliation in two mouse Engrailed-2 mutants. Development 1993, 120, 695–706. [Google Scholar] [CrossRef]
- Joyner, A.L. Engrailed, Wnt and Pax genes regulate midbrain--hindbrain development. Trends Genet. 1996, 12, 15–20. [Google Scholar] [CrossRef]
- Wassarman, K.M.; Lewandoski, M.; Campbell, K.; Joyner, A.L.; Rubenstein, J.L.; Martinez, S.; Martin, G.R. Specification of the anterior hindbrain and establishment of a normal mid/hindbrain organizer is dependent on Gbx2 gene function. Development 1997, 124, 2923–2934. [Google Scholar] [CrossRef]
- Millet, S.; Campbell, K.; Epstein, D.J.; Losos, K.; Harris, E.; Joyner, A.L. A role for Gbx2 in repression of Otx2 and positioning the mid/hindbrain organizer. Nature 1999, 401, 161–164. [Google Scholar] [CrossRef]
- Lecaudey, V.; Anselme, I.; Rosa, F.; Schneider-Maunoury, S. The zebrafish Iroquois gene iro7 positions the r4/r5 boundary and controls neurogenesis in the rostral hindbrain. Development 2004, 131, 3121–3131. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Giudicelli, F.; Taillebourg, E.; Charnay, P.; Gilardi-Hebenstreit, P. Krox-20 patterns the hindbrain through both cell-autonomous and non cell-autonomous mechanisms. Genes Dev. 2001, 15, 567–580. [Google Scholar] [CrossRef] [Green Version]
- Labalette, C.; Wassef, M.A.; Desmarquet-Trin Dinh, C.; Bouchoucha, Y.X.; Le Men, J.; Charnay, P.; Gilardi-Hebenstreit, P. Molecular dissection of segment formation in the developing hindbrain. Development 2015, 142, 185–195. [Google Scholar] [CrossRef] [Green Version]
- Vitobello, A.; Ferretti, E.; Lampe, X.; Vilain, N.; Ducret, S.; Ori, M.; Spetz, J.F.; Selleri, L.; Rijli, F.M. Hox and Pbx factors control retinoic acid synthesis during hindbrain segmentation. Dev. Cell 2011, 20, 469–482. [Google Scholar] [CrossRef] [Green Version]
- Serpente, P.; Tumpel, S.; Ghyselinck, N.B.; Niederreither, K.; Wiedemann, L.M.; Dolle, P.; Chambon, P.; Krumlauf, R.; Gould, A.P. Direct crossregulation between retinoic acid receptor {beta} and Hox genes during hindbrain segmentation. Development 2005, 132, 503–513. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Torbey, P.; Thierion, E.; Collombet, S.; de Cian, A.; Desmarquet-Trin-Dinh, C.; Dura, M.; Concordet, J.P.; Charnay, P.; Gilardi-Hebenstreit, P. Cooperation, cis-interactions, versatility and evolutionary plasticity of multiple cis-acting elements underlie krox20 hindbrain regulation. PLoS Genet. 2018, 14, e1007581. [Google Scholar] [CrossRef] [PubMed]
- Tümpel, S.; Cambronero, F.; Ferretti, E.; Blasi, F.; Wiedemann, L.M.; Krumlauf, R. Expression of Hoxa2 in rhombomere 4 is regulated by a conserved cross-regulatory mechanism dependent upon Hoxb1. Dev. Biol. 2007, 302, 646–660. [Google Scholar] [CrossRef] [Green Version]
- Pouilhe, M.; Gilardi-Hebenstreit, P.; Desmarquet-Trin Dinh, C.; Charnay, P. Direct regulation of vHnf1 by retinoic acid signaling and MAF-related factors in the neural tube. Dev. Biol. 2007, 309, 344–357. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shimeld, S.M.; Donoghue, P.C. Evolutionary crossroads in developmental biology: Cyclostomes (lamprey and hagfish). Development 2012, 139, 2091–2099. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Miyashita, T.; Gess, R.W.; Tietjen, K.; Coates, M.I. Non-ammocoete larvae of Palaeozoic stem lampreys. Nature 2021, 591, 408–412. [Google Scholar] [CrossRef]
