Diagnosis of Liver Fibrosis Using Artificial Intelligence: A Systematic Review
Abstract
1. Introduction
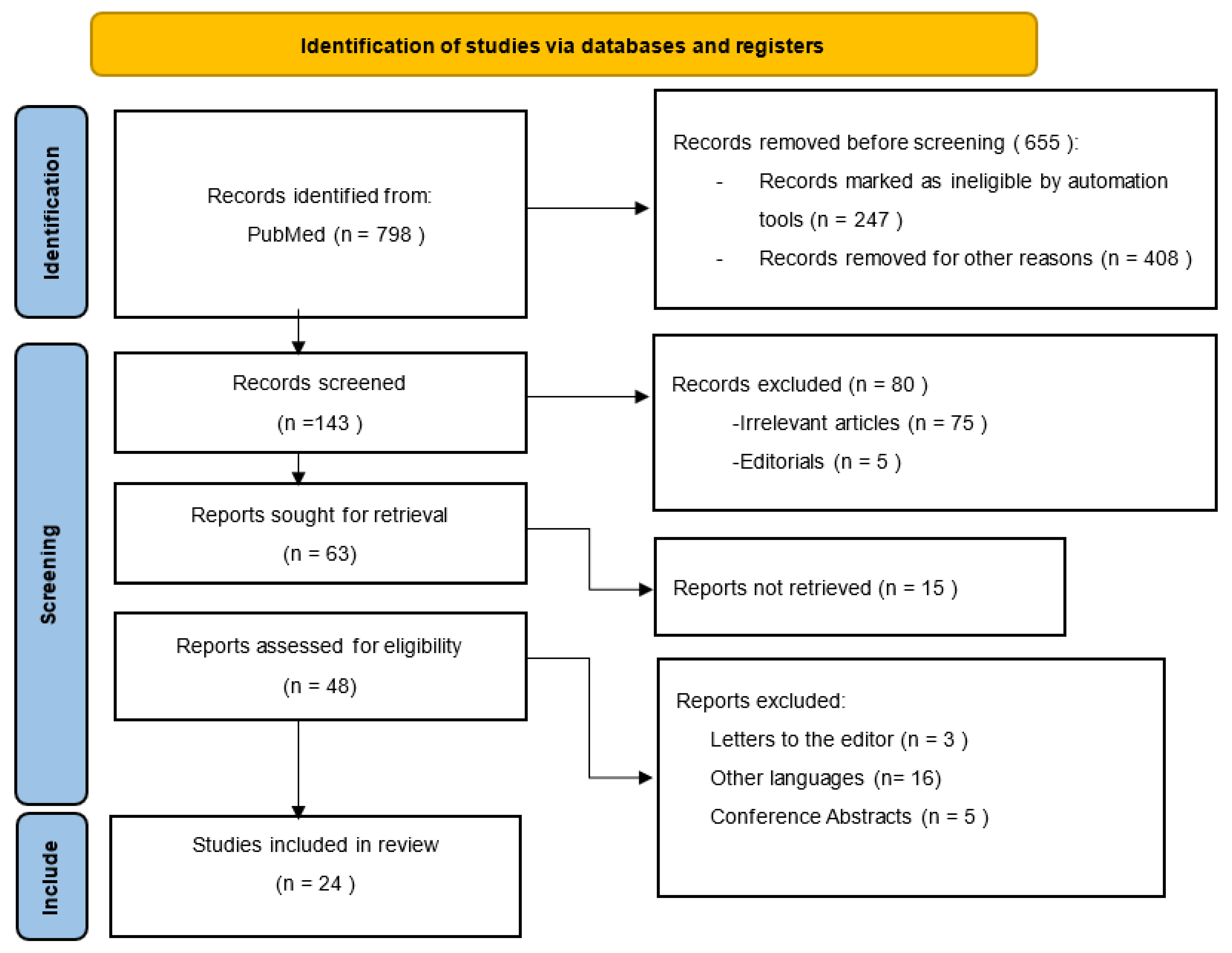
2. Materials and Methods
3. Results
3.1. Artificial Intelligence Techniques and CT Imaging
3.2. Artificial Intelligence Techniques and MRI Imaging
3.3. Artificial Intelligence Techniques and Ultrasonography
3.4. Artificial Intelligence Techniques and Liver Biopsy
4. Discussion
5. Conclusions
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Asrani, S.K.; Devarbhavi, H.; Eaton, J.; Kamath, P.S. Burden of liver diseases in the world. J. Hepatol. 2019, 70, 151–171. [Google Scholar] [CrossRef] [PubMed]
- Marcellin, P.; Kutala, B.K. Liver diseases: A major, neglected global public problem requiring urgent actions and large-scale screening. Liver Int. 2018, 38, 2–6. [Google Scholar] [CrossRef] [PubMed]
- European Association for the Study of the Liver. EASL Clinical Practice Guidelines on non-invasive tests for evaluation of liver disease severity and prognosis—2021 update. J. Hepatol. 2021, 75, 659–689. [Google Scholar] [CrossRef] [PubMed]
- Parola, M.; Pinzani, M. Liver fibrosis: Pathophysiology, pathogenetic targets and clinical issues. Mol. Asp. Med. 2019, 65, 37–55. [Google Scholar] [CrossRef]
- Lambrecht, J.; van Grunsven, L.A.; Tacke, F. Current and emerging pharmacotherapeutic interventions for the treatment of liver fibrosis. Expert Opin. Pharm. 2020, 21, 1637–1650. [Google Scholar] [CrossRef]
- Wang, F.D.; Zhou, J.; Chen, E.Q. Molecular Mechanisms and Potential New Therapeutic Drugs for Liver Fibrosis. Front. Pharm. 2022, 13, 787748. [Google Scholar] [CrossRef]
- Lai, M.; Afdhal, N.H. Liver fibrosis determination. Gastroenterol. Clin. N. Am. 2019, 48, 281–289. [Google Scholar] [CrossRef]
- Friedman, S.L.; Pinzani, M. Hepatic Fibrosis 2022: Unmet Needs and a Blueprint for the Future. Pathology 2022, 75, 473–488. [Google Scholar] [CrossRef]
- Dana, J.; Venkatasami, A.; Saviano, A.; Lupberger, J.; Hoshida, Y.; Vilgrain, V.; Nahon, P.; Reinhold, C.; Gallix, B.; Baumert, T.F. Conventional and artificial intelligence-based imaging for biomarker discovery in chronic liver disease. Hepatol. Int. 2022, 16, 509–522. [Google Scholar] [CrossRef]
- Marozas, M.; Zykus, R.; Sakalauskas, A.; Kupčinskas, L.; Lukoševičius, A. Noninvasive Evaluation of Portal Hypertension Using a Supervised Learning Technique. J. Healthc. Eng. 2017, 2017, 6183714. [Google Scholar] [CrossRef]
- Bayani, A.; Hosseini, A.; Asadi, F.; Hatami, B.; Kavousi, K.; Aria, M.; Zali, M.R. Identifying predictors of varices grading in patients with cirrhosis using ensemble learning. Clin. Chem. Lab. Med. 2022, 60, 1938–1945. [Google Scholar] [CrossRef] [PubMed]
