S-Adenosylmethionine: A Multifaceted Regulator in Cancer Pathogenesis and Therapy
Simple Summary
Abstract
1. Introduction
1.1. Importance of SAMe in Cellular Processes
1.1.1. Methylation Reactions
Nucleic Acids Methylation
Protein Methylation
Methylation of Small Molecules
Lipid Methylation
1.1.2. Polyamine Synthesis
1.1.3. Transsulfuration Pathway
1.1.4. Folate Cycle
2. S-Adenosylmethionine Connections with Different Types of Cancer
2.1. Liver Cancer
2.2. Breast Cancer
2.3. Lung Cancer
2.4. Colorectal Cancer
2.5. Gastric Cancer
2.6. Prostate Cancer
2.7. Other Types of Cancers
3. S-Adenosylmethionine as Biomarker and Therapy
| Type of Cancer | Type of Role | Effect/Mechanism | References |
|---|---|---|---|
| Liver cancer (HCC, cholangiocarcinoma) | Biomarker | Reduction in MAT1A expression and activity | [61] |
| Depletion of SAMe, reduction of promoter methylation, MAT2A and MAT2B increase | [61,121,123,239,240] | ||
| Biomarker lncRNAs are modulated by SAMe | [241] | ||
| Prevention | SAMe supplementation reduces HCC foci occurrence and establishment (animal model) | [117] | |
| Treatment | MAT1A forced expression reduces HCC growth and angiogenesis, and increases apoptosis in vitro and in vivo | [250] | |
| Breast cancer | Treatment | SAMe modulates miRNA-34a, miRNA-34c and miRNA-486-5p leading to apoptosis and autophagy | [251] |
| SAMe hypermethylates uPA and MMP-2 genes inhibiting cellular invasion and growth | [147,252,253] | ||
| Combined therapy | SAMe + Decitabine: targets DNA hypermethylation and hypomethylation reducing reduced tumor volume and metastasis to the lung | [254] | |
| SAMe + doxorubicin: enhances apoptotic cell death through Fas/FasL-dependent caspase 8 and 3 activation | [255] | ||
| SAMe + chloroquine: inhibition of autophagy potentiates SAMe-induced apoptosis | [256] | ||
| Sensitization | SAMe sensitizes cancer to radiation-induced apoptosis | [146] | |
| Colorectal carcinoma | Biomarker | Promoter DNA methylation status serves as diagnosis and prognosis | [243] |
| Treatment | MTAP and MAT2A inhibition promotes lethality in CRC blocking PRMT5 | [259] | |
| Sensitization | SAMe treatment bypasses uL3-mediated drug resistance | [260] | |
| Lung cancer | Biomarker | Elevated SAMe plasma level as early detection biomarker | [242] |
| Sensitization | MAT2A inhibition sensitizes to cisplatin treatment | [177] | |
| Head and neck squamous cancer | Combined therapy | SAMe + cisplatin: promotes ER-stress leading to apoptosis and reduced proliferation and migration | [257,258] |
| Gastric cancer | Treatment | SAMe hypermethylates uPA, c-myc and H-ras inhibiting growth | [198,261] |
| Glioblastoma | Treatment | SAMe induces cell cycle arrest and apoptosis and mitotic catastrophe-induced death | [221] |
| Osteosarcoma and prostate | Treatment | SAMe downregulates ERK1/2 and STAT3 inducing apoptosis and blocking invasion | [226,227,262] |
| Retinoblastoma | Treatment | SAMe inhibits Wnt2/β-catenin pathway reducing tumor growth | [263] |
| Various | Chemoprotection | SAMe protects against chemotherapy-induced liver injury and reduces cancer-related fatigue | [264,265,266] |
4. Conclusions and Future Perspectives
Author Contributions
Funding
Conflicts of Interest
Abbreviations
| 5-MTHF | 5-methyltetrahydrofolate |
| 5,10-CH2THF | 5,10-methylene-THF |
| TOP | 5′-terminal oligopyrimidine |
| ACSL3 | acyl-CoA synthetase long chain family member 3 |
| AFP | alpha-fetoprotein |
| AK4 | adenylate kinase 4 |
| AKT | protein kinase B |
| ALT | alanine transaminase |
| AMD | SAMe decarboxylase proenzyme |
| AMD1 | SAMe decarboxylase proenzyme 1 |
| AMPK | AMP-activated protein kinase |
| APEX1 | apurinic/apyrimidinic endonuclease 1 |
| AST | aspartate transaminase |
| ATF3 | activating transcription factor-3 |
| ATP | adenosine triphosphate |
| BHMT | betaine Hcy methyltransferase |
| CBS | cystathionine beta-synthase |
| CDAA | choline-deficient L-amino acid-defined |
| CDK2 | cyclin-dependent kinase 2 |
| cFLIP | cellular FLICE inhibitory protein |
| CIMP | CpG island methylator phenotype |
| CRC | colorectal cancer |
| CTH | cystathionase |
| dcSAMe | decarboxylated SAMe |
| DILI | drug-induced liver injury |
| DNMTs | DNA methyltransferases |
| DNMT1 | DNA methyltransferase 1 |
| DOK7 | downstream of kinase 7 |
| DUSP1 | dual-specificity MAPK phosphatase |
| EGF | epidermal growth factor |
| eIF5A | eukaryotic initiation factor 5 A isoform 1 |
| ELOVL2 | elongation of very long chain fatty acids-like |
| eNOS | endothelial nitric oxide synthase |
| ERα | estrogen receptor α |
| ERK | extracellular signal regulated kinase |
| ERRFI1 | ERBB receptor feedback inhibitor 1 |
| ESR1 | estrogen receptor 1 |
| GIT1 | G Protein Coupled Receptor Kinase Interacting ArfGAP 1 |
| GNMT | glycine N-methyltrasferase |
| GSH | glutathione |
| GSTπ | glutathione-S-transferase |
| H3K4 | histone H3 fourth lysine |
| H3K27 | histone H3 27th lysine |
| H-Ras | HRas proto-oncogene |
| HCC | hepatocellular carcinoma |
| HFD | high fat diet |
| HGF | hepatocyte growth factor |
| HuR | Hu antigen R |
| ID4 | inhibitor of differentiation 4 |
| IGF | insulin-like growth factor |
| IL-1β | interleukin-1β |
| IHS | isolated hepatic steatosis |
| JAK | Janus kinase |
| KTMs | lysine-specific methyltransferases |
| LARP1 | La-Related Protein 1 |
| LDH | lactate dehydrogenase |
| LINE-1 | long interspersed nuclear element 1 |
| LKB1 | serine/threonine protein kinase 11 |
| m6A | N6-methyladenosine |
| MAPK | mitogen-activated protein kinase |
| MASH | metabolic disfunction-associated steatohepatitis |
| MASLD | metabolic disfunction-associated steatotic liver disease |
| MAT | methionine adenosyltransferase |
| MAT I/III | methionine Adenosyltransferase I/III |
| MAT1A | methionine adenosyltransferase 1A |
| MAT2A | methionine adenosyltransferase 2A |
| MAT2B | methionine adenosyltransferase 2 non-catalytic beta subunit |
| MCD | methionine and choline deficient |
| MDR1 | multidrug resistance 1 |
| MeCP2 | methyl-CpG-binding protein 2 |
| MEK | mitogen-activated protein kinase kinase |
| METTL3 | methyltransferase-like protein 3 |
| METTL14 | methyltransferase-like protein 14 |
| MGMT | O(6)-methylguanine DNA methyltransferase |
| MMP-2 | matrix metalloproteinase-2 |
| MS | methionine synthase |
| mSAMC | mitochondrial S-adenosylmethionine carrier |
| MTs | methyltransferases |
| MTA | methylthioadenosine |
| MTAP | MTA phosphorylase |
| MTHFR | methylenetetrahydrofolate reductase |
| mTOR | Mammalian target of rapamycin |
| MLH1 | MutL Homolog 1 |
| NAFLD | non-alcoholic fatty liver disease |
| NAM | nicotinamide |
| NAT1 | N-acetyltransferase type 1 |
| NNMT | nicotinamide methyltransferase |
| ODC | ornithine decarboxylase |
| PC | phosphatidylcholine |
| PE | phosphatidylethanolamine |
| PEMT | phosphatidylethanolamine N-methyltransferase |
| PI3K | Phosphoinositide 3-kinase |
| PITX2 | paired-like homeodomain transcription factor 2 |
| PRA | progesterone receptor α |
| PRMTs | protein arginine methyl transferases |
| PTEN | Phosphatase and tensin homolog |
| RASSF | Ras-association domain family/tumor suppressor |
| RIP1 | receptor-interacting Protein 1 |
| ROS | reactive oxygen species |
| SAH | S-adenosylhomocysteine |
| SAHH | S-adenosylhomocysteine hydrolase |
| SAMe | S-adenosylmethionine |
| SLD | steatotic liver disease |