- McCauley, D.W.; Bronner-Fraser, M. Neural crest contributions to the lamprey head. Development 2003, 130, 2317–2327. [Google Scholar] [CrossRef] [Green Version]
- Green, S.A.; Simoes-Costa, M.; Bronner, M.E. Evolution of vertebrates as viewed from the crest. Nature 2015, 520, 474–482. [Google Scholar] [CrossRef] [Green Version]
- Square, T.; Jandzik, D.; Romasek, M.; Cerny, R.; Medeiros, D.M. The origin and diversification of the developmental mechanisms that pattern the vertebrate head skeleton. Dev. Biol. 2017, 427, 219–229. [Google Scholar] [CrossRef]
- Martik, M.L.; Gandhi, S.; Uy, B.R.; Gillis, J.A.; Green, S.A.; Simoes-Costa, M.; Bronner, M.E. Evolution of the new head by gradual acquisition of neural crest regulatory circuits. Nature 2019, 574, 675–678. [Google Scholar] [CrossRef] [Green Version]
- Smith, J.J.; Kuraku, S.; Holt, C.; Sauka-Spengler, T.; Jiang, N.; Campbell, M.S.; Yandell, M.D.; Manousaki, T.; Meyer, A.; Bloom, O.E.; et al. Sequencing of the sea lamprey (Petromyzon marinus) genome provides insights into vertebrate evolution. Nat. Genet. 2013, 45, 415–421. [Google Scholar] [CrossRef]
- Murakami, Y.; Pasqualetti, M.; Takio, Y.; Hirano, S.; Rijli, F.M.; Kuratani, S. Segmental development of reticulospinal and branchiomotor neurons in lamprey: Insights into the evolution of the vertebrate hindbrain. Development 2004, 131, 983–995. [Google Scholar] [CrossRef] [Green Version]
- Kuratani, S.; Horigome, N.; Ueki, T.; Aizawa, S.; Hirano, S. Stereotyped axonal bundle formation and neuromeric patterns in embryos of a cyclostome, Lampetra japonica. J. Comp. Neurol. 1998, 391, 99–114. [Google Scholar] [CrossRef]
- Murakami, Y.; Uchida, K.; Rijli, F.M.; Kuratani, S. Evolution of the brain developmental plan: Insights from agnathans. Dev. Biol. 2005, 280, 249–259. [Google Scholar] [CrossRef]
- Kuratani, S.; Horigome, N.; Hirano, S. Developmental morphology of the head mesoderm and reevaluation of segmental theories of the vertebrate head: Evidence from embryos of an agnathan vertebrate, Lampetra japonica. Dev. Biol. 1999, 210, 381–400. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kuratani, S.; Ueki, T.; Hirano, S.; Aizawa, S. Rostral truncation of a cyclostome, Lampetra japonica, induced by all-trans retinoic acid defines the head/trunk interface of the vertebrate body. Dev. Dyn. 1998, 211, 35–51. [Google Scholar] [CrossRef]
- Jimenez-Guri, E.; Pujades, C. An ancient mechanism of hindbrain patterning has been conserved in vertebrate evolution. Evol. Dev. 2011, 13, 38–46. [Google Scholar] [CrossRef]
- Arcas, A.; Wilkinson, D.G.; Nieto, M.A. The Evolutionary History of Ephs and Ephrins: Toward Multicellular Organisms. Mol. Biol. Evol. 2020, 37, 379–394. [Google Scholar] [CrossRef]
- Takio, Y.; Kuraku, S.; Murakami, Y.; Pasqualetti, M.; Rijli, F.M.; Narita, Y.; Kuratani, S.; Kusakabe, R. Hox gene expression patterns in Lethenteron japonicum embryos--insights into the evolution of the vertebrate Hox code. Dev. Biol. 2007, 308, 606–620. [Google Scholar] [CrossRef]
- Handberg-Thorsager, M.; Gutierrez-Mazariegos, J.; Arold, S.T.; Kumar Nadendla, E.; Bertucci, P.Y.; Germain, P.; Tomancak, P.; Pierzchalski, K.; Jones, J.W.; Albalat, R.; et al. The ancestral retinoic acid receptor was a low-affinity sensor triggering neuronal differentiation. Sci. Adv. 2018, 4, eaao1261. [Google Scholar] [CrossRef] [Green Version]
