Outcome Prediction Using Multi-Modal Information: Integrating Large Language Model-Extracted Clinical Information and Image Analysis
Abstract
:Simple Summary
Abstract
1. Introduction
2. Materials and Methods
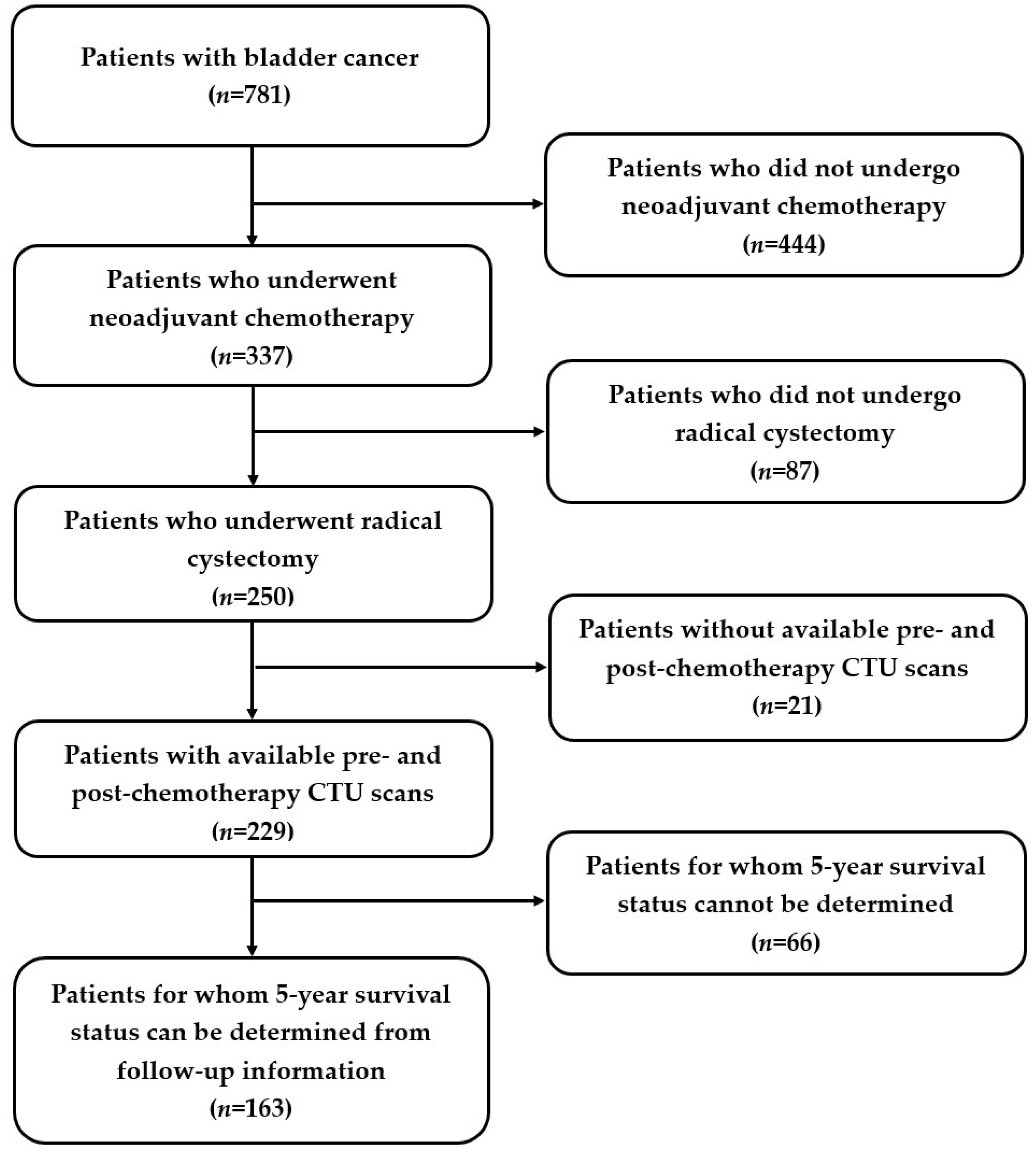
2.1. Patient Cohorts
2.2. Predictive Models
2.2.1. Clinical (C) Descriptors
- User1 prompt: “Please find the pathologic stage (revealed by cystectomy or in the assessment section), Pathologic node stage, lymphovascular invasion (LVI), Angiolymphatic invasion”.
- User2 prompt: “Using only the following information, find the pathologic stage (revealed by cystectomy or in the assessment section), node stage, angiolymphatic or lymphovascular invasion (LVI), and write the answers in the format of a list. If the answers are not in the information, write “unspecified””.