| SOCS | suppressor of cytokine signaling |
| SPD | spermidine |
| SPM | spermine |
| STAT | signal transducer and activator of transcription |
| TCF4 | transcription factor 4 |
| TET | translocation methylcytosine dioxygenase |
| TG | triglyceride |
| TGF-β | transforming growth factor-β |
| THF | tetrahydrofolate |
| TNF | tumor necrosis factor |
| TROP2 | trophoblast surface antigen 2 |
| UCHL1 | ubiquitin C-terminal hydrolase L1 |
| uPA | urokinase-type plasminogen activator |
| VLDL | very low density lipoproteins |
| WT1 | Wilms’ tumor 1 |
| ZEB1 | zinc finger E-box-binding homeobox 1 |
References
- Cantoni, G.L. S-Adenosylmethionine; a New Intermediate Formed Enzymatically from L-Methionine and Adenosinetriphosphate. J. Biol. Chem. 1953, 204, 403–416. [Google Scholar] [CrossRef] [PubMed]
- Cantoni, G.L. Activation of Methionine for Transmethylation. J. Biol. Chem. 1951, 189, 745–754. [Google Scholar] [CrossRef] [PubMed]
- Bridwell-Rabb, J.; Grell, T.A.J.; Drennan, C.L. A Rich Man, Poor Man Story of S-Adenosylmethionine and Cobalamin Revisited. Annu. Rev. Biochem. 2018, 87, 555–584. [Google Scholar] [CrossRef] [PubMed]
- Finkelstein, J.D. Methionine Metabolism in Mammals. J. Nutr. Biochem. 1990, 1, 228–237. [Google Scholar] [CrossRef]
- Mudd, S.H.; Poole, J.R. Labile Methyl Balances for Normal Humans on Various Dietary Regimens. Metabolism 1975, 24, 721–735. [Google Scholar] [CrossRef]
- Mato, J.M.; Corrales, F.J.; Lu, S.C.; Avila, M.A. S-Adenosylmethionine: A Control Switch That Regulates Liver Function. FASEB J. 2002, 16, 15–26. [Google Scholar] [CrossRef]
- Gil, B.; Casado, M.; Pajares, M.A.; Bosca, L.; Mato, J.M.; Martin-Sanz, P.; Alvarez, L. Differential Expression Pattern of S-Adenosylmethionine Synthetase Isoenzymes during Rat Liver Development. Hepatology 1996, 24, 876–881. [Google Scholar] [CrossRef]
- Kotb, M.; Mudd, S.H.; Mato, J.M.; Geller, A.M.; Kredich, N.M.; Chou, J.Y.; Cantoni, G.L. Consensus Nomenclature for the Mammalian Methionine Adenosyltransferase Genes and Gene Products. Trends Genet. 1997, 13, 51–52. [Google Scholar] [CrossRef]
- Lu, S.C.; Mato, J.M. Role of Methionine Adenosyltransferase and S-Adenosylmethionine in Alcohol-Associated Liver Cancer. Alcohol. 2005, 35, 227–234. [Google Scholar] [CrossRef]
- Cabrero, C.; Puerta, J.; Metabolismo, S.A.; Hormonas, N.Y.; Diaz, J. Purification and Comparison of Two Forms of S-Adenosyl-l-Methionine Synthetase from Rat Liver. Eur. J. Biochem. 1987, 170, 299–304. [Google Scholar] [CrossRef]
- Sullivan, D.M.; Hoffman, J.L. Fractionation and Kinetic Properties of Rat Liver and Kidney Methionine Adenosyltransferase Isozymes. Biochemistry 1983, 22, 1636–1641. [Google Scholar] [CrossRef]
- Pascale, R.M.; Simile, M.M.; Calvisi, D.F.; Feo, C.F.; Feo, F. S-Adenosylmethionine: From the Discovery of Its Inhibition of Tumorigenesis to Its Use as a Therapeutic Agent. Cells 2022, 11, 409. [Google Scholar] [CrossRef]
- Lu, S.C.; Mato, J.M. S-Adenosylmethionine in Liver Health, Injury, and Cancer. Physiol. Rev. 2012, 92, 1515–1542. [Google Scholar] [CrossRef] [PubMed]
- Long, M.D.; Smiraglia, D.J.; Campbell, M.J. The Genomic Impact of DNA CpG Methylation on Gene Expression; Relationships in Prostate Cancer. Biomolecules 2017, 7, 15. [Google Scholar] [CrossRef]
- Jones, M.J.; Goodman, S.J.; Kobor, M.S. DNA Methylation and Healthy Human Aging. Aging Cell 2015, 14, 924–932. [Google Scholar] [CrossRef]
- Nicuiescu, M.D.; Lupu, D.S. Nutritional Influence on Epigenetics and Effects on Longevity. Curr. Opin. Clin. Nutr. Metab. Care 2011, 14, 35–40. [Google Scholar] [CrossRef] [PubMed]
- Esteller, M. Cancer Epigenomics: DNA Methylomes and Histone-Modification Maps. Nat. Rev. Genet. 2007, 8, 286–298. [Google Scholar] [CrossRef] [PubMed]
- Zaccara, S.; Ries, R.J.; Jaffrey, S.R. Reading, Writing and Erasing MRNA Methylation. Nat. Rev. Mol. Cell Biol. 2019, 20, 608–624. [Google Scholar] [CrossRef]
- Shi, H.; Wei, J.; He, C. Where, When and How: Context-Dependent Functions of RNA Methylation Writers, Readers, and Erasers. Mol. Cell 2019, 74, 640. [Google Scholar] [CrossRef]
- Ianniello, Z.; Fatica, A. N6-Methyladenosine Role in Acute Myeloid Leukaemia. Int. J. Mol. Sci. 2018, 19, 2345. [Google Scholar] [CrossRef]
- Deng, X.; Su, R.; Weng, H.; Huang, H.; Li, Z.; Chen, J. RNA N6-Methyladenosine Modification in Cancers: Current Status and Perspectives. Cell Res. 2018, 28, 507. [Google Scholar] [CrossRef] [PubMed]
- Chen, X.Y.; Zhang, J.; Zhu, J.S. The Role of M6A RNA Methylation in Human Cancer. Mol. Cancer 2019, 18, 103. [Google Scholar] [CrossRef] [PubMed]
- Sun, Y.; Gong, W.; Zhang, S. METTL3 Promotes Colorectal Cancer Progression through Activating JAK1/STAT3 Signaling Pathway. Cell Death Dis. 2023, 14, 765. [Google Scholar] [CrossRef] [PubMed]
- Grillo, M.A.; Colombatto, S. S-Adenosylmethionine and Protein Methylation. Amino Acids 2005, 28, 357–362. [Google Scholar] [CrossRef]
- Hwang, J.W.; Cho, Y.; Bae, G.U.; Kim, S.N.; Kim, Y.K. Protein Arginine Methyltransferases: Promising Targets for Cancer Therapy. Exp. Mol. Med. 2021, 53, 788–808. [Google Scholar] [CrossRef]
- Falnes, P.; Jakobsson, M.E.; Davydova, E.; Ho, A.; Malecki, J. Protein Lysine Methylation by Seven-β-Strand Methyltransferases. Biochem. J. 2016, 473, 1995–2009. [Google Scholar] [CrossRef]
- Falnes, P.; Małecki, J.M.; Herrera, M.C.; Bengtsen, M.; Davydova, E. Human Seven-β-Strand (METTL) Methyltransferases—Conquering the Universe of Protein Lysine Methylation. J. Biol. Chem. 2023, 299, 104661. [Google Scholar] [CrossRef]
- Luka, Z.; Mudd, S.H.; Wagner, C. Glycine N-Methyltransferase and Regulation of S-Adenosylmethionine Levels. J. Biol. Chem. 2009, 284, 22507–22511. [Google Scholar] [CrossRef]
- Wang, W.; Yang, C.; Wang, T.; Deng, H. Complex Roles of Nicotinamide N-Methyltransferase in Cancer Progression. Cell Death Dis. 2022, 13, 267. [Google Scholar] [CrossRef]
- Da Silva, R.P.; Nissim, I.; Brosnan, M.E.; Brosnan, J.T. Creatine Synthesis: Hepatic Metabolism of Guanidinoacetate and Creatine in the Rat in Vitro and in Vivo. Am. J. Physiol. Endocrinol. Metab. 2009, 296, E256. [Google Scholar] [CrossRef]
- Drinkwater, N.; Gee, C.L.; Puri, M.; Criscione, K.R.; McLeish, M.J.; Grunewald, G.L.; Martin, J.L. Molecular Recognition of Physiological Substrate Noradrenaline by the Adrenaline Synthesising Enzyme PNMT and Factors Influencing Its Methyltransferase Activity. Biochem. J. 2009, 422, 463. [Google Scholar] [CrossRef]
- Li, J.; Sun, C.; Cai, W.; Li, J.; Rosen, B.P.; Chen, J. Insights into S-Adenosyl-l-Methionine (SAM)-Dependent Methyltransferase Related Diseases and Genetic Polymorphisms. Mutat. Res. Rev. Mutat. Res. 2021, 788, 108396. [Google Scholar] [CrossRef] [PubMed]
- Drobna, Z.; Styblo, M.; Thomas, D.J. An Overview of Arsenic Metabolism and Toxicity. Curr. Protoc. Toxicol. 2009, 42, 4.31.1–4.31.6. [Google Scholar] [CrossRef] [PubMed]
- Varela-Rey, M.; Fernández-Ramos, D.; Martínez-López, N.; Embade, N.; Gómez-Santos, L.; Beraza, N.; Vázquez-Chantada, M.; Rodríguez, J.; Luka, Z.; Wagner, C.; et al. Impaired Liver Regeneration in Mice Lacking Glycine N-Methyltransferase. Hepatology 2009, 50, 443–452. [Google Scholar] [CrossRef]
- Martínez–López, N.; García–Rodríguez, J.L.; Varela–Rey, M.; Gutiérrez, V.; Fernández–Ramos, D.; Beraza, N.; Aransay, A.M.; Schlangen, K.; Lozano, J.J.; Aspichueta, P.; et al. Hepatoma Cells From Mice Deficient in Glycine N-Methyltransferase Have Increased RAS Signaling and Activation of Liver Kinase B1. Gastroenterology 2012, 143, 787–798.e13. [Google Scholar] [CrossRef]