- Kim, H.Y.; Lampertico, P.; Nam, J.Y.; Lee, H.C.; Kim, S.U.; Sinn, D.H.; Seo, Y.S.; Lee, H.A.; Park, S.Y.; Lim, Y.S.; et al. An artificial intelligence model to predict hepatocellular carcinoma risk in Korean and Caucasian patients with chronic hepatitis B. J. Hepatol. 2022, 76, 311–318. [Google Scholar] [CrossRef] [PubMed]
- Popa, S.L.; Grad, S.; Chiarioni, G.; Masier, A.; Peserico, G.; Brata, V.D.; Dumitrascu, D.I.; Fantin, A. Applications of Artificial Intelligence in the Automatic Diagnosis of Focal Liver Lesions: A Systematic Review. J. Gastrointest. Liver Dis. 2023, 32, 77–85. [Google Scholar] [CrossRef] [PubMed]
- Xu, X.; Zhang, H.-L.; Liu, Q.-P.; Sun, S.-W.; Zhang, J.; Zhu, F.-P.; Yang, G.; Yan, X.; Zhang, Y.-D.; Liu, X.-S. Radiomic analysis of contrast-enhanced CT predicts microvascular invasion and outcome in hepatocellular carcinoma. J. Hepatol. 2019, 70, 1133–1144. [Google Scholar] [CrossRef]
- Abajian, A.; Murali, N.; Savic, L.J.; Laage-Gaupp, F.M.; Nezami, N.; Duncan, J.S.; Schlachter, T.; Lin, M.; Geschwind, J.-F.; Chapiro, J. Predicting treatment response to intra-arterial therapies for hepatocellular carcinoma with the use of supervised machine learning-an artificial intelligence concept. J. Vasc. Interv. Radiol. 2018, 29, 850–857.e1. [Google Scholar] [CrossRef] [PubMed]
- Page, M.J.; McKenzie, J.E.; Bossuyt, P.M.; Boutron, I.; Hoffmann, T.C.; Mulrow, C.D.; Shamseer, L.; Tetzlaff, J.M.; Akl, E.A.; Brennan, S.E.; et al. The PRISMA 2020 statement: An updated guideline for reporting systematic reviews. BMJ 2021, 372, n71. [Google Scholar] [CrossRef]
- Lurie, Y.; Webb, M.; Cytter-Kuint, R.; Shteingart, S.; Lederkremer, G.Z. Non-invasive diagnosis of liver fibrosis and cirrhosis. World J. Gastroenterol. 2015, 21, 11567–11583. [Google Scholar] [CrossRef]
- Tsochatzis, E.A.; Bosch, J.; Burroughs, A.K. Liver cirrhosis. Lancet 2014, 383, 1749–1761. [Google Scholar] [CrossRef]
- Li, L.; Duan, M.; Chen, W.; Jiang, A.; Li, X.; Yang, J.; Li, Z. The spleen in liver cirrhosis: Revisiting an old enemy with novel targets. J. Transl. Med. 2017, 15, 111. [Google Scholar] [CrossRef]
- Yasaka, K.; Akai, H.; Kunimatsu, A.; Abe, O.; Kiryu, S. Deep learning for staging liver fibrosis on CT: A pilot study. Eur. Radiol. 2018, 28, 4578–4585. [Google Scholar] [CrossRef]
- Li, Q.; Yu, B.; Tian, X.; Cui, X.; Zhang, R.; Guo, Q. Deep residual nets model for staging liver fibrosis on plain CT images. Int. J. CARS 2020, 5, 1399–1406. [Google Scholar] [CrossRef] [PubMed]
- Choi, K.J.; Jang, J.K.; Lee, S.S.; Sung, Y.S.; Shim, W.H.; Kim, H.S.; Yun, J.; Choi, J.-Y.; Lee, Y.; Kang, B.-K.; et al. Development and Validation of a Deep Learning System for Staging Liver Fibrosis by Using Contrast Agent-enhanced CT Images in the Liver. Radiology 2018, 289, 688–697. [Google Scholar] [CrossRef] [PubMed]