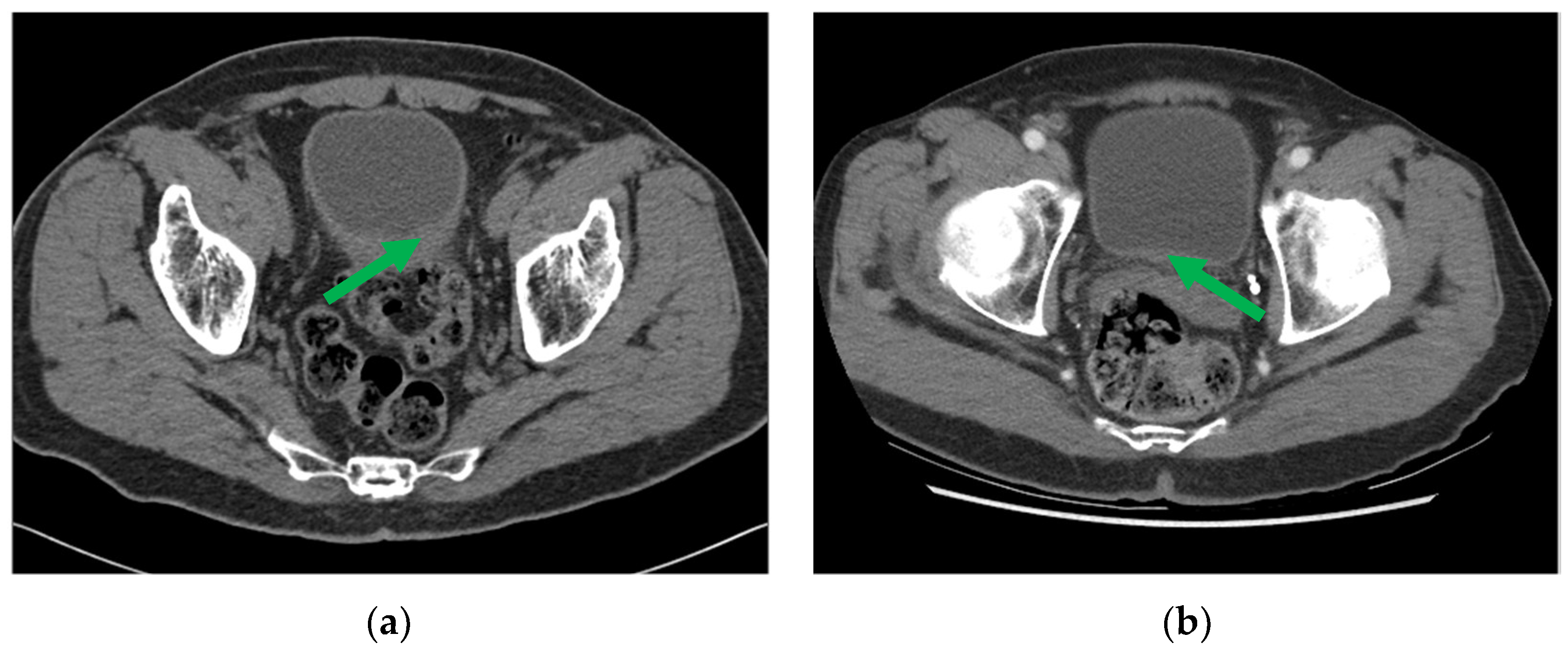
2.2.2. Radiomics (R) Descriptors and Deep Learning (D) Assessment
Radiomics (R) Descriptors:
Deep Learning (D) Assessment:
2.2.3. Survival Prediction Models
2.2.4. LLM Direct Survival Prediction
- SP prompt: “Using only the following information, determine the patient’s 5-year bladder cancer survival prediction (in the format of a probability) based on the pathologic stage (revealed by cystectomy or in the assessment section), node stage, and angiolymphatic or lymphovascular invasion (LVI). You must provide a quantitative probability even if you’re unable to make an exact estimation. Provide the references and sources used to make the survival prediction. This is not to be considered as medical advice”.