- Varela-Rey, M.; Martínez-López, N.; Fernández-Ramos, D.; Embade, N.; Calvisi, D.F.; Woodhoo, A.; Rodríguez, J.; Fraga, M.F.; Julve, J.; Rodríguez-Millán, E.; et al. Fatty Liver and Fibrosis in Glycine N-Methyltransferase Knockout Mice Is Prevented by Nicotinamide. Hepatology 2010, 52, 105–114. [Google Scholar] [CrossRef]
- Martínez-Chantar, M.L.; Vázquez-Chantada, M.; Ariz, U.; Martínez, N.; Varela, M.; Luka, Z.; Capdevila, A.; Rodríguez, J.; Aransay, A.M.; Matthiesen, R.; et al. Loss of the Glycine N-Methyltransferase Gene Leads to Steatosis and Hepatocellular Carcinoma in Mice. Hepatology 2008, 47, 1191–1199. [Google Scholar] [CrossRef]
- Vance, D.E. Phospholipid Methylation in Mammals: From Biochemistry to Physiological Function. Biochim. Et Biophys. Acta BBA Biomembr. 2014, 1838, 1477–1487. [Google Scholar] [CrossRef]
- van der Veen, J.N.; Kennelly, J.P.; Wan, S.; Vance, J.E.; Vance, D.E.; Jacobs, R.L. The Critical Role of Phosphatidylcholine and Phosphatidylethanolamine Metabolism in Health and Disease. Biochim. Biophys. Acta Biomembr. 2017, 1859, 1558–1572. [Google Scholar] [CrossRef]
- Martínez-Uña, M.; Varela-Rey, M.; Cano, A.; Fernández-Ares, L.; Beraza, N.; Aurrekoetxea, I.; Martínez-Arranz, I.; García-Rodríguez, J.L.; Buqué, X.; Mestre, D.; et al. Excess S-Adenosylmethionine Reroutes Phosphatidylethanolamine towards Phosphatidylcholine and Triglyceride Synthesis. Hepatology 2013, 58, 1296–1305. [Google Scholar] [CrossRef]
- Perez-Leal, O.; Merali, S. Regulation of Polyamine Metabolism by Translational Control. Amino Acids 2012, 42, 611–617. [Google Scholar] [CrossRef] [PubMed]
- Pegg, A.E. Mammalian Polyamine Metabolism and Function. IUBMB Life 2009, 61, 880–894. [Google Scholar] [CrossRef] [PubMed]
- Casero, R.A.; Marton, L.J. Targeting Polyamine Metabolism and Function in Cancer and Other Hyperproliferative Diseases. Nat. Rev. Drug Discov. 2007, 6, 373–390. [Google Scholar] [CrossRef] [PubMed]
- Childs, A.C.; Mehta, D.J.; Gerner, E.W. Polyamine-Dependent Gene Expression. Cell Mol. Life Sci. 2003, 60, 1394. [Google Scholar] [CrossRef]
- Igarashi, K.; Kashiwagi, K. Modulation of Cellular Function by Polyamines. Int. J. Biochem. Cell Biol. 2010, 42, 39–51. [Google Scholar] [CrossRef]
- Schuber, F. Influence of Polyamines on Membrane Functions. Biochem. J. 1989, 260, 1–10. [Google Scholar] [CrossRef]
- Seiler, N.; Raul, F. Polyamines and Apoptosis. J. Cell Mol. Med. 2005, 9, 623–642. [Google Scholar] [CrossRef]
- Pompella, A.; Visvikis, A.; Paolicchi, A.; De Tata, V.; Casini, A.F. The Changing Faces of Glutathione, a Cellular Protagonist. Biochem. Pharmacol. 2003, 66, 1499–1503. [Google Scholar] [CrossRef]
- Zhang, H.F.; Klein Geltink, R.I.; Parker, S.J.; Sorensen, P.H. Transsulfuration, Minor Player or Critical for Cysteine Homeostasis in Cancer. Trends Cell Biol. 2022, 32, 800. [Google Scholar] [CrossRef]
- Zuhra, K.; Augsburger, F.; Majtan, T.; Szabo, C. Cystathionine-β-Synthase: Molecular Regulation and Pharmacological Inhibition. Biomolecules 2020, 10, 697. [Google Scholar] [CrossRef]
- Prudova, A.; Bauman, Z.; Braun, A.; Vitvitsky, V.; Lu, S.C.; Banerjee, R. S-Adenosylmethionine Stabilizes Cystathionine Beta-Synthase and Modulates Redox Capacity. Proc. Natl. Acad. Sci. USA 2006, 103, 6489–6494. [Google Scholar] [CrossRef] [PubMed]
- Jacobs, R.L.; Jiang, H.; Kennelly, J.P.; Orlicky, D.J.; Allen, R.H.; Stabler, S.P.; Maclean, K.N. Cystathionine Beta-Synthase Deficiency Alters Hepatic Phospholipid and Choline Metabolism: Post-Translational Repression of Phosphatidylethanolamine N-Methyltransferase Is a Consequence Rather than a Cause of Liver Injury in Homocystinuria. Mol. Genet. Metab. 2017, 120, 325–336. [Google Scholar] [CrossRef] [PubMed]
- Choumenkovitch, S.F.; Selhub, J.; Bagley, P.J.; Maeda, N.; Nadeau, M.R.; Smith, D.E.; Choi, S.W. In the Cystathionine Beta-Synthase Knockout Mouse, Elevations in Total Plasma Homocysteine Increase Tissue S-Adenosylhomocysteine, but Responses of S-Adenosylmethionine and DNA Methylation Are Tissue Specific. J. Nutr. 2002, 132, 2157–2160. [Google Scholar] [CrossRef] [PubMed]
- Kruger, W.D. Cystathionine β-Synthase Deficiency: Of Mice and Men. Mol. Genet. Metab. 2017, 121, 199–205. [Google Scholar] [CrossRef]
- Harvey Mudd, S.; Finkelstein, J.D.; Irreverre, F.; Laster, L. Homocystinuria: An Enzymatic Defect. Science 1964, 143, 1443–1445. [Google Scholar] [CrossRef]
- Serpa, J. Cysteine as a Carbon Source, a Hot Spot in Cancer Cells Survival. Front. Oncol. 2020, 10, 533316. [Google Scholar] [CrossRef]
- Zhu, J.; Berisa, M.; Schwörer, S.; Qin, W.; Cross, J.R.; Thompson, C.B. Transsulfuration Activity Can Support Cell Growth upon Extracellular Cysteine Limitation. Cell Metab. 2019, 30, 865–876.e5. [Google Scholar] [CrossRef]
- Pajares, M.A.; Pérez-Sala, D. Betaine Homocysteine S-Methyltransferase: Just a Regulator of Homocysteine Metabolism? Cell. Mol. Life Sci. 2006, 63, 2792–2803. [Google Scholar] [CrossRef]
- Ducker, G.S.; Rabinowitz, J.D. Cell Metabolism Review One-Carbon Metabolism in Health and Disease. Cell Metab. 2017, 25, 27–42. [Google Scholar] [CrossRef]
- Fernández-Ramos, D.; Lopitz-Otsoa, F.; Millet, O.; Alonso, C.; Lu, S.C.; Mato, J.M. One Carbon Metabolism and S-Adenosylmethionine in Non-Alcoholic Fatty Liver Disease Pathogenesis and Subtypes. Livers 2022, 2, 243–257. [Google Scholar] [CrossRef]
- Maldonado, L.Y.; Arsene, D.; Mato, J.M.; Lu, S.C. Methionine Adenosyltransferases in Cancers: Mechanisms of Dysregulation and Implications for Therapy. Exp. Biol. Med. 2018, 243, 107–117. [Google Scholar] [CrossRef] [PubMed]
- Haws, S.A.; Yu, D.; Ye, C.; Wille, C.K.; Nguyen, L.C.; Krautkramer, K.A.; Tomasiewicz, J.L.; Yang, S.E.; Miller, B.R.; Liu, W.H.; et al. Methyl-Metabolite Depletion Elicits Adaptive Responses to Support Heterochromatin Stability and Epigenetic Persistence. Mol. Cell 2020, 78, 210–223.e8. [Google Scholar] [CrossRef] [PubMed]
- Caudill, M.A.; Wang, J.C.; Melnyk, S.; Pogribny, I.P.; Jernigan, S.; Collins, M.D.; Santos-Guzman, J.; Swendseid, M.E.; Cogger, E.A.; James, S.J. Intracellular S-Adenosylhomocysteine Concentrations Predict Global DNA Hypomethylation in Tissues of Methyl-Deficient Cystathionine β-Synthase Heterozygous Mice. J. Nutr. 2001, 131, 2811–2818. [Google Scholar] [CrossRef] [PubMed]
- Ding, W.; Smulan, L.J.; Hou, N.S.; Taubert, S.; Watts, J.L.; Walker, A.K. S-Adenosylmethionine Levels Govern Innate Immunity through Distinct Methylation-Dependent Pathways. Cell Metab. 2015, 22, 633–645. [Google Scholar] [CrossRef] [PubMed]
- Mosca, L.; Vitiello, F.; Borzacchiello, L.; Coppola, A.; Tranchese, R.V.; Pagano, M.; Caraglia, M.; Cacciapuoti, G.; Porcelli, M. Mutual Correlation between Non-Coding RNA and S-Adenosylmethionine in Human Cancer: Roles and Therapeutic Opportunities. Cancers 2021, 13, 3264. [Google Scholar] [CrossRef]
- Ishiguro, K.; Arai, T.; Suzuki, T. Depletion of S-Adenosylmethionine Impacts on Ribosome Biogenesis through Hypomodification of a Single RRNA Methylation. Nucleic Acids Res. 2019, 47, 4226–4239. [Google Scholar] [CrossRef]