- Yin, Y.; Yakar, D.; Dierckx, R.A.J.O.; Mouridsen, K.B.; Kwee, T.C.; de Haas, R.J. Liver fibrosis staging by deep learning: A visual-based explanation of diagnostic decisions of the model. Eur. Radiol. 2021, 31, 9620–9627. [Google Scholar] [CrossRef] [PubMed]
- Yin, Y.; Yakar, D.; Dierckx, R.A.J.O.; Mouridsen, K.B.; Kwee, T.C.; de Haas, R.J. Combining Hepatic and Splenic CT Radiomic Features Improves Radiomic Analysis Performance for Liver Fibrosis Staging. Diagnostics 2022, 12, 550. [Google Scholar] [CrossRef]
- Budai, B.K.; Tóth, A.; Borsos, P.; Frank, V.G.; Shariati, S.; Fejér, B.; Folhoffer, A.; Szalay, F.; Bérczi, V.; Kaposi, P.N. Three-dimensional CT texture analysis of anatomic liver segments can differentiate between low-grade and high-grade fibrosis. BMC Med. Imaging 2020, 20, 108. [Google Scholar] [CrossRef]
- Wu, L.; Ning, B.; Yang, J.; Chen, Y.; Zhang, C.; Yan, Y. Diagnosis of Liver Cirrhosis and Liver Fibrosis by Artificial Intelligence Algorithm-Based Multislice Spiral Computed Tomography. Comput. Math. Methods Med. 2022, 2022, 1217003. [Google Scholar] [CrossRef]
- Nowak, S.; Mesropyan, N.; Faron, A.; Block, W.; Reuter, M.; Attenberger, U.I.; Luetkens, J.A.; Sprinkart, A.M. Detection of liver cirrhosis in standard T2-weighted MRI using deep transfer learning. Eur. Radiol. 2021, 31, 8807–8815. [Google Scholar] [CrossRef]
- Kato, H.; Kanematsu, M.; Zhang, X.; Saio, M.; Kondo, H.; Goshima, S.; Fujita, H. Computer-Aided Diagnosis of Hepatic Fibrosis: Preliminary Evaluation of MRI Texture Analysis Using the Finite Difference Method and an Artificial Neural Network. Am. J. Roentgenol. 2007, 189, 117–122. [Google Scholar] [CrossRef]
- Hectors, S.J.; Kennedy, P.; Huang, K.H.; Stocker, D.; Carbonell, G.; Greenspan, H.; Friedman, S.; Taouli, B. Fully automated prediction of liver fibrosis using deep learning analysis of gadoxetic acid–enhanced MRI. Eur. Radiol. 2021, 31, 3805–3814. [Google Scholar] [CrossRef]
- Strotzer, Q.D.; Winther, H.; Utpatel, K.; Scheiter, A.; Fellner, C.; Doppler, M.C.; Ringe, K.I.; Raab, F.; Haimerl, M.; Uller, W.; et al. Application of A U-Net for Map-like Segmentation and Classification of Discontinuous Fibrosis Distribution in Gd-EOB-DTPA-Enhanced Liver MRI. Diagnostics 2022, 12, 1938. [Google Scholar] [CrossRef]
- Soufi, M.; Otake, Y.; Hori, M.; Moriguchi, K.; Imai, Y.; Sawai, Y.; Ota, T.; Tomiyama, N.; Sato, Y. Liver shape analysis using partial least squares regression-based statistical shape model: Application for understanding and staging of liver fibrosis. Int. J. CARS 2019, 14, 2083–2093. [Google Scholar] [CrossRef] [PubMed]
- Brattain, L.J.; Telfer, B.A.; Dhyani, M.; Grajo, J.R.; Samir, A.E. Objective liver fibrosis estimation from shear wave elastography. Annu. Int. Conf. IEEE Eng. Med. Biol. Soc. 2018, 2018, 1–5. [Google Scholar] [CrossRef] [PubMed]
- Li, W.; Huang, Y.; Zhuang, B.W.; Liu, G.J.; Hu, H.T.; Li, X.; Liang, J.-Y.; Wang, Z.; Huang, X.-W.; Zhang, C.-Q.; et al. Multiparametric ultrasomics of significant liver fibrosis: A machine learning-based analysis. Eur. Radiol. 2019, 29, 1496–1506. [Google Scholar] [CrossRef]
- Xie, Y.; Chen, S.; Jia, D.; Li, B.; Zheng, Y.; Yu, X. Artificial Intelligence-Based Feature Analysis of Ultrasound Images of Liver Fibrosis. Comput. Intell. Neurosci. 2022, 2022, 2859987. [Google Scholar] [CrossRef] [PubMed]
- Zhang, L.; Li, Q.Y.; Duan, Y.Y.; Yan, G.Z.; Yang, Y.L.; Yang, R.J. Artificial neural network aided non-invasive grading evaluation of hepatic fibrosis by duplex ultrasonography. BMC Med. Inform. Decis. Mak. 2012, 12, 55. [Google Scholar] [CrossRef]
- Lee, J.H.; Joo, I.; Kang, T.W.; Paik, Y.H.; Sinn, D.H.; Ha, S.Y.; Kim, K.; Choi, C.; Lee, G.; Yi, J.; et al. Deep learning with ultrasonography: Automated classification of liver fibrosis using a deep convolutional neural network. Eur. Radiol. 2020, 30, 1264–1273. [Google Scholar] [CrossRef]
- Gatos, I.; Tsantis, S.; Spiliopoulos, S.; Karnabatidis, D.; Theotokas, I.; Zoumpoulis, P.; Loupas, T.; Hazle, J.D.; Kagadis, G.C. A Machine-Learning Algorithm Toward Color Analysis for Chronic Liver Disease Classification, Employing Ultrasound Shear Wave Elastography. Ultrasound Med. Biol. 2017, 43, 1797–1810. [Google Scholar] [CrossRef]
- Astbury, S.; Grove, J.I.; Dorward, D.A.; Guha, I.N.; Fallowfield, J.A.; Kendall, T.J. Reliable computational quantification of liver fibrosis is compromised by inherent staining variation. J. Pathol. Clin. Res. 2021, 7, 471–481. [Google Scholar] [CrossRef]
- Sarvestany, S.S.; Kwong, J.C.; Azhie, A.; Dong, V.; Cerocchi, O.; Ali, A.F.; Karnam, R.S.; Kuriry, H.; Shengir, M.; Candido, E.; et al. Development and validation of an ensemble machine learning framework for detection of all-cause advanced hepatic fibrosis: A retrospective cohort study. Lancet Digit. Health 2022, 4, e188–e199. [Google Scholar] [CrossRef]
- Matalka, I.I.; Al-Jarrah, O.M.; Manasrah, T.M. Quantitative assessment of liver fibrosis: A novel automated image analysis method. Liver Int. 2006, 26, 1054–1064. [Google Scholar] [CrossRef]
- Qiu, Q.T.; Zhang, J.; Duan, J.H.; Wu, S.Z.; Ding, J.L.; Yin, Y. Development and validation of radiomics model built by incorporating machine learning for identifying liver fibrosis and early-stage cirrhosis. Chin. Med. J. 2020, 133, 2653–2659. [Google Scholar] [CrossRef] [PubMed]
- Wei, W.; Wu, X.; Zhou, J.; Sun, Y.; Kong, Y.; Yang, X. Noninvasive Evaluation of Liver Fibrosis Reverse Using Artificial Neural Network Model for Chronic Hepatitis B Patients. Comput. Math. Methods Med. 2019, 2019, 7239780. [Google Scholar] [CrossRef] [PubMed]
- Wang, K.; Lu, X.; Zhou, H.; Gao, Y.; Zheng, J.; Tong, M.; Wu, C.; Liu, C.; Huang, L.; Jiang, T.; et al. Deep learning Radiomics of shear wave elastography significantly improved diagnostic performance for assessing liver fibrosis in chronic hepatitis B: A prospective multicentre study. Gut 2019, 68, 729–741. [Google Scholar] [CrossRef]
- Eslam, M.; Newsome, P.N.; Sarin, S.K.; Anstee, Q.M.; Targher, G.; Romero-Gomez, M.; Zelber-Sagi, S.; Wong, V.W.-S.; Dufour, J.-F.; Schattenberg, J.M.; et al. A new definition for metabolic dysfunction-associated fatty liver disease: An international expert consensus statement. J. Hepatol. 2020, 73, 202–209. [Google Scholar] [CrossRef] [PubMed]
- Lefebvre, T.; Wartelle-Bladou, C.; Wong, P.; Sebastiani, G.; Giard, J.M.; Castel, H.; Murphy-Lavallée, J.; Olivié, D.; Ilinca, A.; Sylvestre, M.-P.; et al. Prospective comparison of transient, point shear wave, and magnetic resonance elastography for staging liver fibrosis. Eur. Radiol. 2019, 29, 6477–6488. [Google Scholar] [CrossRef]
- Durot, I.; Akhbardeh, A.; Sagreiya, H.; Loening, A.M.; Rubin, D.L. A New Multimodel Machine Learning Framework to Improve Hepatic Fibrosis Grading Using Ultrasound Elastography Systems from Different Vendors. Ultrasound Med. Biol. 2020, 46, 26–33. [Google Scholar] [CrossRef] [PubMed]
- Kagadis, G.C.; Drazinos, P.; Gatos, I.; Tsantis, S.; Papadimitroulas, P.; Spiliopoulos, S.; Karnabatidis, D.; Theotokas, I.; Zoumpoulis, P.; Hazle, J.D. Deep learning networks on chronic liver disease assessment with fine-tuning of shear wave elastography image sequences. Phys. Med. Biol. 2020, 65, 215027. [Google Scholar] [CrossRef] [PubMed]
- Decharatanachart, P.; Chaiteerakij, R.; Tiyarattanachai, T.; Treeprasertsuk, S. Application of artificial intelligence in non-alcoholic fatty liver disease and liver fibrosis: A systematic review and meta-analysis. Therap. Adv. Gastroenterol. 2021, 14, 17562848211062807. [Google Scholar] [CrossRef]
- Popa, S.L.; Ismaiel, A.; Cristina, P.; Cristina, M.; Chiarioni, G.; David, L.; Dumitrascu, D.L. Non-Alcoholic Fatty Liver Disease: Implementing Complete Automated Diagnosis and Staging. A Systematic Review. Diagnostics 2021, 11, 1078. [Google Scholar] [CrossRef]
| First Author | Year | Total Number of Images | Diagnosis | Main Findings |
|---|---|---|---|---|
| Yasaka et al. [20] | 2018 | 496 | Liver fibrosis | Magnified CT images were analyzed by deep learning to diagnose and stage liver fibrosis, revealing a moderate correlation with histopathological staging. |
| Li et al. [21] | 2020 | 1041 | Liver fibrosis | The residual neural network (ResNet) is an efficient non-invasive diagnostic method for diagnosing liver fibrosis using plain CT images. |
| Choi et al. [22] | 2018 | 7461 | Liver fibrosis | The deep learning system was able to diagnose and stage live fibrosis with high accuracy (79.4%). |
| Yin et al. [23] | 2021 | 252 | Liver fibrosis | By using contrast-enhanced CT images and deep learning algorithms, liver fibrosis can be successfully diagnosed and staged. |
| Yin et al. [24] | 2022 | 252 | Liver fibrosis | Splenic radiomic features are an important and useful addition to hepatic radiomic features when staging liver fibrosis. |
| Budai et al. [25] | 2020 | 354 | Liver fibrosis | In order to differentiate between low- and high-grade fibrosis, CT texture analysis can be used for prognosis calculations of chronic liver disease. |
| Wu et al. [26] | 2022 | 112 | Liver cirrhosis and liver fibrosis | AI segmentation algorithms can be used to diagnose liver fibrosis in a clinical context. |
| First Author | Year | Total Number of Images | Diagnosis | Main Findings |
|---|---|---|---|---|
| Nowak et al. [27] | 2021 | 713 | Liver cirrhosis | Two pre-trained convolutional neural networks were successfully used to detect liver cirrhosis on standard T2-weighted MRIs. |
| Kato et al. [28] | 2007 | 52 | Liver fibrosis | The computer algorithm revealed a potential usefulness for the diagnosis of hepatic fibrosis. |
| Hectors et al. [29] | 2021 | 355 | Liver fibrosis | Deep learning algorithm, based on gadoxetic acid-enhanced MRI data, was comparable to MR elastography analysis. |
| Strotzer et al. [30] | 2022 | 112 | Liver cirrhosis and liver fibrosis | A multiphase Gd-EOB-DTPA-enhanced liver MRI was used to diagnose fibrosis stage or cirrhosis. |
| Soufi et al. [31] | 2019 | 51 | Liver fibrosis | PLSR-based SSM could help to better understand the variations associated with liver fibrosis staging and diagnosis. |
| First Author | Year | Total Number of Images | Diagnosis | Main Findings |
|---|---|---|---|---|
| Brattain et al. [32] | 2018 | 3392 | Liver fibrosis | A new method of diagnosis for liver fibrosis that is based on a single image per decision compared to previous methods which used 10 images per decision. |
| Li et al. [33] | 2019 | 144 | Chronic hepatitis B | Machine-learning-based analysis of ultrasonography images can help stage liver fibrosis. |
| Xie et al. [34] | 2022 | 640 | Chronic hepatitis B and cirrhosis | The GoogLeNet model shows promising results in terms of recognition of lesions and diagnosis. |
| Zhang et al. [35] | 2012 | 239 | Liver fibrosis or cirrhosis | The ANN model presented high sensitivity and specificity for the non-invasive diagnosis of liver fibrosis. |
| Lee et al. [36] | 2020 | 13,608 | Liver fibrosis | Deep convolutional neural network accurately classified the ultrasonography images for cirrhosis diagnosis. |
| Gatos et al. [37] | 2017 | 126 | chronic liver disease | Color information quantification, from SWE images, by machine-learning can dissociate between chronic liver disease and healthy patients. |
| First Author | Year | Total Number of Images | Diagnosis | Main Findings |
|---|---|---|---|---|
| Astbury et al. [38] | 2021 | 20 | Liver cirrhosis | Standardization between staining methods is still very important, as computational tools cannot yet normalize samples when performing analysis. |
| Sarvestany et al. [39] | 2022 | 1703 | Liver fibrosis | MLAs are able to help differentiate between patients with different prognoses concerning chronic liver disease. |
| Matalka et al. [40] | 2006 | 260 | Liver fibrosis | The automated quantification system differentiated between normal biopsies and samples with liver fibrosis, with an accuracy of 98.46%, and classified each sample with fibrosis according to the Ishak scoring system, with a precision of 94.69%. |
| Qiu et al. [41] | 2020 | 369 | Liver fibrosis | Radiomics analysis of liver images can accurately diagnose liver disease, resulting in a superior diagnosis tool compared to liver biopsy. |
| Wei et al. [42] | 2019 | 141 | Liver fibrosis | The multi-variable model developed can be useful for the evaluation of the clinical evolution of patients with chronic HBV-induced liver fibrosis. |
| Wang et al. [43] | 2018 | 1990 | Chronic hepatitis B | Deep learning Radiomics of elastography (DLRE) is useful for the non-invasive staging of liver fibrosis in patients infected with HBV. |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Popa, S.L.; Ismaiel, A.; Abenavoli, L.; Padureanu, A.M.; Dita, M.O.; Bolchis, R.; Munteanu, M.A.; Brata, V.D.; Pop, C.; Bosneag, A.; et al. Diagnosis of Liver Fibrosis Using Artificial Intelligence: A Systematic Review. Medicina 2023, 59, 992. https://doi.org/10.3390/medicina59050992
Popa SL, Ismaiel A, Abenavoli L, Padureanu AM, Dita MO, Bolchis R, Munteanu MA, Brata VD, Pop C, Bosneag A, et al. Diagnosis of Liver Fibrosis Using Artificial Intelligence: A Systematic Review. Medicina. 2023; 59(5):992. https://doi.org/10.3390/medicina59050992
Chicago/Turabian StylePopa, Stefan Lucian, Abdulrahman Ismaiel, Ludovico Abenavoli, Alexandru Marius Padureanu, Miruna Oana Dita, Roxana Bolchis, Mihai Alexandru Munteanu, Vlad Dumitru Brata, Cristina Pop, Andrei Bosneag, and et al. 2023. "Diagnosis of Liver Fibrosis Using Artificial Intelligence: A Systematic Review" Medicina 59, no. 5: 992. https://doi.org/10.3390/medicina59050992
APA StylePopa, S. L., Ismaiel, A., Abenavoli, L., Padureanu, A. M., Dita, M. O., Bolchis, R., Munteanu, M. A., Brata, V. D., Pop, C., Bosneag, A., Dumitrascu, D. I., Barsan, M., & David, L. (2023). Diagnosis of Liver Fibrosis Using Artificial Intelligence: A Systematic Review. Medicina, 59(5), 992. https://doi.org/10.3390/medicina59050992