- Bertrand, S.; Escriva, H. Evolutionary crossroads in developmental biology: Amphioxus. Development 2011, 138, 4819–4830. [Google Scholar] [CrossRef] [Green Version]
- Lemaire, P. Evolutionary crossroads in developmental biology: The tunicates. Development 2011, 138, 2143–2152. [Google Scholar] [CrossRef] [Green Version]
- Schilling, T.F.; Knight, R.D. Origins of anteroposterior patterning and Hox gene regulation during chordate evolution. Philos. Trans. R. Soc. Lond. B Biol. Sci. 2001, 356, 1599–1613. [Google Scholar] [CrossRef] [Green Version]
- Albuixech-Crespo, B.; Lopez-Blanch, L.; Burguera, D.; Maeso, I.; Sanchez-Arrones, L.; Moreno-Bravo, J.A.; Somorjai, I.; Pascual-Anaya, J.; Puelles, E.; Bovolenta, P.; et al. Molecular regionalization of the developing amphioxus neural tube challenges major partitions of the vertebrate brain. PLoS Biol. 2017, 15, e2001573. [Google Scholar] [CrossRef] [Green Version]
- Williams, N.A.; Holland, P.W. Gene and domain duplication in the chordate Otx gene family: Insights from amphioxus Otx. Mol. Biol. Evol. 1998, 15, 600–607. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Imai, K.S.; Stolfi, A.; Levine, M.; Satou, Y. Gene regulatory networks underlying the compartmentalization of the Ciona central nervous system. Development 2009, 136, 285–293. [Google Scholar] [CrossRef] [Green Version]
- Onai, T.; Lin, H.C.; Schubert, M.; Koop, D.; Osborne, P.W.; Alvarez, S.; Alvarez, R.; Holland, N.D.; Holland, L.Z. Retinoic acid and Wnt/beta-catenin have complementary roles in anterior/posterior patterning embryos of the basal chordate amphioxus. Dev. Biol. 2009, 332, 223–233. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Koop, D.; Holland, N.D.; Semon, M.; Alvarez, S.; de Lera, A.R.; Laudet, V.; Holland, L.Z.; Schubert, M. Retinoic acid signaling targets Hox genes during the amphioxus gastrula stage: Insights into early anterior-posterior patterning of the chordate body plan. Dev. Biol. 2010, 338, 98–106. [Google Scholar] [CrossRef] [Green Version]
- Delsuc, F.; Brinkmann, H.; Chourrout, D.; Philippe, H. Tunicates and not cephalochordates are the closest living relatives of vertebrates. Nature 2006, 439, 965–968. [Google Scholar] [CrossRef] [PubMed]
- Spagnuolo, A.; Ristoratore, F.; Di Gregorio, A.; Aniello, F.; Branno, M.; Di Lauro, R. Unusual number and genomic organization of Hox genes in the tunicate Ciona intestinalis. Gene 2003, 309, 71–79. [Google Scholar] [CrossRef]
- Ishibashi, T.; Usami, T.; Fujie, M.; Azumi, K.; Satoh, N.; Fujiwara, S. Oligonucleotide-based microarray analysis of retinoic acid target genes in the protochordate, Ciona intestinalis. Dev. Dyn. 2005, 233, 1571–1578. [Google Scholar] [CrossRef]
- Kanda, M.; Ikeda, T.; Fujiwara, S. Identification of a retinoic acid-responsive neural enhancer in the Ciona intestinalis Hox1 gene. Dev. Growth Differ. 2013, 55, 260–269. [Google Scholar] [CrossRef]
- Kanda, M.; Wada, H.; Fujiwara, S. Epidermal expression of Hox1 is directly activated by retinoic acid in the Ciona intestinalis embryo. Dev. Biol. 2009, 335, 454–463. [Google Scholar] [CrossRef] [Green Version]
- Canestro, C.; Postlethwait, J.H. Development of a chordate anterior-posterior axis without classical retinoic acid signaling. Dev. Biol. 2007, 305, 522–538. [Google Scholar] [CrossRef] [Green Version]
- Garcia-Fernandez, J.; Holland, P.W. Archetypal organization of the amphioxus Hox gene cluster. Nature 1994, 370, 563–566. [Google Scholar] [CrossRef] [PubMed]