- Rinella, M.E.; Lazarus, J.V.; Ratziu, V.; Francque, S.M.; Sanyal, A.J.; Kanwal, F.; Romero, D.; Abdelmalek, M.F.; Anstee, Q.M.; Arab, J.P.; et al. A Multisociety Delphi Consensus Statement on New Fatty Liver Disease Nomenclature. J. Hepatol. 2023, 79, 1542–1556. [Google Scholar] [CrossRef]
- Alonso, C.; Fernández-Ramos, D.; Varela-Rey, M.; Martínez-Arranz, I.; Navasa, N.; Van Liempd, S.M.; Lavín Trueba, J.L.; Mayo, R.; Ilisso, C.P.; de Juan, V.G.; et al. Metabolomic Identification of Subtypes of Nonalcoholic Steatohepatitis. Gastroenterology 2017, 152, 1449–1461.e7. [Google Scholar] [CrossRef]
- Lu, S.C.; Alvarez, L.; Huang, Z.-Z.; Chen, L.; An, W.; Corrales, F.J.; Avila, M.A.; Kanel, G.; Mato, J.M. Methionine Adenosyltransferase 1A Knockout Mice Are Predisposed to Liver Injury and Exhibit Increased Expression of Genes Involved in Proliferation. Proc. Natl. Acad. Sci. USA 2001, 98, 5560–5565. [Google Scholar] [CrossRef]
- Rinella, M.E.; Lazarus, J.V.; Ratziu, V.; Francque, S.M.; Sanyal, A.J.; Kanwal, F.; Romero, D.; Abdelmalek, M.F.; Anstee, Q.M.; Arab, J.P.; et al. A Multisociety Delphi Consensus Statement on New Fatty Liver Disease Nomenclature. Hepatology 2023, 78, 1966. [Google Scholar] [CrossRef]
- Noureddin, M.; Mato, J.M.; Lu, S.C. Nonalcoholic Fatty Liver Disease: Update on Pathogenesis, Diagnosis, Treatment and the Role of S-Adenosylmethionine. Exp. Biol. Med. 2015, 240, 809–820. [Google Scholar] [CrossRef] [PubMed]
- Williams, C.D.; Stengel, J.; Asike, M.I.; Torres, D.M.; Shaw, J.; Contreras, M.; Landt, C.L.; Harrison, S.A. Prevalence of Nonalcoholic Fatty Liver Disease and Nonalcoholic Steatohepatitis among a Largely Middle-Aged Population Utilizing Ultrasound and Liver Biopsy: A Prospective Study. Gastroenterology 2011, 140, 124–131. [Google Scholar] [CrossRef] [PubMed]
- Labrecque, D.R.; Abbas, Z.; Anania, F.; Ferenci, P.; Khan, A.G.; Goh, K.L.; Hamid, S.S.; Isakov, V.; Lizarzabal, M.; Peñaranda, M.M.; et al. World Gastroenterology Organisation Global Guidelines: Nonalcoholic Fatty Liver Disease and Nonalcoholic Steatohepatitis. J. Clin. Gastroenterol. 2014, 48, 467–473. [Google Scholar] [CrossRef] [PubMed]
- Takuma, Y.; Nouso, K. Nonalcoholic Steatohepatitis-Associated Hepatocellular Carcinoma: Our Case Series and Literature Review. World J. Gastroenterol. WJG 2010, 16, 1436. [Google Scholar] [CrossRef]
- Bray, F.; Laversanne, M.; Sung, H.; Ferlay, J.; Siegel, R.L.; Soerjomataram, I.; Jemal, A. Global Cancer Statistics 2022: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA Cancer J. Clin. 2024, 74, 229–263. [Google Scholar] [CrossRef]
- Hester, D.; Golabi, P.; Paik, J.; Younossi, I.; Mishra, A.; Younossi, Z.M. Among Medicare Patients With Hepatocellular Carcinoma, Non-Alcoholic Fatty Liver Disease Is the Most Common Etiology and Cause of Mortality. J. Clin. Gastroenterol. 2020, 54, 459–467. [Google Scholar] [CrossRef]
- Younossi, Z.; Stepanova, M.; Ong, J.P.; Jacobson, I.M.; Bugianesi, E.; Duseja, A.; Eguchi, Y.; Wong, V.W.; Negro, F.; Yilmaz, Y.; et al. Nonalcoholic Steatohepatitis Is the Fastest Growing Cause of Hepatocellular Carcinoma in Liver Transplant Candidates. Clin. Gastroenterol. Hepatol. 2019, 17, 748–755.e3. [Google Scholar] [CrossRef]
- Wong, R.J.; Cheung, R.; Ahmed, A. Nonalcoholic Steatohepatitis Is the Most Rapidly Growing Indication for Liver Transplantation in Patients with Hepatocellular Carcinoma in the U.S. Hepatology 2014, 59, 2188–2195. [Google Scholar] [CrossRef]
- Ahmad, M.I.; Khan, M.U.; Kodali, S.; Shetty, A.; Bell, S.M.; Victor, D. Hepatocellular Carcinoma Due to Nonalcoholic Fatty Liver Disease: Current Concepts and Future Challenges. J. Hepatocell. Carcinoma 2022, 9, 477. [Google Scholar] [CrossRef]
- Martínez-Chantar, M.L.; Corrales, F.J.; Martínez-Cruz, L.A.; García-Trevijano, E.R.; Huang, Z.-Z.; Chen, L.; Kanel, G.; Avila, M.A.; Mato, J.M.; Lu, S.C. Spontaneous Oxidative Stress and Liver Tumors in Mice Lacking Methionine Adenosyltransferase 1A. FASEB J. 2002, 16, 1292–1294. [Google Scholar] [CrossRef]
- Cai, J.; Sun, W.M.; Hwang, J.J.; Stain, S.C.; Lu, S.C. Changes in S-Adenosylmethionine Synthetase in Human Liver Cancer: Molecular Characterization and Significance. Hepatology 1996, 24, 1090–1097. [Google Scholar] [CrossRef] [PubMed]
- Best, C.H.; Huntsman, M.E. The Effects of the Components of Lecithine upon Deposition of Fat in the Liver. J. Physiol. 1932, 75, 405–412. [Google Scholar] [CrossRef] [PubMed]
- Best, C.H.; Hershey, J.M.; Huntsman, M.E. The Effect of Lecithine on Fat Deposition in the Liver of the Normal Rat. J. Physiol. 1932, 75, 56–66. [Google Scholar] [CrossRef] [PubMed]
- Yamaguchi, K.; Yang, L.; McCall, S.; Huang, J.; Yu, X.X.; Pandey, S.K.; Bhanot, S.; Monia, B.P.; Li, Y.-X.; Diehl, A.M. Inhibiting Triglyceride Synthesis Improves Hepatic Steatosis but Exacerbates Liver Damage and Fibrosis in Obese Mice with Nonalcoholic Steatohepatitis. Hepatology 2007, 45, 1366–1374. [Google Scholar] [CrossRef]
- Yamaguchi, K.; Itoh, Y.; Yokomizo, C.; Nishimura, T.; Niimi, T.; Fujii, H.; Okanoue, T.; Yoshikawa, T. Blockade of Interleukin-6 Signaling Enhances Hepatic Steatosis but Improves Liver Injury in Methionine Choline-Deficient Diet-Fed Mice. Lab. Investig. 2010, 90, 1169–1178. [Google Scholar] [CrossRef]
- Montandon, S.A.; Somm, E.; Loizides-Mangold, U.; de Vito, C.; Dibner, C.; Jornayvaz, F.R. Multi-Technique Comparison of Atherogenic and MCD NASH Models Highlights Changes in Sphingolipid Metabolism. Sci. Rep. 2019, 9, 16810. [Google Scholar] [CrossRef]
- Rizki, G.; Arnaboldi, L.; Gabrielli, B.; Yan, J.; Lee, G.S.; Ng, R.K.; Turner, S.M.; Badger, T.M.; Pitas, R.E.; Maher, J.J. Mice Fed a Lipogenic Methionine-Choline-Deficient Diet Develop Hypermetabolism Coincident with Hepatic Suppression of SCD-1. J. Lipid Res. 2006, 47, 2280–2290. [Google Scholar] [CrossRef]
- Park, H.S.; Jeon, B.H.; Woo, S.H.; Leem, J.; Jang, J.E.; Cho, M.S.; Park, I.S.; Lee, K.U.; Koh, E.H. Time-Dependent Changes in Lipid Metabolism in Mice with Methionine Choline Deficiency-Induced Fatty Liver Disease. Mol. Cells 2011, 32, 571–577. [Google Scholar] [CrossRef]
- Wolf, M.J.; Adili, A.; Piotrowitz, K.; Abdullah, Z.; Boege, Y.; Stemmer, K.; Ringelhan, M.; Simonavicius, N.; Le Egger, M.; Wohlleber, D.; et al. Cancer Cell Metabolic Activation of Intrahepatic CD8 + T Cells and NKT Cells Causes Nonalcoholic Steatohepatitis and Liver Cancer via Cross-Talk with Hepatocytes Cancer Cell CD8 + and NKT Cells Promote NASH and NASH-Induced HCC. Cancer Cell 2014, 26, 549–564. [Google Scholar] [CrossRef]
- Ikawa-Yoshida, A.; Matsuo, S.; Kato, A.; Ohmori, Y.; Higashida, A.; Kaneko, E.; Matsumoto, M. Hepatocellular Carcinoma in a Mouse Model Fed a Choline-Deficient, L-Amino Acid-Defined, High-Fat Diet. Int. J. Exp. Pathol. 2017, 98, 221–233. [Google Scholar] [CrossRef]
- Denda, A.; Kitayama, W.; Kishida, H.; Murata, N.; Tamura, K.; Kusuoka, O.; Tsutsumi, M.; Nishikawa, F.; Kita, E.; Nakae, D.; et al. Expression of Inducible Nitric Oxide (NO) Synthase but Not Prevention by Its Gene Ablation of Hepatocarcinogenesis with Fibrosis Caused by a Choline-Deficient, l-Amino Acid-Defined Diet in Rats and Mice. Nitric Oxide 2007, 16, 164–176. [Google Scholar] [CrossRef] [PubMed]
- Matsumoto, M.; Hada, N.; Sakamaki, Y.; Uno, A.; Shiga, T.; Tanaka, C.; Ito, T.; Katsume, A.; Sudoh, M. An Improved Mouse Model That Rapidly Develops Fibrosis in Non-Alcoholic Steatohepatitis. Int. J. Exp. Pathol. 2013, 94, 93. [Google Scholar] [CrossRef] [PubMed]
