Development and Validation of a Deep Learning Model for Histopathological Slide Analysis in Lung Cancer Diagnosis
Abstract
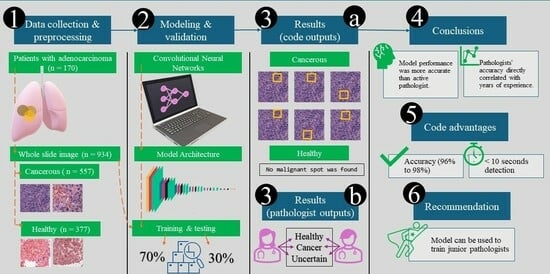
Simple Summary
Abstract
1. Introduction
2. Methodology
2.1. Data Collection and Preprocessing
- The training dataset (71% of total images, n = 662 images) comprised 401 cancerous and 261 healthy images.
- This was split further to training and validation datasets with 90% (n = 596 images) and 10% (n = 66 images). The training dataset was used to train the model’s parameters. In contrast, the validation dataset was used to fine-tune the model and optimize hyperparameters. We followed National Cancer Institute criteria to differentiate between normal and cancer cells [7].
- The test dataset (29% of total images, n = 272) included 156 cancerous and 116 healthy images. It was employed to assess the model’s final performance and generalization capability.
2.2. Model Selection
2.3. Separable Convolutional Neural Networks
2.4. Residual Blocks
2.5. Model Architecture
2.6. Model Optimization and Training
2.7. Statistical Analysis
3. Results
3.1. Accuracy and Validation
3.2. Model Application
3.3. Comparison of the Model Results with the Pathologist’s Decision
4. Discussion
4.1. Main Findings
4.2. Comparison with Existing ML Models in Lung Cancer Detection
4.3. Clinical Implications
4.4. Model Limitations
4.5. Ethical Considerations and Potential Social Impacts
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Luo, G.; Zhang, Y.; Etxeberria, J.; Arnold, M.; Cai, X.; Hao, Y.; Zou, H. Projections of Lung Cancer Incidence by 2035 in 40 Countries Worldwide: Population-Based Study. JMIR Public Health Surveill 2023, 9, e43651. [Google Scholar] [CrossRef] [PubMed]
- Lung Cancer Statistics—World Cancer Research Fund International. Mhtml. Available online: https://www.wcrf.org/cancer-trends/lung-cancer-statistics/ (accessed on 5 March 2024).
- Ahmed, A.A.; Abouzid, M.; Kaczmarek, E. Deep Learning Approaches in Histopathology. Cancers 2022, 14, 5264. [Google Scholar] [CrossRef] [PubMed]
- Ahmed, A.; Brychcy, A.; Abouzid, M.; Witt, M.; Kaczmarek, E. Perception of Pathologists in Poland of Artificial Intelligence and Machine Learning in Medical Diagnosis—A Cross-Sectional Study. JPM 2023, 13, 962. [Google Scholar] [CrossRef] [PubMed]
- Sebastian, A.M.; Peter, D. Artificial Intelligence in Cancer Research: Trends, Challenges and Future Directions. Life 2022, 12, 1991. [Google Scholar] [CrossRef] [PubMed]
- Gholamy, A.; Kreinovich, V.; Kosheleva, O. Why 70/30 or 80/20 Relation Between Training and Testing Sets: A Pedagogical Explanation. Int. J. Intell. Technol. Appl. Stat. 2018, 11, 105–111. [Google Scholar]
- What Is Cancer?—NCI. Available online: https://www.cancer.gov/about-cancer/understanding/what-is-cancer (accessed on 25 February 2024).
- Ketkar, N.; Moolayil, J. Convolutional Neural Networks. In Deep Learning with Python: Learn Best Practices of Deep Learning Models with PyTorch; Ketkar, N., Moolayil, J., Eds.; Apress: Berkeley, CA, USA, 2021; pp. 197–242. ISBN 978-1-4842-5364-9. [Google Scholar]
- Zhang, Z. Improved Adam Optimizer for Deep Neural Networks. In Proceedings of the 2018 IEEE/ACM 26th International Symposium on Quality of Service (IWQoS), Banff, AB, Canada, 4–6 June 2018; pp. 1–2. [Google Scholar]
- Wei, J.W.; Tafe, L.J.; Linnik, Y.A.; Vaickus, L.J.; Tomita, N.; Hassanpour, S. Pathologist-Level Classification of Histologic Patterns on Resected Lung Adenocarcinoma Slides with Deep Neural Networks. Sci. Rep. 2019, 9, 3358. [Google Scholar] [CrossRef] [PubMed]
- Gürsoy Çoruh, A.; Yenigün, B.; Uzun, Ç.; Kahya, Y.; Büyükceran, E.U.; Elhan, A.; Orhan, K.; Kayı Cangır, A. A Comparison of the Fusion Model of Deep Learning Neural Networks with Human Observation for Lung Nodule Detection and Classification. BJR 2021, 94, 20210222. [Google Scholar] [CrossRef] [PubMed]
- Wu, J.; Liu, C.; Liu, X.; Sun, W.; Li, L.; Gao, N.; Zhang, Y.; Yang, X.; Zhang, J.; Wang, H.; et al. Artificial Intelligence-Assisted System for Precision Diagnosis of PD-L1 Expression in Non-Small Cell Lung Cancer. Mod. Pathol. 2022, 35, 403–411. [Google Scholar] [CrossRef] [PubMed]
- Xie, Y.; Meng, W.-Y.; Li, R.-Z.; Wang, Y.-W.; Qian, X.; Chan, C.; Yu, Z.-F.; Fan, X.-X.; Pan, H.-D.; Xie, C.; et al. Early lung cancer diagnostic biomarker discovery by machine learning methods. Transl Oncol. 2021, 14, 100907. [Google Scholar] [CrossRef] [PubMed]
- Dritsas, E.; Trigka, M. Lung Cancer Risk Prediction with Machine Learning Models. BDCC 2022, 6, 139. [Google Scholar] [CrossRef]
- Ji, X.; Chen, J.; Ye, J.; Xu, S.; Lin, B.; Hou, K. Epidemiological Analysis of Global and Regional Lung Cancer Mortality: Based on 30-Year Data Analysis of Global Burden Disease Database. Healthcare 2023, 11, 2920. [Google Scholar] [CrossRef] [PubMed]
- Prof. Marszałek: Braki Kadrowe w Patomorfologii nie są Jedynie Polskim Problemem. Available online: https://pulsmedycyny.pl/prof-marszalek-braki-kadrowe-w-patomorfologii-nie-sa-jedynie-polskim-problemem-1149337 (accessed on 4 April 2024).