- Ferrier, D.E.; Minguillon, C.; Holland, P.W.; Garcia-Fernandez, J. The amphioxus Hox cluster: Deuterostome posterior flexibility and Hox14. Evol. Dev. 2000, 2, 284–293. [Google Scholar] [CrossRef] [Green Version]
- Wada, H.; Garcia-Fernandez, J.; Holland, P.W. Colinear and segmental expression of amphioxus Hox genes. Dev. Biol. 1999, 213, 131–141. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pascual-Anaya, J.; Adachi, N.; Alvarez, S.; Kuratani, S.; D’Aniello, S.; Garcia-Fernandez, J. Broken colinearity of the amphioxus Hox cluster. EvoDevo 2012, 3, 28. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Schubert, M.; Holland, N.D.; Laudet, V.; Holland, L.Z. A retinoic acid-Hox hierarchy controls both anterior/posterior patterning and neuronal specification in the developing central nervous system of the cephalochordate amphioxus. Dev. Biol. 2006, 296, 190–202. [Google Scholar] [CrossRef] [PubMed]
- Schubert, M.; Yu, J.K.; Holland, N.D.; Escriva, H.; Laudet, V.; Holland, L.Z. Retinoic acid signaling acts via Hox1 to establish the posterior limit of the pharynx in the chordate amphioxus. Development 2005, 132, 61–73. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Holland, L.Z.; Holland, N.D. Expression of AmphiHox-1 and AmphiPax-1 in amphioxus embryos treated with retinoic acid: Insights into evolution and patterning of the chordate nerve cord and pharynx. Development 1996, 122, 1829–1838. [Google Scholar] [CrossRef]
- Zieger, E.; Candiani, S.; Garbarino, G.; Croce, J.C.; Schubert, M. Roles of Retinoic Acid Signaling in Shaping the Neuronal Architecture of the Developing Amphioxus Nervous System. Mol. Neurobiol. 2018, 55, 5210–5229. [Google Scholar] [CrossRef]
- Manzanares, M.; Wada, H.; Itasaki, N.; Trainor, P.A.; Krumlauf, R.; Holland, P.W. Conservation and elaboration of Hox gene regulation during evolution of the vertebrate head. Nature 2000, 408, 854–857. [Google Scholar] [CrossRef]
- Wada, H.; Escriva, H.; Zhang, S.; Laudet, V. Conserved RARE localization in amphioxus Hox clusters and implications for Hox code evolution in the vertebrate neural crest. Dev. Dyn. 2006, 235, 1522–1531. [Google Scholar] [CrossRef]
- Studer, M.; Gavalas, A.; Marshall, H.; Ariza-McNaughton, L.; Rijli, F.M.; Chambon, P.; Krumlauf, R. Genetic interactions between Hoxa1 and Hoxb1 reveal new roles in regulation of early hindbrain patterning. Development 1998, 125, 1025–1036. [Google Scholar] [CrossRef]
- Knight, R.D.; Panopoulou, G.D.; Holland, P.W.; Shimeld, S.M. An amphioxus Krox gene: Insights into vertebrate hindbrain evolution. Dev. Genes Evol. 2000, 210, 518–521. [Google Scholar] [CrossRef]
- Jackman, W.R.; Kimmel, C.B. Coincident iterated gene expression in the amphioxus neural tube. Evol. Dev. 2002, 4, 366–374. [Google Scholar] [CrossRef] [PubMed]
- Natale, A.; Sims, C.; Chiusano, M.L.; Amoroso, A.; D’Aniello, E.; Fucci, L.; Krumlauf, R.; Branno, M.; Locascio, A. Evolution of anterior Hox regulatory elements among chordates. BMC Evol. Biol. 2011, 11, 330. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Locascio, A.; Aniello, F.; Amoroso, A.; Manzanares, M.; Krumlauf, R.; Branno, M. Patterning the ascidian nervous system: Structure, expression and transgenic analysis of the CiHox3 gene. Development 1999, 126, 4737–4748. [Google Scholar] [CrossRef] [PubMed]