- Rinella, M.E.; Elias, M.S.; Smolak, R.R.; Fu, T.; Borensztajn, J.; Green, R.M. Mechanisms of Hepatic Steatosis in Mice Fed a Lipogenic Methionine Choline-Deficient Diet. J. Lipid Res. 2008, 49, 1068–1076. [Google Scholar] [CrossRef] [PubMed]
- Gallage, S.; Avila, J.E.B.; Ramadori, P.; Focaccia, E.; Rahbari, M.; Ali, A.; Malek, N.P.; Anstee, Q.M.; Heikenwalder, M. A Researcher’s Guide to Preclinical Mouse NASH Models. Nat. Metab. 2022, 4, 1632–1649. [Google Scholar] [CrossRef]
- Itagaki, H.; Shimizu, K.; Morikawa, S.; Ogawa, K.; Ezaki, T. Morphological and Functional Characterization of Non-Alcoholic Fatty Liver Disease Induced by a Methionine-Choline-Deficient Diet in C57BL/6 Mice. Int. J. Clin. Exp. Pathol. 2013, 6, 2683–2696. [Google Scholar]
- Kucsera, D.; Tóth, V.E.; Gergő, D.; Vörös, I.; Onódi, Z.; Görbe, A.; Ferdinandy, P.; Varga, Z.V. Characterization of the CDAA Diet-Induced Non-Alcoholic Steatohepatitis Model: Sex-Specific Differences in Inflammation, Fibrosis, and Cholesterol Metabolism in Middle-Aged Mice. Front. Physiol. 2021, 12, 609465. [Google Scholar] [CrossRef]
- Kodama, Y.; Kisseleva, T.; Iwaisako, K.; Miura, K.; Taura, K.; De Minicis, S.; Österreicher, C.H.; Schnabl, B.; Seki, E.; Brenner, D.A. C-Jun N-Terminal Kinase-1 From Hematopoietic Cells Mediates Progression From Hepatic Steatosis to Steatohepatitis and Fibrosis in Mice. Gastroenterology 2009, 137, 1467–1477.e5. [Google Scholar] [CrossRef]
- Yoshiji, H.; Nakae, D.; Mizumoto, Y.; Horiguchi, K.; Tamura, K.; Denda, A.; Tsujii, T.; Konishi, Y. Inhibitory Effect of Dietary Iron Deficiency on Inductions of Putative Preneoplastic Lesions as Well as 8-Hydroxydeoxyguanosine in DNA and Lipid Peroxidation in the Livers of Rats Caused by Exposure to a Choline-Deficient L-Amino Acid Defined Diet. Carcinogenesis 1992, 13, 1227–1233. [Google Scholar] [CrossRef][Green Version]
- Iruarrizaga-Lejarreta, M.; Varela-Rey, M.; Fernández-Ramos, D.; Martínez-Arranz, I.; Delgado, T.C.; Simon, J.; Gutiérrez-de Juan, V.; delaCruz-Villar, L.; Azkargorta, M.; Lavin, J.L.; et al. Role of Aramchol in Steatohepatitis and Fibrosis in Mice. Hepatol. Commun. 2017, 1, 911–927. [Google Scholar] [CrossRef]
- Ramani, K.; Mato, J.M.; Lu, S.C. Role of Methionine Adenosyltransferase Genes in Hepatocarcinogenesis. Cancers 2011, 3, 1480–1497. [Google Scholar] [CrossRef]
- Murphy, S.K.; Yang, H.; Moylan, C.A.; Pang, H.; Dellinger, A.; Abdelmalek, M.F.; Garrett, M.E.; Ashley–Koch, A.; Suzuki, A.; Tillmann, H.L.; et al. Relationship Between Methylome and Transcriptome in Patients With Nonalcoholic Fatty Liver Disease. Gastroenterology 2013, 145, 1076–1087. [Google Scholar] [CrossRef]
- Calvisi, D.F.; Simile, M.M.; Ladu, S.; Pellegrino, R.; De Murtas, V.; Pinna, F.; Tomasi, M.L.; Frau, M.; Virdis, P.; De Miglio, M.R.; et al. Altered Methionine Metabolism and Global DNA Methylation in Liver Cancer: Relationship with Genomic Instability and Prognosis. Int. J. Cancer 2007, 121, 2410–2420. [Google Scholar] [CrossRef] [PubMed]
- Jelnes, P.; Santoni-Rugiu, E.; Rasmussen, M.; Friis, S.L.; Nielsen, J.H.; Tygstrup, N.; Bisgaard, H.C. Remarkable Heterogeneity Displayed by Oval Cells in Rat and Mouse Models of Stem Cell–Mediated Liver Regeneration. Hepatology 2007, 45, 1462–1470. [Google Scholar] [CrossRef] [PubMed]
- Dumble, M.L.; Croager, E.J.; Yeoh, G.C.T.; Quail, E.A. Generation and Characterization of P53 Null Transformed Hepatic Progenitor Cells: Oval Cells Give Rise to Hepatocellular Carcinoma. Carcinogenesis 2002, 23, 435–445. [Google Scholar] [CrossRef]
- Nguyen, L.N.; Furuya, M.H.; Wolfraim, L.A.; Nguyen, A.P.; Holdren, M.S.; Campbell, J.S.; Knight, B.; Yeoh, G.C.T.; Fausto, N.; Parks, W.T. Transforming Growth Factor-Beta Differentially Regulates Oval Cell and Hepatocyte Proliferation. Hepatology 2007, 45, 31–41. [Google Scholar] [CrossRef] [PubMed]
- Ding, W.; Mouzaki, M.; You, H.; Laird, J.C.; Mato, J.; Lu, S.C.; Rountree, C.B. CD133+ Liver Cancer Stem Cells from Methionine Adenosyl Transferase 1A–Deficient Mice Demonstrate Resistance to Transforming Growth Factor (TGF)-β–Induced Apoptosis. Hepatology 2009, 49, 1277–1286. [Google Scholar] [CrossRef] [PubMed]
- Rountree, C.B.; Senadheera, S.; Mato, J.M.; Crooks, G.M.; Lu, S.C. Expansion of Liver Cancer Stem Cells during Aging in Methionine Adenosyltransferase 1A–Deficient Mice. Hepatology 2008, 47, 1288–1297. [Google Scholar] [CrossRef]
- Tomasi, M.L.; Ramani, K.; Lopitz-Otsoa, F.; Rodríguez, M.S.; Li, T.W.H.; Ko, K.; Yang, H.; Bardag-Gorce, F.; Iglesias-Ara, A.; Feo, F.; et al. S-Adenosylmethionine Regulates Dual-Specificity Mitogen-Activated Protein Kinase Phosphatase Expression in Mouse and Human Hepatocytes. Hepatology 2010, 51, 2152–2161. [Google Scholar] [CrossRef]
- Calvisi, D.F.; Pinna, F.; Meloni, F.; Ladu, S.; Pellegrino, R.; Sini, M.; Daino, L.; Simile, M.M.; De Miglio, M.R.; Virdis, P.; et al. Dual-Specificity Phosphatase 1 Ubiquitination in Extracellular Signal-Regulated Kinase–Mediated Control of Growth in Human Hepatocellular Carcinoma. Cancer Res. 2008, 68, 4192–4200. [Google Scholar] [CrossRef]
- Vázquez-Chantada, M.; Ariz, U.; Varela-Rey, M.; Embade, N.; Martínez-Lopez, N.; Fernández-Ramos, D.; Gómez-Santos, L.; Lamas, S.; Lu, S.C.; Martínez-Chantar, M.L.; et al. Evidence for LKB1/AMP-Activated Protein Kinase/Endothelial Nitric Oxide Synthase Cascade Regulated by Hepatocyte Growth Factor, S-Adenosylmethionine, and Nitric Oxide in Hepatocyte Proliferation. Hepatology 2009, 49, 608–617. [Google Scholar] [CrossRef]
- Martínez-Chantar, M.L.; Vázquez-Chantada, M.; Garnacho, M.; Latasa, M.U.; Varela-Rey, M.; Dotor, J.; Santamaria, M.; Martínez-Cruz, L.A.; Parada, L.A.; Lu, S.C.; et al. S-Adenosylmethionine Regulates Cytoplasmic HuR via AMP-Activated Kinase. Gastroenterology 2006, 131, 223–232. [Google Scholar] [CrossRef] [PubMed]
- Martínez-López, N.; Varela-Rey, M.; Fernández-Ramos, D.; Woodhoo, A.; Vázquez-Chantada, M.; Embade, N.; Espinosa-Hevia, L.; Bustamante, F.J.; Parada, L.A.; Rodriguez, M.S.; et al. Activation of LKB1-Akt Pathway Independent of Phosphoinositide 3-Kinase Plays a Critical Role in the Proliferation of Hepatocellular Carcinoma from Nonalcoholic Steatohepatitis. Hepatology 2010, 52, 1621–1631. [Google Scholar] [CrossRef] [PubMed]
- Ramani, K.; Robinson, A.E.; Berlind, J.; Fan, W.; Abeynayake, A.; Binek, A.; Barbier-Torres, L.; Noureddin, M.; Nissen, N.N.; Yildirim, Z.; et al. S-Adenosylmethionine Inhibits La Ribonucleoprotein Domain Family Member 1 in Murine Liver and Human Liver Cancer Cells. Hepatology 2021, 75, 280. [Google Scholar] [CrossRef] [PubMed]
- Mudd, S.H.; Brosnan, J.T.; Brosnan, M.E.; Jacobs, R.L.; Stabler, S.P.; Allen, R.H.; Vance, D.E.; Wagner, C. Methyl Balance and Transmethylation Fluxes in Humans. Am. J. Clin. Nutr. 2007, 85, 19–25. [Google Scholar] [CrossRef]
- Chen, Y.M.A.; Shiu, J.Y.A.; Tzeng, S.J.; Shih, L.S.; Chen, Y.J.; Lut, W.Y.; Chen, P.H. Characterization of Glycine-N-Methyltransferase-Gene Expression in Human Hepatocellular Carcinoma. Int. J. Cancer 1998, 75, 787–793. [Google Scholar] [CrossRef]
- Avila, M.A.; Berasain, C.; Torres, L.; Martín-Duce, A.; Corrales, F.J.; Yang, H.; Prieto, J.; Lu, S.C.; Caballería, J.; Rodés, J.; et al. Reduced MRNA Abundance of the Main Enzymes Involved in Methionine Metabolism in Human Liver Cirrhosis and Hepatocellular Carcinoma. J. Hepatol. 2000, 33, 907–914. [Google Scholar] [CrossRef]
