Methanogenesis—General Principles and Application in Wastewater Remediation
Abstract
:1. Introduction
2. Hydrogen for Methane Formation
3. Global Methane Cycle
4. Methanogens
Hydrogenases from Methanogenic Origin and Their Function
5. Methanogenic Pathways—Dependence on Substrates and Mechanism
6. Factors Effecting Methanogenesis
6.1. Hydrogen Concentration
6.2. Oxygen and Sulfate Presence
6.3. Environmental Salinity
6.4. Temperature
6.5. pH Values
6.6. Pressure
7. Application of Methanogenesis in Wastewater Remediation
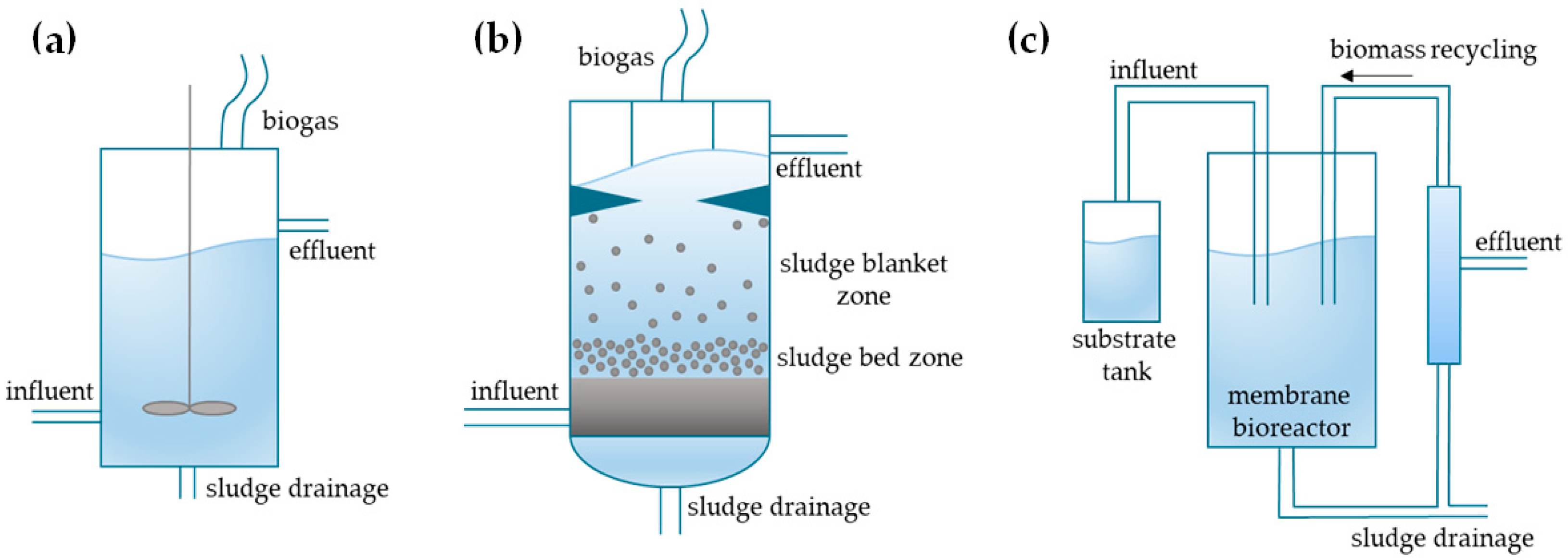
7.1. Classification of Anaerobic Reactors
7.2. Microbial Interactions Within Reactors
7.3. Wastewater Types and the Presence of Heavy Metals
7.4. Effect of Methanogenesis on Wastewater Treatments
8. Methanogenesis for Renewable Energy Production
8.1. Methane for Hydrogen Storage
8.2. Power-to-Gas
8.3. Bioelectrochemical Methane Production
8.4. Micro Biogas Plants
8.5. Methanogens in the Agricultural Sector
9. Conclusions
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Getabalew, M.; Alemneh, T.; Bzuneh, E. Review on Methanogenesis and its Role. World J. Agric. Soil Sci. 2020, 6, 1–7. [Google Scholar]
- Schink, B.; Montag, D.; Keller, A.; Müller, N. Hydrogen or formate: Alternative key players in methanogenic degradation. Environ. Microbiol. Rep. 2017, 9, 189–202. [Google Scholar] [CrossRef] [PubMed]
- Tabatabaei, M.; Rahim, R.A.; Abdullah, N.; Wright, A.D.G.; Shirai, Y.; Sakai, K.; Sulaiman, A.; Hassan, M.A. Importance of the methanogenic archaea populations in anaerobic wastewater treatments. Process Biochem. 2010, 45, 1214–1225. [Google Scholar] [CrossRef]
- Thauer, R.K.; Kaster, A.K.; Seedorf, H.; Buckel, W.; Hedderich, R. Methanogenic archaea: Ecologically relevant differences in energy conservation. Nat. Rev. Microbiol. 2008, 6, 579–591. [Google Scholar] [CrossRef]
- Schuchmann, K.; Chowdhury, N.P.; Müller, V. Complex Multimeric [FeFe] Hydrogenases: Biochemistry, Physiology and New Opportunities for the Hydrogen Economy. Front. Microbiol. 2018, 9, 2911. [Google Scholar] [CrossRef]
- Conrad, R. Contribution of hydrogen to methane production and control of hydrogen concentrations in methanogenic soils and sediments. FEMS Microbiol. Ecol. 1999, 28, 193–202. [Google Scholar] [CrossRef]
- Thauer, R.K. Hydrogenases and the global H2 cycle. Eur. J. Inorg. Chem. 2011, 2011, 919–921. [Google Scholar] [CrossRef]
- Thauer, R.K.; Kaster, A.K.; Goenrich, M.; Schick, M.; Hiromoto, T.; Shima, S. Hydrogenases from methanogenic archaea, nickel, a novel cofactor, and H2 storage. Annu. Rev. Biochem. 2010, 79, 507–536. [Google Scholar] [CrossRef]
- Katz, B.J. Microbial Processes and Natural Gas Accumulations. Open Geol. J. 2011, 5, 75–83. [Google Scholar] [CrossRef]
- Buan, N.R. Methanogens: Pushing the boundaries of biology. Emerg. Top. Life Sci. 2018, 2, 629–646. [Google Scholar]
- Zieliński, M.; Kazimierowicz, J.; Dębowski, M. Advantages and Limitations of Anaerobic Wastewater Treatment—Technological Basics, Development Directions, and Technological Innovations. Energies 2023, 16, 83. [Google Scholar] [CrossRef]
- Woese, C.R.; Fox, G.E. Phylogenetic structure of the prokaryotic domain: The primary kingdoms. Proc. Nat. Acad. Sci. USA 1977, 74, 5088–5090. [Google Scholar] [CrossRef] [PubMed]
- Mueller, J.; Booth, J.G. Managing Excessive Methanogenesis During ERD/ISCR Remedial. Remediat. J. 2016, 26, 53–71. [Google Scholar] [CrossRef]
- Kietäväinen, R.; Purkamo, L. The origin, source, and cycling of methane in deep crystalline rock biosphere. Front. Microbiol. 2015, 6, 725. [Google Scholar] [CrossRef]
- Liu, Y.; Whitman, W.B. Metabolic, Phylogenetic, and Ecological Diversity of the Methanogenic Archaea. Ann. N. Y. Acad. Sci. 2008, 1125, 171–189. [Google Scholar] [CrossRef]
- Mand, T.D.; Metcalf, W.W. Energy Conservation and Hydrogenase Function in Methanogenic Archaea, in Particular the Genus Methanosarcina. Microbiol. Mol. Biol. Rev. 2019, 83, e00020-19. [Google Scholar] [CrossRef]
- Xuan, J.; He, L.; Wen, W.; Feng, Y. Hydrogenase and Nitrogenase: Key Catalysts in Biohydrogen Production. Molecules 2023, 28, 1392. [Google Scholar] [CrossRef]
- Stephenson, M.; Stickland, L.H. Hydrogenase: A bacterial enzyme activating molecular hydrogen: The properties of the enzyme. Biochem. J. 1931, 25, 205–214. [Google Scholar] [CrossRef]
- Stephenson, M.; Stickland, L.H. Hydrogenase: The reduction of sulphate to sulphide by molecular hydrogen. Biochem. J. 1931, 25, 215–220. [Google Scholar] [CrossRef]
- Stephenson, M.; Stickland, L.H. Hydrogenase: The bacterial formation of methane by the reduction of one-carbon compounds by molecular hydrogen. Biochem. J. 1933, 27, 1517–1527. [Google Scholar] [CrossRef]
- Mand, T.D.; Kulkarni, G.; Metcalf, W.W. Genetic, Biochemical, and Molecular Characterization of Methanosarcina barkeri Mutants Lacking Three Distinct Classes of Hydrogenase. J. Bacteriol. 2018, 200, e00342-18. [Google Scholar] [CrossRef] [PubMed]
- Qiu, S.; Li, Q.; Xu, Y.; Shen, S.; Sun, C. Learning from nature: Understanding hydrogenase enzyme using computational approach. WIREs Comp. Mol. Sci. 2019, 10, e1422. [Google Scholar] [CrossRef]
- Cozannet, M.; Le Guellec, S.; Alain, K. A variety of substrates for methanogenesis. Case Stud. Chem. Environ. Eng. 2023, 8, 100533. [Google Scholar] [CrossRef]
- Kulkarni, G.; Kridelbaugh, D.M.; Guss, A.M.; Metcalf, W.W. Hydrogen is a preferred intermediate in the energy-conserving electron transport chain of Methanosarcina barkeri. Proc. Nat. Acad. Sci. USA 2009, 106, 15915–15920. [Google Scholar] [CrossRef] [PubMed]
- Hoehler, T.; Losey, N.A.; Gunsalus, R.P.; McInerney, M.J. Environmental Constraints that Limit Methanogenesis. In Biogenesis of Hydrocarbons; Stams, A., Sousa, D., Eds.; Handbook of Hydrocarbon and Lipid Microbiology; Springer: Cham, Switzerland, 2018; pp. 1–26. [Google Scholar]
- Eberly, J.O.; Ely, E.L. Thermotolerant Hydrogenases: Biological Diversity, Properties, and Biotechnological Applications. Crit. Rev. Microbiol. 2008, 34, 117–130. [Google Scholar] [CrossRef]
- Ogata, H.; Lubitz, W.; Higuchi, Q. Structure and function of [NiFe] hydrogenases. J. Biochem. 2016, 106, 251–258. [Google Scholar] [CrossRef]
- Vítězová, M.; Kohoutová, A.; Vítěz, T.; Hanišáková, N.; Kushkevych, I. Methanogenic Microorganisms in Industrial Wastewater Anaerobic Treatment. Processes 2020, 8, 1546. [Google Scholar] [CrossRef]
- Chen, S.; Cheng, H.; Wyckoff, K.N.; He, Q. Linkages of Firmicutes and Bacteroidetes popu-lations to methanogenic process performance. J. Ind. Microbiol. Biotechnol. 2016, 43, 771–781. [Google Scholar] [CrossRef]
- Harirchi, S.; Wainaina, S.; Sar, T.; Nojoumi, S.A.; Parchami, M.; Parchami, M.; Varjani, S.; Khanal, S.K.; Wong, J.; Awasthi, M.K.; et al. Microbiological insights into an-aerobic digestion for biogas, hydrogen or volatile fatty acids (VFAs): A review. Bioengineered 2022, 13, 6521–6557. [Google Scholar] [CrossRef]
- Li, Y.; Meng, Z.; Xu, Y.; Shi, Q.; Ma, Y.; Aung, M.; Cheng, Y.; Zhu, W. Interactions between Anaerobic Fungi and Methanogens in the Rumen and Their Biotechnological Potential in Biogas Production from Lignocellulosic Materials. Microorganisms 2021, 9, 190. [Google Scholar] [CrossRef]
- Rao, A.G.; Reddy, T.S.K.; Parakash, S.S.; Vanajakshi, J.; Joseph, J.; Sarma, P.N. pH regulation of alkaline wastewater with carbon dioxide: A case study of treatment of brewery wastewater in UASB reactor coupled with absorber. Bioresour. Technol. 2007, 98, 2131–2136. [Google Scholar] [CrossRef] [PubMed]
- Paulo, L.M.; Stams, A.J.M.; Sousa, D.Z. Methanogens, sulphate and heavy metals: A complex system. Rev. Environ. Sci. Biotechnol. 2015, 14, 537–553. [Google Scholar] [CrossRef]
- Martin, M.R.; Fornero, J.J.; Stark, R.; Mets, L.; Angenent, L.T. A Single-Culture Bioprocess of Methanothermobacter thermautotrophicus to Upgrade Digester Biogas by CO2-to-CH4 Conversion with H2. Archaea 2013, 2013, 157529. [Google Scholar] [CrossRef] [PubMed]
- Contreras, G.; Thomsen, J.; Pfitzer, M.; Hafenbradl, D.; Kostner, D.; Holtmann, D.; Scmitz, R.A.; Rother, M.; Molitor, B. New perspectives for biotechnological applications of methanogens. Curr. Res. Biotechnol. 2022, 4, 468–474. [Google Scholar] [CrossRef]
- Wulf, C.; Zapp, P.; Schreiber, A. Review of Power-to-X Demonstration Projects in Europe. Front. Energy Res. 2020, 8, 191. [Google Scholar] [CrossRef]
- Enzmann, F.; Mayer, F.; Rother, M.; Holtmann, D. Methanogens: Biochemical background and biotechnological applications. ABM Express 2018, 8, 1. [Google Scholar] [CrossRef]

| Methanogens with Cytochromes | Methanogens Without Cytochromes | Ref. |
|---|---|---|
| contain methanophenazine | do not contain methanophenazine | [4] |
| only some grow on CO2 and H2; most grow on methylamines and acetate; cannot grow on formate | grow on CO2 and H2; cannot grow on methylamines and acetate; most can grow on formate | |
| growth yields on CO2 and H2 ≤ 7 g/mol CH4 | growth yields on CO2 and H2 ≤ 3 g/mol CH4 | |
| H2 partial pressure threshold is >10 Pa | H2 partial pressure threshold is <10 Pa | |
| not hyperthermophilic | hyperthermophilic |
| Substrate | Methanogen Order | Ref. |
|---|---|---|
| pyruvate | Methanococcales, Methanosarcinales | [23] |
| carbon monoxide | Methanosarcinales | |
| primary and secondary alcohols (ethanol, 2-propanol and 2-butanol) | Methanomicrobiales | |
| methylated amine compounds (monomethylamine, dimethylamine, trimethylamine, choline and glycine betaine) | Methanosarcinales | |
| organosulfur compounds (methanethiol and dimethylsulfide) | Methanosarcinales | |
| metoxylated aromatic compounds | Methanosarcinales, Methanomicrobiales |
| Coenzymes | Functional Role | Ref. |
|---|---|---|
| methanofuran (MF) | carriers of carbon moiety for generation of methane | [1] |
| tetrahydromethanopterin (H4MPT) | ||
| tetrahydrosarcinapterin (H4SPT) | ||
| coenzyme M (HS-CoM) | ||
| coenzyme B (HS-CoB) | transferring of electrons for carbon reduction | |
| coenzyme F420 | ||
| coenzyme F430 | ||
| methanophenazine |
| Reactor Type | Retention Times | Examples | Ref. |
|---|---|---|---|
| Type A | RTm = RTs = RTl | closed digester tank, continuously stirred tank reactor | [3] |
| Type B | RTm, RTs > RTl | upflow anaerobic sludge bed reactor, closed digester tank with solid recycle | |
| Type C | RTm > RTs, RTl | membrane bioreactor, upflow anaerobic sludge fixed film reactor |
| Industry Branch | Amount of Waste Produced | Characteristics of Wastewater | Methanogens | Reactor Type | Ref. |
|---|---|---|---|---|---|
| Beer industry | 3–10 L of wastewater per 1 L of beer | high level of oxygen demand pH from 4.5 to 12 temperatures from 18–40 °C | Methanosaeta concilii Methanosarcina mazei | B | [3,32] |
| Oil extraction | 0.5–0.75 t of palm oil mill effluent per 1 t of oil palm branch | high level of oxygen demand pH from 4 to 5 temperatures from 80 to 90 °C | Methanosaeta concilii | A, B, C | [3] |
| 1200–1800 L of olive mill wastewater per 1 t of olives | high level of oxygen demand pH around 5 high temperatures | Methanosaeta concilii Methanobacterium formicicum | B | [3] | |
| Paper industry | at least 30,000 L of wastewater per 1 t of paper pulp | high level of oxygen demand fluctuating pH temperatures from 50 to 60 °C | Methanosarcina barkeri | B | [3] |
| Dairy industry | 500–2000 L of wastewater per 1 L of milk | high level of oxygen demand pH from 5.7 to 7.8 temperatures from 30 to 40 °C | Methanosaeta spp. | B | [3,11,28] |
| Fruit and vegetable processing industry | n.a. | low level of oxygen demand low pH temperature range—n.a. | Methanothrix spp. | B | [28] |
| Slaughterhouse wastewater | 90–140 L of wastewater per 1 slaughtered pig; although variable with type of animal | high level of oxygen demand fluctuating pH temperature range—n.a. | Methanobacterium Methanosarcinales | n.a. | [28] |
| Chemical industry | variable with the type of chemical produced | variable with the type of chemical produced | Methanothrix spp. | B, C | [28] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Marić, A.-K.; Sudar, M.; Findrik Blažević, Z.; Vuković Domanovac, M. Methanogenesis—General Principles and Application in Wastewater Remediation. Energies 2024, 17, 5374. https://doi.org/10.3390/en17215374
Marić A-K, Sudar M, Findrik Blažević Z, Vuković Domanovac M. Methanogenesis—General Principles and Application in Wastewater Remediation. Energies. 2024; 17(21):5374. https://doi.org/10.3390/en17215374
Chicago/Turabian StyleMarić, Ana-Katarina, Martina Sudar, Zvjezdana Findrik Blažević, and Marija Vuković Domanovac. 2024. "Methanogenesis—General Principles and Application in Wastewater Remediation" Energies 17, no. 21: 5374. https://doi.org/10.3390/en17215374
APA StyleMarić, A.-K., Sudar, M., Findrik Blažević, Z., & Vuković Domanovac, M. (2024). Methanogenesis—General Principles and Application in Wastewater Remediation. Energies, 17(21), 5374. https://doi.org/10.3390/en17215374