- Sauerbrei, A.; Kerasidou, A.; Lucivero, F.; Hallowell, N. The Impact of Artificial Intelligence on the Person-Centred, Doctor-Patient Relationship: Some Problems and Solutions. BMC Med. Inform. Decis. Mak. 2023, 23, 73. [Google Scholar] [CrossRef] [PubMed]
- Bajwa, J.; Munir, U.; Nori, A.; Williams, B. Artificial Intelligence in Healthcare: Transforming the Practice of Medicine. Future Healthc. J. 2021, 8, e188–e194. [Google Scholar] [CrossRef] [PubMed]
- Kiseleva, A.; Kotzinos, D.; De Hert, P. Transparency of AI in Healthcare as a Multilayered System of Accountabilities: Between Legal Requirements and Technical Limitations. Front. Artif. Intell. 2022, 5, 879603. [Google Scholar] [CrossRef] [PubMed]
- El-Sherif, D.M.; Abouzid, M.; Elzarif, M.T.; Ahmed, A.A.; Albakri, A.; Alshehri, M.M. Telehealth and Artificial Intelligence Insights into Healthcare during the COVID-19 Pandemic. Healthcare 2022, 10, 385. [Google Scholar] [CrossRef]
- Oh, S.; Kim, J.H.; Choi, S.-W.; Lee, H.J.; Hong, J.; Kwon, S.H. Physician Confidence in Artificial Intelligence: An Online Mobile Survey. J. Med. Internet Res. 2019, 21, e12422. [Google Scholar] [CrossRef]



| Dataset | Accuracy | AUC |
|---|---|---|
| Training dataset | 98% | 99% |
| Validation dataset | 96% | 97% |
| Testing dataset | 97% | 98% |
| Image nr. | Model | Pathologist Decision (n = 10) | Pathologist Slide Accuracy (%) | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Decision | Array | Accuracy(%) | P1 | P2 | P3 | P4 | P5 | P6 | P7 | P8 | P9 | P10 | ||
| 1 | H | <0.001 | 100 | H | 100 | |||||||||
| 2 | <0.001 | 100 | 100 | |||||||||||
| 3 | <0.001 | 100 | 100 | |||||||||||
| 4 | 0.05 | 100 | 100 | |||||||||||
| 5 | 0.004 | 100 | 100 | |||||||||||
| 6 | <0.001 | 100 | H | C | 90 | |||||||||
| 7 | C | 1 | 100 | UC | 0 | |||||||||
| 8 | 1 | 100 | UC | C | UC | C | UC | 20 | ||||||
| 9 | 1 | 100 | C | 100 | ||||||||||
| 10 | 1 | 100 | 100 | |||||||||||
| 11 | 1 | 100 | C | UC | 90 | |||||||||
| 12 | 1 | 100 | UC | C | 70 | |||||||||
| 13 | 1 | 100 | C | UC | C | UC | C | UC | C | 40 | ||||
| 14 | 1 | 100 | C | 100 | ||||||||||
| 15 | 1 | 100 | UC | C | 80 | |||||||||
| Pathologist year of experience | 11 | 11 | 14 | 12 | 12 | 14 | 12 | 11 | 13 | 10 | ||||
| Individual pathologist accuracy (%) | 73.33 | 73.33 | 93.33 | 80.00 | 80.00 | 93.33 | 80.00 | 73.33 | 86.67 | 66.67 | ||||
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ahmed, A.A.; Fawi, M.; Brychcy, A.; Abouzid, M.; Witt, M.; Kaczmarek, E. Development and Validation of a Deep Learning Model for Histopathological Slide Analysis in Lung Cancer Diagnosis. Cancers 2024, 16, 1506. https://doi.org/10.3390/cancers16081506
Ahmed AA, Fawi M, Brychcy A, Abouzid M, Witt M, Kaczmarek E. Development and Validation of a Deep Learning Model for Histopathological Slide Analysis in Lung Cancer Diagnosis. Cancers. 2024; 16(8):1506. https://doi.org/10.3390/cancers16081506
Chicago/Turabian StyleAhmed, Alhassan Ali, Muhammad Fawi, Agnieszka Brychcy, Mohamed Abouzid, Martin Witt, and Elżbieta Kaczmarek. 2024. "Development and Validation of a Deep Learning Model for Histopathological Slide Analysis in Lung Cancer Diagnosis" Cancers 16, no. 8: 1506. https://doi.org/10.3390/cancers16081506
APA StyleAhmed, A. A., Fawi, M., Brychcy, A., Abouzid, M., Witt, M., & Kaczmarek, E. (2024). Development and Validation of a Deep Learning Model for Histopathological Slide Analysis in Lung Cancer Diagnosis. Cancers, 16(8), 1506. https://doi.org/10.3390/cancers16081506