- Lu, S.C.; Ramani, K.; Ou, X.; Lin, M.; Yu, V.; Ko, K.; Park, R.; Bottiglieri, T.; Tsukamoto, H.; Kanel, G.; et al. S-Adenosylmethionine in the Chemoprevention and Treatment of Hepatocellular Carcinoma in a Rat Model. Hepatology 2009, 50, 462–471. [Google Scholar] [CrossRef]
- Mudd, S.H.; Cerone, R.; Schiaffino, M.C.; Fantasia, A.R.; Minniti, G.; Caruso, U.; Lorini, R.; Watkins, D.; Matiaszuk, N.; Rosenblatt, D.S.; et al. Glycine N-methyltransferase Deficiency: A Novel Inborn Error Causing Persistent Isolated Hypermethioninaemia. J. Inherit. Metab. Dis. 2001, 24, 448–464. [Google Scholar] [CrossRef]
- Augoustides-Savvopoulou, P.; Luka, Z.; Karyda, S.; Stabler, S.P.; Allen, R.H.; Patsiaoura, K.; Wagner, C.; Mudd, S.H. Glycine N-Methyltransferase Deficiency: A New Patient with a Novel Mutation. J. Inherit. Metab. Dis. 2003, 26, 745–759. [Google Scholar] [CrossRef]
- Calvisi, D.F.; Ladu, S.; Gorden, A.; Farina, M.; Conner, E.A.; Lee, J.S.; Factor, V.M.; Thorgeirsson, S.S. Ubiquitous Activation of Ras and Jak/Stat Pathways in Human HCC. Gastroenterology 2006, 130, 1117–1128. [Google Scholar] [CrossRef]
- Cai, J.; Mao, Z.; Hwang, J.J.; Lu, S.C. Differential Expression of Methionine Adenosyltransferase Genes Influences the Rate of Growth of Human Hepatocellular Carcinoma Cells. Cancer Res. 1998, 58, 1444–1450. [Google Scholar] [PubMed]
- LeGros, H.L.; Halim, A.-B.; Geller, A.M.; Kotb, M. Cloning, Expression, and Functional Characterization of the β Regulatory Subunit of Human Methionine Adenosyltransferase (MAT II). J. Biol. Chem. 2000, 275, 2359–2366. [Google Scholar] [CrossRef] [PubMed]
- Yang, H.; Ara, A.I.; Magilnick, N.; Xia, M.; Ramani, K.; Chen, H.; Lee, T.D.; Mato, J.M.; Lu, S.C. Expression Pattern, Regulation, and Functions of Methionine Adenosyltransferase 2β Splicing Variants in Hepatoma Cells. Gastroenterology 2008, 134, 281–291. [Google Scholar] [CrossRef] [PubMed]
- Ramani, K.; Yang, H.; Xia, M.; Ara, A.I.; Mato, J.M.; Lu, S.C. Leptin’s Mitogenic Effect in Human Liver Cancer Cells Requires Induction of Both Methionine Adenosyltransferase 2A and 2β. Hepatology 2008, 47, 521–531. [Google Scholar] [CrossRef]
- Tomasi, M.L.; Ryoo, M.; Skay, A.; Tomasi, I.; Giordano, P.; Mato, J.M.; Lu, S.C. Polyamine and Methionine Adenosyltransferase 2A Crosstalk in Human Colon and Liver Cancer. Exp. Cell Res. 2013, 319, 1902–1911. [Google Scholar] [CrossRef]
- Tomasi, M.L.; Ryoo, M.; Ramani, K.; Tomasi, I.; Giordano, P.; Mato, J.M.; Lu, S.C.; Lauda Tomasi, M.; Ryoo, M.; Ramani, K.; et al. Methionine Adenosyltransferase A2 Sumoylation Positively Regulate Bcl-2 Expression in Human Colon and Liver Cancer Cells. Oncotarget 2015, 6, 37706–37723. [Google Scholar] [CrossRef]
- Xia, M.; Chen, Y.; Wang, L.-C.; Zandi, E.; Yang, H.; Bemanian, S.; Martínez-Chantar, M.L.; Mato, J.M.; Lu, S.C. Novel Function and Intracellular Localization of Methionine Adenosyltransferase 2β Splicing Variants. J. Biol. Chem. 2010, 285, 20015–20021. [Google Scholar] [CrossRef]
- Peng, H.; Dara, L.; Li, T.W.H.; Zheng, Y.; Yang, H.; Tomasi, L.M.; Tomasi, I.; Giordano, P.; Mato, J.M.; Lu, S.C. Methionine Adenosyltransferase 2B-GIT1 Interplay Activates MEK1-ERK1/2 to Induce Growth in Human Liver and Colon Cancer. Hepatology 2013, 57, 2299–2313. [Google Scholar] [CrossRef]
- Peng, H.; Li, T.W.H.; Yang, H.; Moyer, M.P.; Mato, J.M.; Lu, S.C. Methionine Adenosyltransferase 2B–GIT1 Complex Serves as a Scaffold to Regulate Ras/Raf/MEK1/2 Activity in Human Liver and Colon Cancer Cells. Am. J. Pathol. 2015, 185, 1135–1144. [Google Scholar] [CrossRef]
- Segovia-Mendoza, M.; Gómez de León, C.T.; García-Becerra, R.; Ambrosio, J.; Nava-Castro, K.E.; Morales-Montor, J. The Chemical Environmental Pollutants BPA and BPS Induce Alterations of the Proteomic Profile of Different Phenotypes of Human Breast Cancer Cells: A Proposed Interactome. Environ. Res. 2020, 191, 109960. [Google Scholar] [CrossRef]
- Koual, M.; Tomkiewicz, C.; Cano-Sancho, G.; Antignac, J.P.; Bats, A.S.; Coumoul, X. Environmental Chemicals, Breast Cancer Progression and Drug Resistance. Environ. Health 2020, 19, 1–25. [Google Scholar] [CrossRef] [PubMed]
- Hvidtfeldt, U.A.; Chen, J.; Rodopoulou, S.; Strak, M.; de Hoogh, K.; Andersen, Z.J.; Bellander, T.; Brandt, J.; Fecht, D.; Forastiere, F.; et al. Breast Cancer Incidence in Relation to Long-Term Low-Level Exposure to Air Pollution in the ELAPSE Pooled Cohort. Cancer Epidemiol. Biomark. Prev. 2023, 32, 105–113. [Google Scholar] [CrossRef] [PubMed]
- Coyle, Y.M. The Effect of Environment on Breast Cancer Risk. Breast Cancer Res. Treat. 2004, 84, 273–288. [Google Scholar] [CrossRef] [PubMed]
- Brody, J.G.; Rudel, R.A. Environmental Pollutants and Breast Cancer. Environ. Health Perspect. 2003, 111, 1007–1019. [Google Scholar] [CrossRef]
- Wolff, M.S.; Weston, A. Breast Cancer Risk and Environmental Exposures. Environ. Health Perspect. 1997, 105, 891–896. [Google Scholar] [CrossRef]
- Laden, F.; Hunter, D.J. ENVIRONMENTAL RISK FACTORS AND FEMALE BREAST CANCER. Annu. Rev. Public Health 1998, 19, 101–123. [Google Scholar] [CrossRef]
- O’Donovan, P.J.; Livingston, D.M. BRCA1 and BRCA2: Breast/Ovarian Cancer Susceptibility Gene Products and Participants in DNA Double-Strand Break Repair. Carcinogenesis 2010, 31, 961–967. [Google Scholar] [CrossRef]
- Metcalfe, K.; Lynch, H.T.; Ghadirian, P.; Tung, N.; Olivotto, I.; Warner, E.; Olopade, O.I.; Eisen, A.; Weber, B.; McLennan, J.; et al. Contralateral Breast Cancer in BRCA1 and BRCA2 Mutation Carriers. J. Clin. Oncol. 2016, 22, 2328–2335. [Google Scholar] [CrossRef]
- Welcsh, P.L.; King, M.C. BRCA1 and BRCA2 and the Genetics of Breast and Ovarian Cancer. Hum. Mol. Genet. 2001, 10, 705–713. [Google Scholar] [CrossRef]
- Arruabarrena-Aristorena, A.; Toska, E. Epigenetic Mechanisms Influencing Therapeutic Response in Breast Cancer. Front. Oncol. 2022, 12, 924808. [Google Scholar] [CrossRef]
- Thang, N.X.; Yoo, S.; La, H.; Lee, H.; Park, C.; Park, K.S.; Hong, K. Epigenetic Factors as Etiological Agents, Diagnostic Markers, and Therapeutic Targets for Luminal Breast Cancer. Biomedicines 2022, 10, 748. [Google Scholar] [CrossRef] [PubMed]
- Garcia-Martinez, L.; Zhang, Y.; Nakata, Y.; Chan, H.L.; Morey, L. Epigenetic Mechanisms in Breast Cancer Therapy and Resistance. Nat. Commun. 2021, 12, 1786. [Google Scholar] [CrossRef] [PubMed]
- Karsli-Ceppioglu, S.; Dagdemir, A.; Judes, G.; Ngollo, M.; Penault-Llorca, F.; Pajon, A.; Bignon, Y.J.; Bernard-Gallon, D. Epigenetic Mechanisms of Breast Cancer: An Update of the Current Knowledge. Epigenomics 2014, 6, 651–664. [Google Scholar] [CrossRef] [PubMed]
- Batra, R.N.; Lifshitz, A.; Vidakovic, A.T.; Chin, S.F.; Sati-Batra, A.; Sammut, S.J.; Provenzano, E.; Ali, H.R.; Dariush, A.; Bruna, A.; et al. DNA Methylation Landscapes of 1538 Breast Cancers Reveal a Replication-Linked Clock, Epigenomic Instability and Cis-Regulation. Nat. Commun. 2021, 12, 5406. [Google Scholar] [CrossRef] [PubMed]
- Chekhun, V.F.; Kulik, G.I.; Yurchenko, O.V.; Tryndyak, V.P.; Todor, I.N.; Luniv, L.S.; Tregubova, N.A.; Pryzimirska, T.V.; Montgomery, B.; Rusetskaya, N.V.; et al. Role of DNA Hypomethylation in the Development of the Resistance to Doxorubicin in Human MCF-7 Breast Adenocarcinoma Cells. Cancer Lett. 2006, 231, 87–93. [Google Scholar] [CrossRef]
- Luzhna, L.; Kovalchuk, O. Modulation of DNA Methylation Levels Sensitizes Doxorubicin-Resistant Breast Adenocarcinoma Cells to Radiation-Induced Apoptosis. Biochem. Biophys. Res. Commun. 2010, 392, 113–117. [Google Scholar] [CrossRef]
- Pakneshan, P.; Szyf, M.; Farias-Eisner, R.; Rabbani, S.A. Reversal of the Hypomethylation Status of Urokinase (UPA) Promoter Blocks Breast Cancer Growth and Metastasis. J. Biol. Chem. 2004, 279, 31735–31744. [Google Scholar] [CrossRef]
- Phuong, N.T.T.; Kim, S.K.; Lim, S.C.; Kim, H.S.; Kim, T.H.; Lee, K.Y.; Ahn, S.-G.; Yoon, J.-H.; Kang, K.W. Role of PTEN Promoter Methylation in Tamoxifen-Resistant Breast Cancer Cells. Breast Cancer Res. Treat. 2011, 130, 73–83. [Google Scholar] [CrossRef]
- Shen, J.; He, Y.; Li, S.; Chen, H. Crosstalk of Methylation and Tamoxifen in Breast Cancer (Review). Mol. Med. Rep. 2024, 30, 180. [Google Scholar] [CrossRef]
- Zhang, J.; Zhou, C.; Jiang, H.; Liang, L.; Shi, W.; Zhang, Q.; Sun, P.; Xiang, R.; Wang, Y.; Yang, S. ZEB1 Induces ER-α Promoter Hypermethylation and Confers Antiestrogen Resistance in Breast Cancer. Cell Death Dis. 2017, 8, e2732. [Google Scholar] [CrossRef]
- Jiménez-Garduño, A.M.; Mendoza-Rodríguez, M.G.; Urrutia-Cabrera, D.; Domínguez-Robles, M.C.; Pérez-Yépez, E.A.; Ayala-Sumuano, J.T.; Meza, I. IL-1β Induced Methylation of the Estrogen Receptor ERα Gene Correlates with EMT and Chemoresistance in Breast Cancer Cells. Biochem. Biophys. Res. Commun. 2017, 490, 780–785. [Google Scholar] [CrossRef] [PubMed]
- Kim, M.R.; Wu, M.J.; Zhang, Y.; Yang, J.Y.; Chang, C.J. TET2 Directs Mammary Luminal Cell Differentiation and Endocrine Response. Nat. Commun. 2020, 11, 4642. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Liang, Y.; Zhou, S.; Chen, J.; Wu, C. UCHL1 Contributes to Insensitivity to Endocrine Therapy in Triple-Negative Breast Cancer by Deubiquitinating and Stabilizing KLF5. Breast Cancer Res. 2024, 26, 44. [Google Scholar] [CrossRef] [PubMed]
- Stone, A.; Zotenko, E.; Locke, W.J.; Korbie, D.; Millar, E.K.A.; Pidsley, R.; Stirzaker, C.; Graham, P.; Trau, M.; Musgrove, E.A.; et al. DNA Methylation of Oestrogen-Regulated Enhancers Defines Endocrine Sensitivity in Breast Cancer. Nat. Commun. 2015, 6, 7758. [Google Scholar] [CrossRef] [PubMed]
- Zou, J.X.; Duan, Z.; Wang, J.; Sokolov, A.; Xu, J.; Chen, C.Z.; Li, J.J.; Chen, H.W. Kinesin Family Deregulation Coordinated by Bromodomain Protein ANCCA and Histone Methyltransferase MLL for Breast Cancer Cell Growth, Survival, and Tamoxifen Resistance. Mol. Cancer Res. 2014, 12, 539–549. [Google Scholar] [CrossRef]
- Zhao, W.; Sun, M.; Li, S.; Chen, Z.; Geng, D. Transcription Factor ATF3 Mediates the Radioresistance of Breast Cancer. J. Cell Mol. Med. 2018, 22, 4664–4675. [Google Scholar] [CrossRef]
- Liu, X.; Gonzalez, G.; Dai, X.; Miao, W.; Yuan, J.; Huang, M.; Bade, D.; Li, L.; Sun, Y.; Wang, Y. Adenylate Kinase 4 Modulates the Resistance of Breast Cancer Cells to Tamoxifen through an M6A-Based Epitranscriptomic Mechanism. Mol. Ther. 2020, 28, 2593. [Google Scholar] [CrossRef]
- Wang, J.; Xie, S.; Yang, J.; Xiong, H.; Jia, Y.; Zhou, Y.; Chen, Y.; Ying, X.; Chen, C.; Ye, C.; et al. The Long Noncoding RNA H19 Promotes Tamoxifen Resistance in Breast Cancer via Autophagy. J. Hematol. Oncol. 2019, 12, 81. [Google Scholar] [CrossRef]
- Phuong, N.T.T.; Kim, S.K.; Im, J.H.; Yang, J.W.; Choi, M.C.; Lim, S.C.; Lee, K.Y.; Kim, Y.M.; Yoon, J.H.; Kang, K.W. Induction of Methionine Adenosyltransferase 2A in Tamoxifen-Resistant Breast Cancer Cells. Oncotarget 2016, 7, 13902. [Google Scholar] [CrossRef]
- Xu, J.; Wu, D.; Wang, S.; Wang, Z. MAT2B Expression Correlates with Poor Prognosis in Triple-Negative Breast Cancer. Cancer Manag. Res. 2019, 11, 5501–5511. [Google Scholar] [CrossRef]
- Wallace, H.M.; Duthie, J.; Evans, D.M.; Lamond, S.; Nicoll, K.M.; Heys, S.D. Alterations in Polyamine Catabolic Enzymes in Human Breast Cancer Tissue. Clin. Cancer Res. 2000, 6, 3657–3661. [Google Scholar] [PubMed]
- Akinyele, O.; Wallace, H.M. Characterising the Response of Human Breast Cancer Cells to Polyamine Modulation. Biomolecules 2021, 11, 743. [Google Scholar] [CrossRef] [PubMed]
- Kingsnorth, A.N.; Wallace, H.M.; Bundred, N.J.; Dixon, J.M.J. Polyamines in Breast Cancer. Br. J. Surg. 2005, 71, 352–356. [Google Scholar] [CrossRef] [PubMed]
- Casero, R.A.; Murray Stewart, T.; Pegg, A.E. Polyamine Metabolism and Cancer: Treatments, Challenges and Opportunities. Nat. Rev. Cancer 2018, 18, 681. [Google Scholar] [CrossRef]
- Pegg, A.E.; Williams-Ashman, H.G. Phosphate-Stimulated Breakdown of 5’-Methylthioadenosine by Rat Ventral Prostate. Biochem. J. 1969, 115, 241–247. [Google Scholar] [CrossRef]
- Schmid, M.; Sen, M.; Rosenbach, M.D.; Carrera, C.J.; Friedman, H.; Carson, D.A. A Methylthioadenosine Phosphorylase (MTAP) Fusion Transcript Identifies a New Gene on Chromosome 9p21 That Is Frequently Deleted in Cancer. Oncogene 2000, 19, 5747–5754. [Google Scholar] [CrossRef]
- Zhang, Y.; Zhang, T.T.; Gao, L.; Tan, Y.N.; Li, Y.T.; Tan, X.Y.; Huang, T.X.; Li, H.H.; Bai, F.; Zou, C.; et al. Downregulation of MTAP Promotes Tumor Growth and Metastasis by Regulating ODC Activity in Breast Cancer. Int. J. Biol. Sci. 2022, 18, 3034. [Google Scholar] [CrossRef]
- Liao, R.; Chen, X.; Cao, Q.; Bai, L.; Ma, C.; Dai, Z.; Dong, C. AMD1 Promotes Breast Cancer Aggressiveness via a Spermidine-EIF5A Hypusination-TCF4 Axis. Breast Cancer Res. 2024, 26, 70. [Google Scholar] [CrossRef]
- Hill, W.; Lim, E.L.; Weeden, C.E.; Lee, C.; Augustine, M.; Chen, K.; Kuan, F.C.; Marongiu, F.; Evans, E.J.; Moore, D.A.; et al. Lung Adenocarcinoma Promotion by Air Pollutants. Nature 2023, 616, 159–167. [Google Scholar] [CrossRef]
- Petrovic, D.; Bodinier, B.; Dagnino, S.; Whitaker, M.; Karimi, M.; Campanella, G.; Haugdahl Nøst, T.; Polidoro, S.; Palli, D.; Krogh, V.; et al. Epigenetic Mechanisms of Lung Carcinogenesis Involve Differentially Methylated CpG Sites beyond Those Associated with Smoking. Eur. J. Epidemiol. 2022, 37, 629. [Google Scholar] [CrossRef]
- Zhu, T.; Bao, X.; Chen, M.; Lin, R.; Zhuyan, J.; Zhen, T.; Xing, K.; Zhou, W.; Zhu, S. Mechanisms and Future of Non-Small Cell Lung Cancer Metastasis. Front. Oncol. 2020, 10, 585284. [Google Scholar] [CrossRef] [PubMed]
- Kaufman, J.M.; Amann, J.M.; Park, K.; Arasada, R.R.; Li, H.; Shyr, Y.; Carbone, D.P. LKB1 Loss Induces Characteristic Patterns of Gene Expression in Human Tumors Associated with NRF2 Activation and Attenuation of PI3K-AKT. J. Thorac. Oncol. 2014, 9, 794. [Google Scholar] [CrossRef]
- Koenig, M.J.; Agana, B.A.; Kaufman, J.M.; Sharpnack, M.F.; Wang, W.Z.; Weigel, C.; Navarro, F.C.P.; Amann, J.M.; Cacciato, N.; Arasada, R.R.; et al. STK11/LKB1 Loss of Function Is Associated with Global DNA Hypomethylation and S-Adenosyl-Methionine Depletion in Human Lung Adenocarcinoma. Cancer Res. 2021, 81, 4194–4204. [Google Scholar] [CrossRef] [PubMed]
- Ulanovskaya, O.A.; Zuhl, A.M.; Cravatt, B.F. NNMT Promotes Epigenetic Remodeling in Cancer by Creating a Metabolic Methylation Sink. Nat. Chem. Biol. 2013, 9, 300. [Google Scholar] [CrossRef] [PubMed]
- Tomida, M.; Mikami, I.; Takeuchi, S.; Nishimura, H.; Akiyama, H. Serum Levels of Nicotinamide N-Methyltransferase in Patients with Lung Cancer. J. Cancer Res. Clin. Oncol. 2009, 135, 1223–1229. [Google Scholar] [CrossRef] [PubMed]