- Albalat, R. The retinoic acid machinery in invertebrates: Ancestral elements and vertebrate innovations. Mol. Cell Endocrinol. 2009, 313, 23–35. [Google Scholar] [CrossRef] [PubMed]
- Gutierrez-Mazariegos, J.; Schubert, M.; Laudet, V. Evolution of Retinoic Acid Receptors and Retinoic Acid Signaling. In The Biochemistry of Retinoic Acid Receptors I: Structure, Activation, and Function at the Molecular Level; Asson-Batres, M.A., Rochette-Egly, C., Eds.; Springer: Dordrecht, The Netherlands, 2014; pp. 55–73. [Google Scholar]
- Escriva, H.; Delaunay, F.; Laudet, V. Ligand binding and nuclear receptor evolution. BioEssays 2000, 22, 717–727. [Google Scholar] [CrossRef]
- Sandell, L.L.; Lynn, M.L.; Inman, K.E.; McDowell, W.; Trainor, P.A. RDH10 oxidation of Vitamin A is a critical control step in synthesis of retinoic acid during mouse embryogenesis. PLoS ONE 2012, 7, e30698. [Google Scholar] [CrossRef]
- Sandell, L.L.; Sanderson, B.W.; Moiseyev, G.; Johnson, T.; Mushegian, A.; Young, K.; Rey, J.P.; Ma, J.X.; Staehling-Hampton, K.; Trainor, P.A. RDH10 is essential for synthesis of embryonic retinoic acid and is required for limb, craniofacial, and organ development. Genes Dev. 2007, 21, 1113–1124. [Google Scholar] [CrossRef] [Green Version]
- Albalat, R.; Brunet, F.; Laudet, V.; Schubert, M. Evolution of retinoid and steroid signaling: Vertebrate diversification from an amphioxus perspective. Genome Biol. Evol. 2011, 3, 985–1005. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Belyaeva, O.V.; Chang, C.; Berlett, M.C.; Kedishvili, N.Y. Evolutionary origins of retinoid active short-chain dehydrogenases/reductases of SDR16C family. Chem. Biol. Interact. 2015, 234, 135–143. [Google Scholar] [CrossRef] [Green Version]
- Niederreither, K.; Vermot, J.; Le Roux, I.; Schuhbaur, B.; Chambon, P.; Dolle, P. The regional pattern of retinoic acid synthesis by RALDH2 is essential for the development of posterior pharyngeal arches and the enteric nervous system. Development 2003, 130, 2525–2534. [Google Scholar] [CrossRef] [Green Version]
- Li, H.; Wagner, E.; McCaffery, P.; Smith, D.; Andreadis, A.; Dräger, U.C. A retinoic acid synthesizing enzyme in ventral retina and telencephalon of the embryonic mouse. Mech. Dev. 2000, 95, 283–289. [Google Scholar] [CrossRef]
- Mic, F.A.; Molotkov, A.; Fan, X.; Cuenca, A.E.; Duester, G. RALDH3, a retinaldehyde dehydrogenase that generates retinoic acid, is expressed in the ventral retina, otic vesicle and olfactory pit during mouse development. Mech. Dev. 2000, 97, 227–230. [Google Scholar] [CrossRef]
- Fan, X.; Molotkov, A.; Manabe, S.; Donmoyer, C.M.; Deltour, L.; Foglio, M.H.; Cuenca, A.E.; Blaner, W.S.; Lipton, S.A.; Duester, G. Targeted disruption of Aldh1a1 (Raldh1) provides evidence for a complex mechanism of retinoic acid synthesis in the developing retina. Mol. Cell Biol. 2003, 23, 4637–4648. [Google Scholar] [CrossRef] [Green Version]
- Sobreira, T.J.; Marletaz, F.; Simoes-Costa, M.; Schechtman, D.; Pereira, A.C.; Brunet, F.; Sweeney, S.; Pani, A.; Aronowicz, J.; Lowe, C.J.; et al. Structural shifts of aldehyde dehydrogenase enzymes were instrumental for the early evolution of retinoid-dependent axial patterning in metazoans. Proc. Natl. Acad. Sci. USA 2011, 108, 226–231. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Canestro, C.; Postlethwait, J.H.; Gonzalez-Duarte, R.; Albalat, R. Is retinoic acid genetic machinery a chordate innovation? Evol. Dev. 2006, 8, 394–406. [Google Scholar] [CrossRef] [PubMed]
- Nagatomo, K.; Fujiwara, S. Expression of Raldh2, Cyp26 and Hox-1 in normal and retinoic acid-treated Ciona intestinalis embryos. Gene Expr. Patterns 2003, 3, 273–277. [Google Scholar] [CrossRef]
- Escriva, H.; Holland, N.D.; Gronemeyer, H.; Laudet, V.; Holland, L.Z. The retinoic acid signaling pathway regulates anterior/posterior patterning in the nerve cord and pharynx of amphioxus, a chordate lacking neural crest. Development 2002, 129, 2905–2916. [Google Scholar] [CrossRef] [PubMed]
- Fujiwara, S.; Kawamura, K. Acquisition of retinoic acid signaling pathway and innovation of the chordate body plan. Zool. Sci. 2003, 20, 809–818. [Google Scholar] [CrossRef]
- Kamimura, M.; Fujiwara, S.; Kawamura, K.; Yubisui, T. Functional retinoid receptors in budding ascidians. Dev. Growth Differ. 2000, 42, 1–8. [Google Scholar] [CrossRef] [PubMed]
- Escriva, H.; Bertrand, S.; Germain, P.; Robinson-Rechavi, M.; Umbhauer, M.; Cartry, J.; Duffraisse, M.; Holland, L.; Gronemeyer, H.; Laudet, V. Neofunctionalization in vertebrates: The example of retinoic acid receptors. PLoS Genet. 2006, 2, e102. [Google Scholar] [CrossRef] [Green Version]
- Carvalho, J.E.; Theodosiou, M.; Chen, J.; Chevret, P.; Alvarez, S.; De Lera, A.R.; Laudet, V.; Croce, J.C.; Schubert, M. Lineage-specific duplication of amphioxus retinoic acid degrading enzymes (CYP26) resulted in sub-functionalization of patterning and homeostatic roles. BMC Evol. Biol. 2017, 17, 24. [Google Scholar] [CrossRef] [Green Version]
- Nomaksteinsky, M.; Rottinger, E.; Dufour, H.D.; Chettouh, Z.; Lowe, C.J.; Martindale, M.Q.; Brunet, J.F. Centralization of the deuterostome nervous system predates chordates. Curr. Biol. 2009, 19, 1264–1269. [Google Scholar] [CrossRef] [PubMed]
- Aronowicz, J.; Lowe, C.J. Hox gene expression in the hemichordate Saccoglossus kowalevskii and the evolution of deuterostome nervous systems. Integr. Comp. Biol. 2006, 46, 890–901. [Google Scholar] [CrossRef] [PubMed]
- Mooi, R.; David, B. Radial Symmetry, the Anterior/Posterior Axis, and Echinoderm Hox Genes. Annu. Rev. Ecol. Evol. Syst. 2008, 39, 43–62. [Google Scholar] [CrossRef]
- He, S.; Del Viso, F.; Chen, C.Y.; Ikmi, A.; Kroesen, A.E.; Gibson, M.C. An axial Hox code controls tissue segmentation and body patterning in Nematostella vectensis. Science 2018, 361, 1377–1380. [Google Scholar] [CrossRef] [Green Version]
- Arendt, D. Hox genes and body segmentation. Science 2018, 361, 1310–1311. [Google Scholar] [CrossRef]
- David, B.; Mooi, R. How Hox genes can shed light on the place of echinoderms among the deuterostomes. EvoDevo 2014, 5, 22. [Google Scholar] [CrossRef] [Green Version]
- Yamakawa, S.; Morino, Y.; Honda, M.; Wada, H. The role of retinoic acid signaling in starfish metamorphosis. EvoDevo 2018, 9, 10. [Google Scholar] [CrossRef] [Green Version]
- Yamakawa, S.; Morino, Y.; Kohtsuka, H.; Wada, H. Retinoic Acid Signaling Regulates the Metamorphosis of Feather Stars (Crinoidea, Echinodermata): Insight into the Evolution of the Animal Life Cycle. Biomolecules 2019, 10, 37. [Google Scholar] [CrossRef] [Green Version]
- Mainguy, G.; In der Rieden, P.M.; Berezikov, E.; Woltering, J.M.; Plasterk, R.H.; Durston, A.J. A position-dependent organisation of retinoid response elements is conserved in the vertebrate Hox clusters. Trends Genet. 2003, 19, 476–479. [Google Scholar] [CrossRef]
- Nolte, C.; De Kumar, B.; Krumlauf, R. Hox genes: Downstream "effectors" of retinoic acid signaling in vertebrate embryogenesis. Genesis 2019, 57, e23306. [Google Scholar] [CrossRef]
- Nolte, C.; Amores, A.; Nagy Kovacs, E.; Postlethwait, J.; Featherstone, M. The role of a retinoic acid response element in establishing the anterior neural expression border of Hoxd4 transgenes. Mech. Dev. 2003, 120, 325–335. [Google Scholar] [CrossRef]
- Nolte, C.; Rastegar, M.; Amores, A.; Bouchard, M.; Grote, D.; Maas, R.; Kovacs, E.N.; Postlethwait, J.; Rambaldi, I.; Rowan, S.; et al. Stereospecificity and PAX6 function direct Hoxd4 neural enhancer activity along the antero-posterior axis. Dev. Biol. 2006, 299, 582–593. [Google Scholar] [CrossRef] [Green Version]
- Morrison, A.; Ariza-McNaughton, L.; Gould, A.; Featherstone, M.; Krumlauf, R. HOXD4 and regulation of the group 4 paralog genes. Development 1997, 124, 3135–3146. [Google Scholar] [CrossRef] [PubMed]
- Oosterveen, T.; Niederreither, K.; Dolle, P.; Chambon, P.; Meijlink, F.; Deschamps, J. Retinoids regulate the anterior expression boundaries of 5’ Hoxb genes in posterior hindbrain. EMBO J. 2003, 22, 262–269. [Google Scholar] [CrossRef] [Green Version]
- Qian, P.; De Kumar, B.; He, X.C.; Nolte, C.; Gogol, M.; Ahn, Y.; Chen, S.; Li, Z.; Xu, H.; Perry, J.M.; et al. Retinoid-Sensitive Epigenetic Regulation of the Hoxb Cluster Maintains Normal Hematopoiesis and Inhibits Leukemogenesis. Cell Stem Cell 2018, 22, 740–754.e7. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gil-Gálvez, A.; Jiménez-Gancedo, S.; Acemel, R.D.; Bertrand, S.; Schubert, M.; Escrivá, H.; Tena, J.J.; Gómez-Skarmeta, J.L. Gain of gene regulatory network interconnectivity at the origin of vertebrates. bioRxiv 2020. [Google Scholar] [CrossRef]
- Carvalho, J.E.; Lahaye, F.; Croce, J.C.; Schubert, M. CYP26 function is required for the tissue-specific modulation of retinoic acid signaling during amphioxus development. Int. J. Dev. Biol. 2017, 61, 733–747. [Google Scholar] [CrossRef]
- Hu, P.; Tian, M.; Bao, J.; Xing, G.; Gu, X.; Gao, X.; Linney, E.; Zhao, Q. Retinoid regulation of the zebrafish cyp26a1 promoter. Dev. Dyn. 2008, 237, 3798–3808. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Loudig, O.; Babichuk, C.; White, J.; Abu-Abed, S.; Mueller, C.; Petkovich, M. Cytochrome P450RAI(CYP26) promoter: A distinct composite retinoic acid response element underlies the complex regulation of retinoic acid metabolism. Mol. Endocrinol. 2000, 14, 1483–1497. [Google Scholar] [CrossRef]
- Dolle, P. Developmental expression of retinoic acid receptors (RARs). Nucl. Recept. Signal. 2009, 7, e006. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hoffmann, B.; Lehmann, J.M.; Zhang, X.K.; Hermann, T.; Husmann, M.; Graupner, G.; Pfahl, M. A retinoic acid receptor-specific element controls the retinoic acid receptor-beta promoter. Mol. Endocrinol. 1990, 4, 1727–1736. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lehmann, J.M.; Zhang, X.K.; Pfahl, M. RAR gamma 2 expression is regulated through a retinoic acid response element embedded in Sp1 sites. Mol. Cell Biol. 1992, 12, 2976–2985. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lyn, S.; Giguere, V. Localisation of CRABP-I and CRABP-II mRNA in the early mouse embryo by whole-mount in situ hybridisation: Implications for teratogenesis and neural development. Dev. Dyn. 1994, 199, 280–291. [Google Scholar] [CrossRef] [PubMed]
- Cai, A.Q.; Radtke, K.; Linville, A.; Lander, A.D.; Nie, Q.; Schilling, T.F. Cellular retinoic acid-binding proteins are essential for hindbrain patterning and signal robustness in zebrafish. Development 2012, 139, 2150–2155. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Aström, A.; Pettersson, U.; Chambon, P.; Voorhees, J.J. Retinoic acid induction of human cellular retinoic acid-binding protein-II gene transcription is mediated by retinoic acid receptor-retinoid X receptor heterodimers bound to one far upstream retinoic acid-responsive element with 5-base pair spacing. J. Biol. Chem. 1994, 269, 22334–22339. [Google Scholar] [CrossRef]
- Durand, B.; Saunders, M.; Leroy, P.; Leid, M.; Chambon, P. All-trans and 9-cis retinoic acid induction of CRABPII transcription is mediated by RAR-RXR heterodimers bound to DR1 and DR2 repeated motifs. Cell 1992, 71, 73–85. [Google Scholar] [CrossRef]
- Mansfield, S.G.; Cammer, S.; Alexander, S.C.; Muehleisen, D.P.; Gray, R.S.; Tropsha, A.; Bollenbacher, W.E. Molecular cloning and characterization of an invertebrate cellular retinoic acid binding protein. Proc. Natl. Acad. Sci. USA 1998, 95, 6825–6830. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zieger, E.; Schubert, M. New Insights Into the Roles of Retinoic Acid Signaling in Nervous System Development and the Establishment of Neurotransmitter Systems. Int. Rev. Cell Mol. Biol. 2017, 330, 1–84. [Google Scholar] [CrossRef] [PubMed]
- Castillo, H.A.; Cravo, R.M.; Azambuja, A.P.; Simoes-Costa, M.S.; Sura-Trueba, S.; Gonzalez, J.; Slonimsky, E.; Almeida, K.; Abreu, J.G.; de Almeida, M.A.; et al. Insights into the organization of dorsal spinal cord pathways from an evolutionarily conserved raldh2 intronic enhancer. Development 2010, 137, 507–518. [Google Scholar] [CrossRef] [Green Version]
- Gutierrez-Mazariegos, J.; Nadendla, E.K.; Studer, R.A.; Alvarez, S.; de Lera, A.R.; Kuraku, S.; Bourguet, W.; Schubert, M.; Laudet, V. Evolutionary diversification of retinoic acid receptor ligand-binding pocket structure by molecular tinkering. R. Soc. Open Sci. 2016, 3, 150484. [Google Scholar] [CrossRef] [PubMed]
- Campo-Paysaa, F.; Jandzik, D.; Takio-Ogawa, Y.; Cattell, M.V.; Neef, H.C.; Langeland, J.A.; Kuratani, S.; Medeiros, D.M.; Mazan, S.; Kuraku, S.; et al. Evolution of retinoic acid receptors in chordates: Insights from three lamprey species, Lampetra fluviatilis, Petromyzon marinus, and Lethenteron japonicum. EvoDevo 2015, 6, 18. [Google Scholar] [CrossRef] [Green Version]


Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Bedois, A.M.H.; Parker, H.J.; Krumlauf, R. Retinoic Acid Signaling in Vertebrate Hindbrain Segmentation: Evolution and Diversification. Diversity 2021, 13, 398. https://doi.org/10.3390/d13080398
Bedois AMH, Parker HJ, Krumlauf R. Retinoic Acid Signaling in Vertebrate Hindbrain Segmentation: Evolution and Diversification. Diversity. 2021; 13(8):398. https://doi.org/10.3390/d13080398
Chicago/Turabian StyleBedois, Alice M. H., Hugo J. Parker, and Robb Krumlauf. 2021. "Retinoic Acid Signaling in Vertebrate Hindbrain Segmentation: Evolution and Diversification" Diversity 13, no. 8: 398. https://doi.org/10.3390/d13080398
APA StyleBedois, A. M. H., Parker, H. J., & Krumlauf, R. (2021). Retinoic Acid Signaling in Vertebrate Hindbrain Segmentation: Evolution and Diversification. Diversity, 13(8), 398. https://doi.org/10.3390/d13080398