2.3. Statistical Analysis
3. Results
3.1. Patient Characteristics
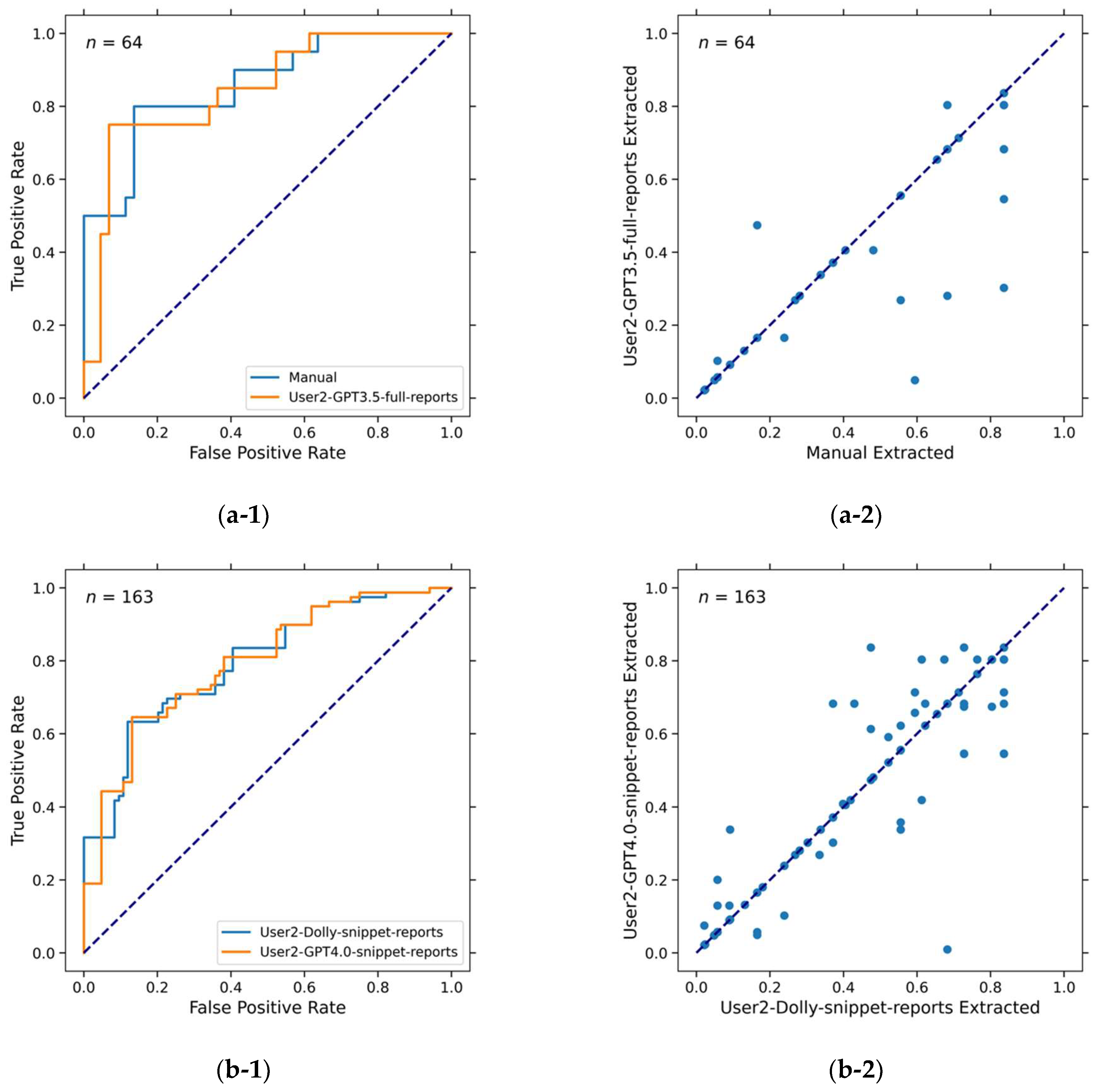
3.2. LLM Accuracy in Extracting Information
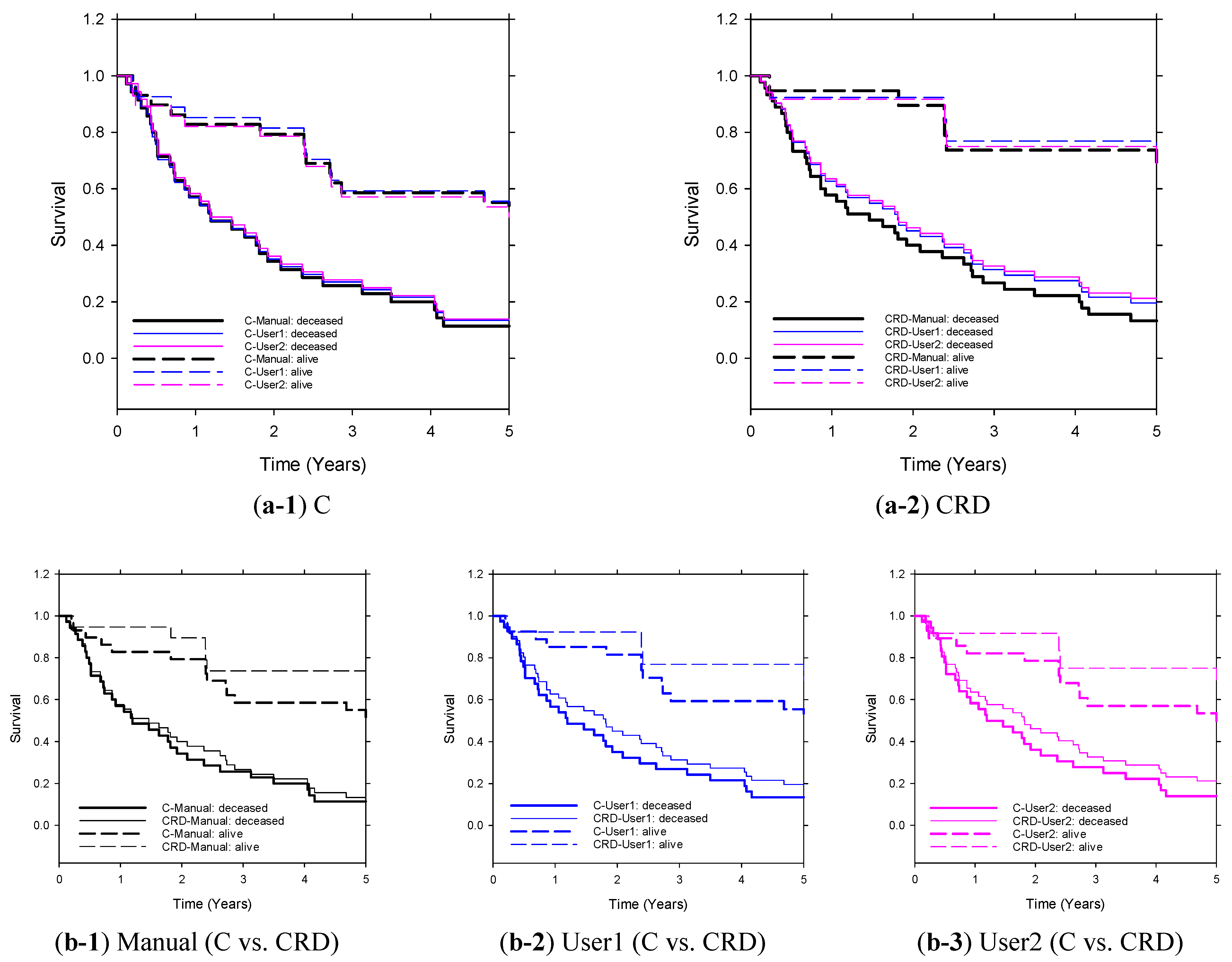
3.3. Five-Year Survival Prediction
3.4. LLM Direct Survival Prediction
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- National Cancer Institute: SEER Cancer Stat Facts: Bladder Cancer. Bethesda, Md: National Cancer Institute. Available online: https://seer.cancer.gov/statfacts/html/urinb.html (accessed on 1 April 2024).
- Sun, D.; Hadjiiski, L.; Gormley, J.; Chan, H.-P.; Caoili, E.M.; Cohan, R.H.; Alva, A.; Gulani, V.; Zhou, C. Survival Prediction of Patients with Bladder Cancer after Cystectomy Based on Clinical, Radiomics, and Deep-Learning Descriptors. Cancers 2023, 15, 4372. [Google Scholar] [CrossRef] [PubMed]
- Eisenberg, M.S.; Boorjian, S.A.; Cheville, J.C.; Thompson, R.H.; Thapa, P.; Kaushik, D.; Frank, I. The SPARC score: A multifactorial outcome prediction model for patients undergoing radical cystectomy for bladder cancer. J. Urol. 2013, 190, 2005–2010. [Google Scholar] [CrossRef] [PubMed]
- Zheng, Q.; Yang, R.; Ni, X.; Yang, S.; Xiong, L.; Yan, D.; Xia, L.; Yuan, J.; Wang, J.; Jiao, P. Accurate Diagnosis and Survival Prediction of Bladder Cancer Using Deep Learning on Histological Slides. Cancers 2022, 14, 5807. [Google Scholar] [CrossRef]
- Riester, M.; Taylor, J.M.; Feifer, A.; Koppie, T.; Rosenberg, J.E.; Downey, R.J.; Bochner, B.H.; Michor, F. Combination of a novel gene expression signature with a clinical nomogram improves the prediction of survival in high-risk bladder cancer. Clin. Cancer Res. 2012, 18, 1323–1333. [Google Scholar] [CrossRef] [PubMed]
- Seiler, R.; Ashab, H.A.D.; Erho, N.; van Rhijn, B.W.; Winters, B.; Douglas, J.; van Kessel, K.E.; van de Putte, E.E.F.; Sommerlad, M.; Wang, N.Q.; et al. Impact of molecular subtypes in muscle-invasive bladder cancer on predicting response and survival after neoadjuvant chemotherapy. Eur. Urol. 2017, 72, 544–554. [Google Scholar] [CrossRef] [PubMed]
- Koga, F.; Fujii, Y.; Masuda, H.; Numao, N.; Yokoyama, M.; Ishioka, J.i.; Saito, K.; Kawakami, S.; Kihara, K. Pathology-based risk stratification of muscle-invasive bladder cancer patients undergoing cystectomy for persistent disease after induction chemoradiotherapy in bladder-sparing approaches. BJU Int. 2012, 110, E203–E208. [Google Scholar] [CrossRef] [PubMed]
- Wang, G.; Lam, K.-M.; Deng, Z.; Choi, K.-S. Prediction of mortality after radical cystectomy for bladder cancer by machine learning techniques. Comput. Biol. Med. 2015, 63, 124–132. [Google Scholar] [CrossRef] [PubMed]
- Xylinas, E.; Cha, E.; Sun, M.; Rink, M.; Trinh, Q.; Novara, G.; Green, D.; Pycha, A.; Fradet, Y.; Daneshmand, S.; et al. Risk stratification of pT1-3N0 patients after radical cystectomy for adjuvant chemotherapy counselling. Br. J. Cancer 2012, 107, 1826–1832. [Google Scholar] [CrossRef]
- Galsky, M.D.; Moshier, E.; Krege, S.; Lin, C.C.; Hahn, N.; Ecke, T.; Sonpavde, G.; Godbold, J.; Oh, W.K.; Bamias, A. Nomogram for predicting survival in patients with unresectable and/or metastatic urothelial cancer who are treated with cisplatin-based chemotherapy. Cancer 2013, 119, 3012–3019. [Google Scholar] [CrossRef]
- Shariat, S.F.; Karakiewicz, P.I.; Palapattu, G.S.; Amiel, G.E.; Lotan, Y.; Rogers, C.G.; Vazina, A.; Bastian, P.J.; Gupta, A.; Sagalowsky, A.I.; et al. Nomograms provide improved accuracy for predicting survival after radical cystectomy. Clin. Cancer Res. 2006, 12, 6663–6676. [Google Scholar] [CrossRef]
- Wang, J.; Wu, Y.; He, W.; Yang, B.; Gou, X. Nomogram for predicting overall survival of patients with bladder cancer: A population-based study. Int. J. Biol. Markers 2020, 35, 29–39. [Google Scholar] [CrossRef] [PubMed]
- Garg, R.K.; Urs, V.L.; Agarwal, A.A.; Chaudhary, S.K.; Paliwal, V.; Kar, S.K. Exploring the role of ChatGPT in patient care (diagnosis and treatment) and medical research: A systematic review. Health Promot. Perspect. 2023, 13, 183. [Google Scholar] [CrossRef] [PubMed]
- Levine, D.M.; Tuwani, R.; Kompa, B.; Varma, A.; Finlayson, S.G.; Mehrotra, A.; Beam, A. The diagnostic and triage accuracy of the GPT-3 artificial intelligence model. medRxiv 2023. [Google Scholar] [CrossRef]
- Eriksen, A.V.; Möller, S.; Ryg, J. Use of GPT-4 to diagnose complex clinical cases. NEJM-AI 2023, 1, AIp2300031. [Google Scholar] [CrossRef]
- Cai, Q.; Wang, H.; Li, Z.; Liu, X. A survey on multimodal data-driven smart healthcare systems: Approaches and applications. IEEE Access 2019, 7, 133583–133599. [Google Scholar] [CrossRef]
- Salvi, M.; Loh, H.W.; Seoni, S.; Barua, P.D.; García, S.; Molinari, F.; Acharya, U.R. Multi-modality approaches for medical support systems: A systematic review of the last decade. Inf. Fusion 2023, 103, 102134. [Google Scholar] [CrossRef]
- Chaudhary, K.; Poirion, O.B.; Lu, L.; Garmire, L.X. Deep learning–based multi-omics integration robustly predicts survival in liver cancer. Clin. Cancer Res. 2018, 24, 1248–1259. [Google Scholar] [CrossRef] [PubMed]
- Lin, P.; Lin, Y.-Q.; Gao, R.-Z.; Wan, W.-J.; He, Y.; Yang, H. Integrative radiomics and transcriptomics analyses reveal subtype characterization of non-small cell lung cancer. Eur. Radiol. 2023, 33, 6414–6425. [Google Scholar] [CrossRef]
- Subramanian, I.; Verma, S.; Kumar, S.; Jere, A.; Anamika, K. Multi-omics data integration, interpretation, and its application. Bioinform. Biol. Insights 2020, 14, 1177932219899051. [Google Scholar] [CrossRef]
- Shao, W.; Wang, T.; Sun, L.; Dong, T.; Han, Z.; Huang, Z.; Zhang, J.; Zhang, D.; Huang, K. Multi-task multi-modal learning for joint diagnosis and prognosis of human cancers. Med. Image Anal. 2020, 65, 101795. [Google Scholar] [CrossRef]
- Li, S.; Zhou, B. A review of radiomics and genomics applications in cancers: The way towards precision medicine. Radiat. Oncol. 2022, 17, 217. [Google Scholar] [CrossRef]
- Conover, M.; Hayes, M.; Mathur, A.; Meng, X.; Xie, J.; Wan, J.; Shah, S.; Ghodsi, A.; Wendell, P.; Zaharia, M. Free Dolly: Introducing the World’s First Truly open Instruction-Tuned Llm...Databricks Blog. Available online: https://www.databricks.com/blog/2023/04/12/dolly-first-open-commercially-viable-instruction-tuned-llm (accessed on 1 April 2024).
- Chiang, W.-L.; Li, Z.; Lin, Z.; Sheng, Y.; Wu, Z.; Zhang, H.; Zheng, L.; Zhuang, S.; Zhuang, Y.; Gonzalez, J.E.; et al. Vicuna: An Open-Source Chatbot Impressing Gpt-4 with 90%* Chatgpt Quality. Available online: https://vicuna.lmsys.org (accessed on 14 April 2023).
- Touvron, H.; Martin, L.; Stone, K.; Albert, P.; Almahairi, A.; Babaei, Y.; Bashlykov, N.; Batra, S.; Bhargava, P.; Bhosale, S.; et al. Llama 2: Open foundation and fine-tuned chat models. arXiv 2023, arXiv:2307.09288. [Google Scholar] [CrossRef]
- Brown, T.; Mann, B.; Ryder, N.; Subbiah, M.; Kaplan, J.D.; Dhariwal, P.; Neelakantan, A.; Shyam, P.; Sastry, G.; Askell, A.; et al. Language models are few-shot learners. Adv. Neural Inf. Process. Syst. 2020, 33, 1877–1901. [Google Scholar]
- OpenAI. GPT-4 Technical Report. 2023. Available online: https://arxiv.org/abs/2303.08774 (accessed on 1 April 2024).
- Hadjiiski, L.; Chan, H.-P.; Caoili, E.M.; Cohan, R.H.; Wei, J.; Zhou, C. Auto-initialized cascaded level set (AI-CALS) segmentation of bladder lesions on multidetector row CT urography. Acad. Radiol. 2013, 20, 148–155. [Google Scholar] [CrossRef] [PubMed]
- Kirby, J.S.; Armato, S.G.; Drukker, K.; Li, F.; Hadjiiski, L.; Tourassi, G.D.; Clarke, L.P.; Engelmann, R.M.; Giger, M.L.; Redmond, G.; et al. LUNGx Challenge for computerized lung nodule classification. J. Med. Imaging 2016, 3, 044506. [Google Scholar] [CrossRef]
- Sun, D.; Hadjiiski, L.; Alva, A.; Zakharia, Y.; Joshi, M.; Chan, H.-P.; Garje, R.; Pomerantz, L.; Elhag, D.; Cohan, R.H.; et al. Computerized decision support for bladder cancer treatment response assessment in CT urography: Effect on diagnostic accuracy in multi-institution multi-specialty study. Tomography. 2022, 8, 644–656. [Google Scholar] [CrossRef]
- Bennasar, M.; Hicks, Y.; Setchi, R. Feature selection using joint mutual information maximisation. Expert Syst. Appl. 2015, 42, 8520–8532. [Google Scholar] [CrossRef]
- Szegedy, C.; Liu, W.; Jia, Y.; Sermanet, P.; Reed, S.; Anguelov, D.; Erhan, D.; Vanhoucke, V.; Rabinovich, A. Going deeper with convolutions. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, Boston, MA, USA, 7–12 June 2015; pp. 1–9. [Google Scholar]
- Goh, A.T. Back-propagation neural networks for modeling complex systems. Artif. Intell. Eng. 1995, 9, 143–151. [Google Scholar] [CrossRef]
- Metz, C.E.; Herman, B.A.; Shen, J.H. Maximum likelihood estimation of receiver operating characteristic (ROC) curves from continuously-distributed data. Stat. Med. 1998, 17, 1033–1053. [Google Scholar] [CrossRef]
- Gallas, B.D.; Stephen, L.H. Generalized Roe and Metz ROC Model: Analytic Link between Simulated Decision Scores and Empirical AUC Variances and Covariances. J. Med. Img. 2014, 1, 031006. [Google Scholar] [CrossRef]
- Bland, J.M.; Altman, D.G. Multiple significance tests: The Bonferroni method. BMJ 1995, 310, 170. [Google Scholar] [CrossRef] [PubMed]
- Altman, D.G. Practical Statistics for Medical Research; CRC Press: Boca Raton, FL, USA, 1990. [Google Scholar]
- Kluth, L.A.; Black, P.C.; Bochner, B.H.; Catto, J.; Lerner, S.P.; Stenzl, A.; Sylvester, R.; Vickers, A.J.; Xylinas, E.; Shariat, S.F. Prognostic and prediction tools in bladder cancer: A comprehensive review of the literature. Eur. Urol. 2015, 68, 238–253. [Google Scholar] [CrossRef] [PubMed]
- Borhani, S.; Borhani, R.; Kajdacsy-Balla, A. Artificial intelligence: A promising frontier in bladder cancer diagnosis and outcome prediction. Crit. Rev. Oncol. Hematol. 2022, 171, 103601. [Google Scholar] [CrossRef] [PubMed]


| Attributes | # of Patients | |
|---|---|---|
| Gender | Male | 131 |
| Female | 32 | |
| Age at surgery | Mean age ± standard deviation | 64 ± 9 |
| Tobacco use | Current | 40 |
| Former | 88 | |
| Never | 34 | |
| Unknown | 1 | |
| Pathologic T stage | pT0 | 35 |
| pTa/pTi/pTis | 15 | |
| pT1 | 16 | |
| pT2 | 36 | |
| pT3 | 45 | |
| pT4 | 16 | |
| Pathologic N stage | N0 | 112 |
| N1 | 24 | |
| N2 | 23 | |
| N3 | 4 | |
| LVI 1 | Yes | 61 |
| No | 102 | |
| Neoadjuvant chemotherapy | Yes | 163 |
| No | 0 | |
| Adjuvant radiotherapy | Yes | 0 |
| No | 163 |
| n = 163 | ||||||||
| Ground Truth | Prompt | Dolly | Vicuna | Llama | GPT-3.5 | GPT-3.5-full-report | GPT-4.0 | GPT-4.0-full-report |
| LLM 1 accuracy | ||||||||
| 100% | User1 | 74% | 76% | 82% | 87% | 85% | 97% | 94% |
| User2 | 87% | 83% | 93% | 91% | 91% | 96% | 95% | |
| AUC 2 ± standard deviation (C model 3) | ||||||||
| 0.78 ± 0.04 | User1 | 0.74 ± 0.04 | 0.70 ± 0.04 | 0.73 ± 0.04 | 0.76 ± 0.04 | 0.74 ± 0.04 | 0.77 ± 0.04 | 0.77 ± 0.04 |
| User2 | 0.77 ± 0.04 | 0.72 ± 0.04 | 0.75 ± 0.04 | 0.75 ± 0.04 | 0.77 ± 0.04 | 0.77 ± 0.04 | 0.77 ± 0.04 | |
| Test set (n = 64) | |||||||||
| Manual | Prompt | Dolly | Vicuna | Llama | GPT-3.5 | GPT-3.5-full-report | GPT-4.0 | GPT-4.0-full-report | |
| LLM 1 accuracy | |||||||||
| Average accuracy of pathologic T stage, N stage, and LVI 2 | - | User1 | 85% | 80% | 88% | 92% | 87% | 95% | 95% |
| - | User2 | 93% | 87% | 93% | 94% | 91% | 94% | 94% | |
| Pathologic T stage | - | User1 | 92% | 88% | 89% | 94% | 88% | 97% | 97% |
| - | User2 | 94% | 84% | 91% | 88% | 86% | 88% | 91% | |
| AUC 3 ± standard deviation | |||||||||
| C 4 | 0.82 ± 0.06 | User1 | 0.85 ± 0.05 | 0.80 ± 0.06 | 0.78 ± 0.06 | 0.84 ± 0.05 | 0.81 ± 0.06 | 0.82 ± 0.05 | 0.83 ± 0.05 |
| User2 | 0.85 ± 0.05 | 0.76 ± 0.06 | 0.80 ± 0.06 | 0.79 ± 0.06 | 0.82 ± 0.05 | 0.80 ± 0.06 | 0.82 ± 0.05 | ||
| CR 5 | 0.88 ± 0.04 | User1 | 0.86 ± 0.05 | 0.82 ± 0.05 | 0.81 ± 0.06 | 0.87 ± 0.04 | 0.81 ± 0.05 | 0.86 ± 0.05 | 0.88 ± 0.04 |
| User2 | 0.89 ± 0.04 | 0.81 ± 0.05 | 0.84 ± 0.05 | 0.84 ± 0.05 | 0.85 ± 0.05 | 0.85 ± 0.05 | 0.84 ± 0.05 | ||
| CD 6 | 0.85 ± 0.05 | User1 | 0.85 ± 0.05 | 0.81 ± 0.05 | 0.79 ± 0.06 | 0.86 ± 0.05 | 0.77 ± 0.06 | 0.83 ± 0.05 | 0.86 ± 0.05 |
| User2 | 0.84 ± 0.05 | 0.77 ± 0.06 | 0.80 ± 0.06 | 0.81 ± 0.05 | 0.82 ± 0.05 | 0.80 ± 0.05 | 0.81 ± 0.05 | ||
| CRD 7 | 0.89 ± 0.04 | User1 | 0.87 ± 0.05 | 0.84 ± 0.05 | 0.81 ± 0.06 | 0.88 ± 0.05 | 0.85 ± 0.05 | 0.87 ± 0.05 | 0.88 ± 0.04 |
| User2 | 0.87 ± 0.05 | 0.83 ± 0.06 | 0.86 ± 0.05 | 0.86 ± 0.05 | 0.86 ± 0.05 | 0.87 ± 0.05 | 0.88 ± 0.05 | ||
| n = 163 | n = 64 | |||
| Snippet report | Full report | Snippet report | Full report | |
| AUC 1 ± standard deviation | 0.76 ± 0.04 | 0.73 ± 0.04 | 0.81 ± 0.05 | 0.75 ± 0.07 |
| Source relevant | 68.0% (111/163) | 82.8% (135/163) | 62.5% (40/64) | 87.5% (56/64) |
| Source irrelevant (hallucination) | 32.0% (52/163) | 17.2% (28/163) | 37.5% (24/64) | 12.5% (8/64) |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Sun, D.; Hadjiiski, L.; Gormley, J.; Chan, H.-P.; Caoili, E.; Cohan, R.; Alva, A.; Bruno, G.; Mihalcea, R.; Zhou, C.; et al. Outcome Prediction Using Multi-Modal Information: Integrating Large Language Model-Extracted Clinical Information and Image Analysis. Cancers 2024, 16, 2402. https://doi.org/10.3390/cancers16132402
Sun D, Hadjiiski L, Gormley J, Chan H-P, Caoili E, Cohan R, Alva A, Bruno G, Mihalcea R, Zhou C, et al. Outcome Prediction Using Multi-Modal Information: Integrating Large Language Model-Extracted Clinical Information and Image Analysis. Cancers. 2024; 16(13):2402. https://doi.org/10.3390/cancers16132402
Chicago/Turabian StyleSun, Di, Lubomir Hadjiiski, John Gormley, Heang-Ping Chan, Elaine Caoili, Richard Cohan, Ajjai Alva, Grace Bruno, Rada Mihalcea, Chuan Zhou, and et al. 2024. "Outcome Prediction Using Multi-Modal Information: Integrating Large Language Model-Extracted Clinical Information and Image Analysis" Cancers 16, no. 13: 2402. https://doi.org/10.3390/cancers16132402
APA StyleSun, D., Hadjiiski, L., Gormley, J., Chan, H.-P., Caoili, E., Cohan, R., Alva, A., Bruno, G., Mihalcea, R., Zhou, C., & Gulani, V. (2024). Outcome Prediction Using Multi-Modal Information: Integrating Large Language Model-Extracted Clinical Information and Image Analysis. Cancers, 16(13), 2402. https://doi.org/10.3390/cancers16132402