- Wang, Z.; Yip, L.Y.; Lee, J.H.J.; Wu, Z.; Chew, H.Y.; Chong, P.K.W.; Teo, C.C.; Ang, H.Y.K.; Peh, K.L.E.; Yuan, J.; et al. Methionine Is a Metabolic Dependency of Tumor-Initiating Cells. Nat. Med. 2019, 25, 825–837. [Google Scholar] [CrossRef]
- Zhao, X.; Wang, L.; Lin, H.; Wang, J.; Fu, J.; Zhu, D.; Xu, W. Inhibition of MAT2A-Related Methionine Metabolism Enhances The Efficacy of Cisplatin on Cisplatin-Resistant Cells in Lung Cancer. Cell J. Yakhteh 2022, 24, 204. [Google Scholar] [CrossRef]
- Liu, C.; Ren, Q.; Deng, J.; Wang, S.; Ren, L. C-MYC/METTL3/LINC01006 Positive Feedback Loop Promotes Migration, Invasion and Proliferation of Non-Small Cell Lung Cancer. Biomed. J. 2024, 47, 100664. [Google Scholar] [CrossRef]
- Xuan, Y.F.; Lu, S.; Ou, Y.J.; Bao, X.B.; Huan, X.J.; Song, S.S.; Miao, Z.H.; Wang, Y.Q. The Combination of Methionine Adenosyltransferase 2A Inhibitor and Methyltransferase like 3 Inhibitor Promotes Apoptosis of Non-Small Cell Lung Cancer Cells and Produces Synergistic Anti-Tumor Activity. Biochem. Biophys. Res. Commun. 2024, 716, 150011. [Google Scholar] [CrossRef]
- Ascenção, K.; Szabo, C. Emerging Roles of Cystathionine β-Synthase in Various Forms of Cancer. Redox Biol. 2022, 53, 102331. [Google Scholar] [CrossRef]
- Qiu, L.; Hu, L.; Wang, H.; Li, J.; Ruan, X.; Sun, B.; Zhi, J.; Zheng, X.; Gu, L.; Gao, M.; et al. FATS Regulates Polyamine Biosynthesis by Promoting ODC Degradation in an ERβ-Dependent Manner in Non-Small-Cell Lung Cancer. Cell Death Dis. 2020, 11, 839. [Google Scholar] [CrossRef] [PubMed]
- Basu, I.; Locker, J.; Cassera, M.B.; Belbin, T.J.; Merino, E.F.; Dong, X.; Hemeon, I.; Evans, G.B.; Guha, C.; Schramm, V.L. Growth and Metastases of Human Lung Cancer Are Inhibited in Mouse Xenografts by a Transition State Analogue of 5’-Methylthioadenosine Phosphorylase. J. Biol. Chem. 2011, 286, 4902–4911. [Google Scholar] [CrossRef] [PubMed]
- Xu, Y.; Hong, Z.; Yu, S.; Huang, R.; Li, K.; Li, M.; Xie, S.; Zhu, L. Fresh Insights Into SLC25A26: Potential New Therapeutic Target for Cancers: A Review. Oncol. Rev. 2024, 18, 1379323. [Google Scholar] [CrossRef] [PubMed]
- Ye, Q.; Singh, S.; Qian, P.R.; Guo, N.L. Immune-Omics Networks of Cd27, Pd1, and Pdl1 in Non-Small Cell Lung Cancer. Cancers 2021, 13, 4296. [Google Scholar] [CrossRef] [PubMed]
- Winawer, S.J. Natural History of Colorectal Cancer. Am. J. Med. 1999, 106, 3–6. [Google Scholar] [CrossRef]
- Nazemalhosseini Mojarad, E.; Kuppen, P.J.; Aghdaei, H.A.; Zali, M.R. The CpG Island Methylator Phenotype (CIMP) in Colorectal Cancer. Gastroenterol. Hepatol. Bed Bench 2013, 6, 120–128. [Google Scholar]
- Advani, S.M.; Advani, P.; DeSantis, S.M.; Brown, D.; VonVille, H.M.; Lam, M.; Loree, J.M.; Sarshekeh, A.M.; Bressler, J.; Lopez, D.S.; et al. Clinical, Pathological, and Molecular Characteristics of CpG Island Methylator Phenotype in Colorectal Cancer: A Systematic Review and Meta-Analysis. Transl. Oncol. 2018, 11, 1188–1201. [Google Scholar] [CrossRef]
- Ewing, I.; Hurley, J.J.; Josephides, E.; Millar, A. The Molecular Genetics of Colorectal Cancer. Frontline Gastroenterol. 2014, 5, 26–30. [Google Scholar] [CrossRef]
- Helderman, N.C.; Andini, K.D.; van Leerdam, M.E.; van Hest, L.P.; Hoekman, D.R.; Ahadova, A.; Bajwa-ten Broeke, S.W.; Bosse, T.; van der Logt, E.M.J.; Imhann, F.; et al. MLH1 Promotor Hypermethylation in Colorectal and Endometrial Carcinomas from Patients with Lynch Syndrome. J. Mol. Diagn. 2024, 26, 106–114. [Google Scholar] [CrossRef]
- Levine, A.J.; Phipps, A.I.; Baron, J.A.; Buchanan, D.D.; Ahnen, D.J.; Cohen, S.A.; Lindor, N.M.; Newcomb, P.A.; Rosty, C.; Haile, R.W.; et al. Clinicopathologic Risk Factor Distributions for MLH1 Promoter Region Methylation in CIMP-Positive Tumors. Cancer Epidemiol. Biomark. Prev. 2016, 25, 68–75. [Google Scholar] [CrossRef]
- Suter, C.M.; Martin, D.I.; Ward, R.I. Hypomethylation of L1 Retrotransposons in Colorectal Cancer and Adjacent Normal Tissue. Int. J. Colorectal Dis. 2004, 19, 95–101. [Google Scholar] [CrossRef] [PubMed]
- Rodriguez, J.; Frigola, J.; Vendrell, E.; Risques, R.A.; Fraga, M.F.; Morales, C.; Moreno, V.; Esteller, M.; Capellà, G.; Ribas, M.; et al. Chromosomal Instability Correlates with Genome-Wide DNA Demethylation in Human Primary Colorectal Cancers. Cancer Res. 2006, 66, 8462–8468. [Google Scholar] [CrossRef] [PubMed]
- Szigeti, K.A.; Kalmár, A.; Galamb, O.; Valcz, G.; Barták, B.K.; Nagy, Z.B.; Zsigrai, S.; Felletár, I.; Patai, Á.V.; Micsik, T.; et al. Global DNA Hypomethylation of Colorectal Tumours Detected in Tissue and Liquid Biopsies May Be Related to Decreased Methyl-Donor Content. BMC Cancer 2022, 22, 605. [Google Scholar] [CrossRef] [PubMed]
- Mima, K.; Nowak, J.A.; Qian, Z.R.; Cao, Y.; Song, M.; Masugi, Y.; Shi, Y.; da Silva, A.; Gu, M.; Li, W.; et al. Tumor LINE-1 Methylation Level and Colorectal Cancer Location in Relation to Patient Survival. Oncotarget 2016, 7, 55098. [Google Scholar] [CrossRef]
- Antelo, M.; Balaguer, F.; Shia, J.; Shen, Y.; Hur, K.; Moreira, L.; Cuatrecasas, M.; Bujanda, L.; Giraldez, M.D.; Takahashi, M.; et al. A High Degree of LINE-1 Hypomethylation Is a Unique Feature of Early-Onset Colorectal Cancer. PLoS ONE 2012, 7, e45357. [Google Scholar] [CrossRef]
- Alonso-Aperte, E.; González, M.P.; Póo-Prieto, R.; Varela-Moreiras, G. Folate Status and S-Adenosylmethionine/S-Adenosylhomocysteine Ratio in Colorectal Adenocarcinoma in Humans. Eur. J. Clin. Nutr. 2007, 62, 295–298. [Google Scholar] [CrossRef]
- Sibani, S.; Melnyk, S.; Pogribny, I.P.; Wang, W.; Hiou-Tim, F.; Deng, L.; Trasler, J.; Jill James, S.; Rozen, R. Studies of Methionine Cycle Intermediates (SAM, SAH), DNA Methylation and the Impact of Folate Deficiency on Tumor Numbers in Min Mice. Carcinogenesis 2002, 23, 61–65. [Google Scholar] [CrossRef]
- Luo, J.; Li, Y.N.; Wang, F.; Zhang, W.M.; Geng, X. S-Adenosylmethionine Inhibits the Growth of Cancer Cells by Reversing the Hypomethylation Status of c-Myc and H-Ras in Human Gastric Cancer and Colon Cancer. Int. J. Biol. Sci. 2010, 6, 784–795. [Google Scholar] [CrossRef]
- Ito, K.; Ikeda, S.; Kojima, N.; Miura, M.; Shimizu-Saito, K.; Yamaguchi, I.; Katsuyama, I.; Sanada, K.; Iwai, T.; Senoo, H.; et al. Correlation between the Expression of Methionine Adenosyltransferase and the Stages of Human Colorectal Carcinoma. Surg. Today 2000, 30, 706–710. [Google Scholar] [CrossRef]
- Chen, H.; Xia, M.; Lin, M.; Yang, H.; Kuhlenkamp, J.; Li, T.; Sodir, N.M.; Chen, Y.H.; Josef-Lenz, H.; Laird, P.W.; et al. Role of Methionine Adenosyltransferase 2A and S-Adenosylmethionine in Mitogen-Induced Growth of Human Colon Cancer Cells. Gastroenterology 2007, 133, 207–218. [Google Scholar] [CrossRef]
- Li, T.W.H.; Zhang, Q.; Oh, P.; Xia, M.; Chen, H.; Bemanian, S.; Lastra, N.; Circ, M.; Moyer, M.P.; Mato, J.M.; et al. S-Adenosylmethionine and Methylthioadenosine Inhibit Cellular FLICE Inhibitory Protein Expression and Induce Apoptosis in Colon Cancer Cells. Mol. Pharmacol. 2009, 76, 192. [Google Scholar] [CrossRef] [PubMed]
- Tomasi, M.L.; Cossu, C.; Spissu, Y.; Floris, A.; Ryoo, M.; Iglesias-Ara, A.; Wang, Q.; Pandol, S.J.; Bhowmick, N.A.; Seki, E.; et al. S-Adenosylmethionine and Methylthioadenosine Inhibit Cancer Metastasis by Targeting MicroRNA 34a/b-Methionine Adenosyltransferase 2A/2B Axis. Oncotarget 2017, 8, 78851–78869. [Google Scholar] [CrossRef] [PubMed]
- Borzacchiello, L.; Veglia Tranchese, R.; Grillo, R.; