1. Introduction
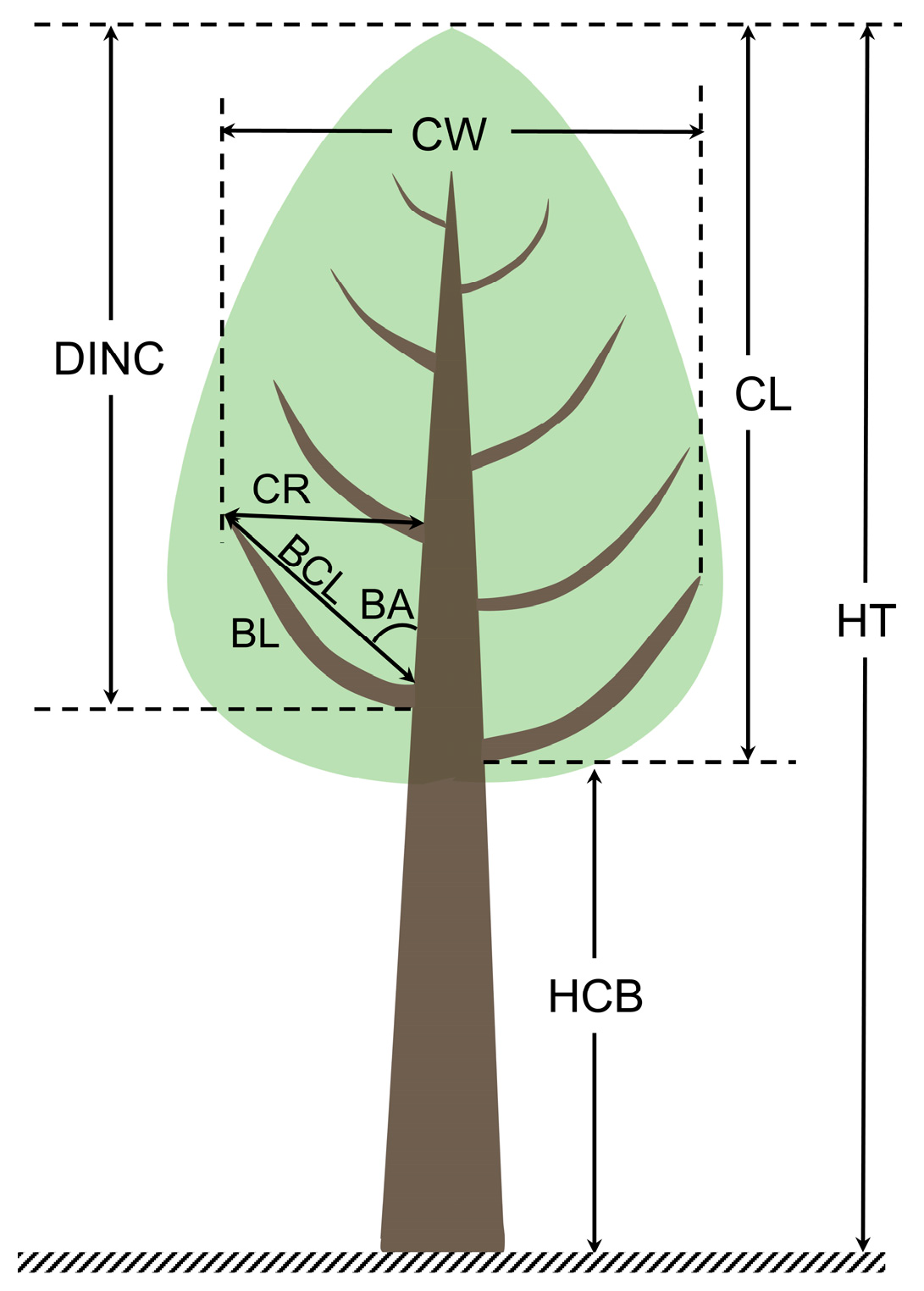
The crown vertical profile (CVP) of a tree is the minimum boundary surrounding the crown that connects the tree top and the branch vertices [
1]. It reflects the shape and size of the crown and directly describes peoples’ overall impression of a tree. The CVP has become one of the most critical forest structure parameters, both at the sample plot and tree levels, and it lays the foundation for exploring the spatial attributes of tree crowns, such as the crown volume and surface area [
2,
3]. It can reflect the horizontal distribution of the leaf quantity and biomass in the tree crown [
4] and can also function as an indicator for forest fire prediction models [
5,
6,
7]. Therefore, the accurate simulation of the trees’ CVP holds great significance for forest management and fire prediction.
Numerous models have been developed to describe trees’ CVPs [
8]. The crown radius (CR) is an essential variable for developing CVP models; however, the challenge of accurately obtaining CR information remains unresolved. In the past, it was frequently necessary to destructively harvest trees first, and then measure the branch attributes, and finally, use trigonometric relationships to compute the crown radius [
9,
10]. This field measurement allows the accurate acquisition of crown radius values for any height and is undoubtedly one of the most reliable and efficient methods [
1]. However, field measurements can be labor- and material-intensive and time-consuming [
11], and even natural forests are not allowed to be destructively harvested due to the natural forest protection policy [
12] in China. Remote sensing technology has been developed to provide a new solution for detecting forest parameters with better time efficiency and cost efficiency, and it is regarded as an alternative way to solve the limitations of field surveys [
13].
As an active remote sensing approach, LiDAR has emerged as a feasible tool for extracting crucial forest parameters [
14] since it can penetrate the forest canopy to reach the ground and provide vertical structure and topography information [
15]. Over the past two decades, LiDAR platforms have been rapidly developed, including satellite platforms (CALIOP), airborne platforms (airborne laser scanning (ALS)), and near-ground platforms (terrestrial laser scanning (TLS) and backpack laser scanning (BLS)), which provide technical support for monitoring a wide range of forest dynamics changes. LiDAR enables the capture of sensitive and detailed forest structure information, consisting of sample plot and tree level crown and trunk attribute measurements [
16,
17,
18,
19,
20,
21]. Furthermore, many efficient individual tree detection (ITD) [
22,
23,
24,
25,
26] and individual tree crown detection (ITCD) algorithms [
27,
28,
29,
30,
31,
32] have been gradually developed to improve the ability to describe individual tree structures in detail and to lay the foundation for extracting CVPs.
Terrestrial laser scanning (TLS) can provide high-accuracy and high-density point cloud data, which have been used to restore the 3D structure of individual trees in detail [
33]. TLS has been utilized by researchers to extract common individual tree attribute metrics, such as diameter at the breast height (DBH), tree height, and position of individual trees [
34,
35,
36,
37,
38]. It also enables the description of crown attributes, such as the crown width, crown volume, crown surface area [
39,
40,
41,
42], and other crown metrics [
43], which provide direct evidence to rationalize the effects caused by competition on the crown. Additionally, TLS also allows the calculation of individual tree volume [
44,
45] and biomass [
46,
47]. Recent research has demonstrated that the detailed structure of tree trunks and branches can be reconstructed using TLS data and quantitative structural models (QSM) [
48,
49,
50,
51,
52,
53]. As a result, QSM can be utilized to examine whole-tree topology and indirectly determine the tree wood volume and aboveground biomass [
47,
54]. However, surprisingly, a few studies have used TLS to develop CVP models, even though TLS can accurately characterize tree metrics.
To date, researchers have mainly used the ‘cumulative width percentile’ in layered point cloud data to determine the outer contour points of the crown point cloud, and then defined the horizontal distance from the outer contour points to the center of the trunk as the CR for developing the CVP model. For instance, Ferrarese [
8] developed a regional CVP model for three conifer species in the interior of the north-western United States by using a point cloud at the 95th percentile in a point cloud height bin of increments of 0.25 m as the outer CVP points. This is the first documented report on the application of TLS to develop a CVP model. Quan [
1] et al. developed a CVP model for larch in north-eastern China based on UAV-LS data. The findings of the comparison between the prediction results and those of previously published CVP models developed using field measurement data showed that there was strong agreement between them. Both research findings imply that developing tree CVP models based on point cloud data have great potential. However, none of the above studies provide the specific accuracy of extracting outer crown contour points (or crown radius) based on point clouds, and strong evidence for using point cloud data instead of actual measurement data for individual tree attribute studies is not provided.
Previously developed CVP models mainly used ordinary least squares (OLS) regression to estimate the model parameters [
55,
56,
57,
58], which ignored random errors arising from different levels due to the usual hierarchical nested structure of the modeling data, resulting in the poor prediction accuracy of the models. The mixed-effects model consists of two parts, fixed effects and random effects [
59,
60], which can not only estimate the average trend of the overall sample, but also reflect the differences between different ‘levels’, thus significantly improving the prediction accuracy of the model. Gao [
61] developed a mixed-effects model of the plot-level CVP using field-measured branch data from 49 larch plants to explain the differences in CVPs between different sites. Zhao [
62] et al. developed a two-level CVP mixed-effects model using field-measured branch data from 58 northern Chinese larch plants. The results showed that the prediction effect of the two-level CVP mixed-effects model was significantly more effective than that of the one-level mixed-effects model. However, the above researchers developed mixed-effects models for CVPs by selecting numerous sample trees from the sample plots for destructive harvesting to obtain modeling data. We have no idea whether these selected sample trees can adequately represent the growth conditions of all trees in the sample plots; therefore, the mixed-effects models they developed are limited in terms of representativeness.
Overall, due to the expensive acquisition of data, previous studies on CVP models have primarily focused on a limited number of trees, which has resulted in an inability to adequately characterize the crown at the regional level. Data from the TLS can produce a larger, more detailed sample, which makes it possible to develop regional-level CVP models. Therefore, in this paper, the main objective is to develop a CVP model using TLS at the sample plot scale to complement an individual tree-based CVP model and improve its applicability and prediction accuracy. The specific objectives are (1) to propose a method for the automatic extraction of crown radii at different heights using TLS point cloud data; (2) to develop a two-level mixed-effects model for the CVP using the extracted crown radii. This study provides an efficient method to accurately access crown information and enrich the means of conducting forest surveys. At the same time, our results can contribute theoretical support to accurately characterize crown characteristics at the regional level, which is beneficial for forest managers to develop reasonable forest management strategies.
4. Discussion
In the traditional approach, destructive harvesting is often required to obtain branch factors to simulate trends in the shape of the CVP [
9,
57,
58,
61,
89]. TLS, as an active remote sensing technique, can truly and effectively restore the 3D structure of individual trees using massive point cloud data [
33,
48,
73,
93], which provides a new perspective for describing the trees’ CVP. In this study, we propose a method to automatically extract crown radii at different heights based on TLS point cloud data. The validity of the method was then verified using field measurement data from destructive harvesting. Finally, we developed a two-level nonlinear mixed-effects model for simulating the changing trend of planted Korean pine CVPs using the CRs extracted from point cloud data.
The focus of the study in this paper is on the crown portion of the tree; so, it is essential to accurately determine the HCBs. Numerous studies have demonstrated the effectiveness of automatic HCB detection using ALS point cloud data at both stand and individual tree scales [
11,
71,
72]. In contrast, the automatic detection of HCBs using TLS point cloud data has not been reported very often [
94]. In this study, HCBs were automatically detected in the plantation based on TLS point cloud data, and the results of the study showed a good correlation between the detected HCBs and the field measurements (
Figure 9), which is consistent with the reported results in the literature [
11]; however, we produced better R
2 and RMSE values. This is expected because TLS is more suitable for the accurate extraction of HCB information from forest stands because of its ability to produce a higher point cloud density in the lower forest canopy compared to that of ALS [
95]. The results of HCB detection in sample plots with different stand densities (
Table 3) indicated that the extraction method was not affected by stand density and was highly applicable and robust. The detection of HCBs via TLS underestimated the HCBs overall, with a bias of −0.19 m. One reasonable explanation is interference from the presence of dead branches on individual trees [
8]. Wang [
96] and Béland [
97] have conducted research and reported that geometric or radiometric intensity features can be used to distinguish between leaves and branches on coniferous and broadleaf trees. Therefore, in future studies, considering the automatic detection of HCBs using geometric [
96,
98] and intensity [
97,
99] information will greatly improve the accuracy of HCB extraction.
In previous literature that used point cloud data to study CVP models, CRs were calculated as the horizontal distance from the crown’s outermost point cloud to the vertical line where the apex of the tree is located [
1]. This ignores the effect of the degree of tree tilt and results in a large error between the calculated CR values and the actual CR values. In this study, we simulated the measurement of destructive harvesting in the field [
9,
58,
61], in which the CRs were calculated as the distance from the crown’s outermost point cloud to the trunk surface. We detected circles from the crown point cloud projection using the RANSAC algorithm and treated them as tree trunks; the RANSAC algorithm has been demonstrated to be very suitable for extracting trunk information [
74,
75,
100]. This reduced the error caused by the large degree of tree tilt to a certain extent and, at the same time, it can be better compared with the field measurement data to analyze the accuracy of CR extraction.
The results of this paper indicate a strong correlation between the CRs extracted using TLS data and actual field measurement data (
Figure 11). Although TLS overestimates CRs in general, such a bias is within acceptable limits considering the time and effort required to obtain CRs in the field. The extraction results of the maximum CR in different RDINC ranges were generally better, but there were still large differences in the extraction accuracies between the crown top and the crown bottom, where the extraction accuracy of the crown bottom was better than that of the crown top (
Figure 10). Quan [
101] and Xu [
102] et al. used UAV-LS point cloud data to extract CRs at different heights of
Larix olgensis and
Cunninghamia lanceolata (Lamb.) Hook., respectively, but they did not provide specific extraction accuracies because they did not have actual field measurement data for reference. Li [
86] et al. extracted the trunk diameters of
Larix olgensis at different heights in the vertical direction based on TLS data, and they found that the accuracy of the trunk diameter extraction decreased sharply when the relative height exceeded 0.9. In this study, the extraction accuracy of CRs at different heights was analyzed explicitly with reference to the branch factor data measured in a field containing 30 planted Korean pine trees harvested via destructive harvesting (
Table 2). The extraction accuracy was low in the RDINC range of 0–0.15, stabilized from an RDINC of 0.15, and it remained at approximately 91%, which is consistent with the findings of Li [
86]. Although trunk diameter extraction was not performed in this study, it was studied in the vertical direction using point cloud data; so, it can be compared with the results of Li’s study for mutual verification. TLS applies a bottom-up scanning method [
95,
103], which gradually becomes less effective as the trees become taller. Due to the mutual occlusion of tree crowns, it further increases the noise of the top crown point cloud, which eventually leads to a decrease in the accuracy of the CRs extracted at the top of the tree crown. It has been demonstrated that the fusion of UAV-LS and TLS data can restore the crown structure more clearly and completely [
104]. Therefore, it will be our future goal to improve the extraction accuracy of tree crown structure information by combining LiDAR data from multiple platforms in future research.
Most of the data for forest stand growth modeling have a hierarchical structure, and nonlinear mixed-effects models are an effective approach to address this problem [
90,
91] and have been widely used in the development of tree CVP models [
61]. The subject of this paper was 283 individual trees from six sample plots with different attributes, which makes it very suitable for developing a mixed-effects model. With the aim of adequately explaining the random disturbances caused by sample plots and sample trees on the trees’ CVP, we developed tree-level, plot-level, and two-level mixed-effects models. In the process of fitting a mixed-effects model, the model can have difficulty converging as the random effects increase [
105]. We have adequately considered all combinations of random effect parameters, and the results show that the model does not converge when there are more than three random effect parameters. The mixed-effects model has a significantly better fit than the base model does (
Table 8), demonstrating that the introduction of random effects can greatly improve the prediction accuracy of the model, which is consistent with the research results of Gao [
61] and Liu [
105] et al. Ferrarese [
8] et al. reported that the CVP was not strongly influenced by the tree size and sample site conditions. This is because their study samples were biased toward more open stand conditions and bare canopies. The plot-level mixed-effects model we developed reflects differences between the sample plots, such as the stand density, indicating that all sample plot factors have some effect on the CVP. The tree-level mixed-effects model outperformed the sample plot-level mixed-effects model, which indicates that the differences in the CVP models were mainly due to random disturbances caused by different size sample trees. The two-level mixed-effects model had the best performance compared to those of the plot-level mixed-effects model and the tree-level mixed-effects model, which demonstrates that considering both the plot-level and tree-level effects was more effective at adequately explaining the random influence on the tree crown outer profile model.
Roeh [
106] and Cluzeau [
58] chose to use an indirect method to describe the CVP, as they considered that there were many difficulties in directly measuring the crown radius at different heights. TLS can provide detailed information regarding the crown structure, which is very suitable for directly describing the CVP [
8], and thus, successfully overcomes the abovementioned difficulties. The previously developed CVP models based on field measurements usually require the destructive harvesting of trees, and as a result, the study sample is very limited, e.g., (Marshall [
9], 36 western hemlock; Doruska [
107], 34 loblolly pine; Linnell Nemec [
49], 45 amabilis fir, 60 lodgepole pine, and 60 white spruce). In this study, we automatically extracted the crown radii of different heights from 283 individual trees in six sample plots using TLS without destructive sampling. It produced a large sample size and a representative dataset. A two-level mixed-effects model for the CVP was then developed based on this dataset, which greatly improved the predictive capability of the model. Overall, the results of this paper demonstrate that TLS can replace the traditional method of measuring crown structures and improve the ability to describe the CVP.
It is worth emphasizing that our proposed method is more applicable to complete crown point clouds. The subject of this study is a planted Korean pine forest, where there is less crossover between crowns; so, the crown point cloud data we obtained are complete. Using the proposed method, the crown radius of each individual tree can be extracted more successfully, which is conducive to the development of high-precision crown vertical profile models. In more complex mixed forests, crowns are difficult to be completely segmented due to the serious crossover phenomenon between the crowns. Since the crown point cloud itself is incomplete, the extracted crown radius also does not match the reality, which will seriously affect the prediction accuracy of the developed model. Therefore, a more robust crown segmentation algorithm suitable for high-density mixed forests would improve the applicability of the proposed approach. It is also an interesting topic for our future research.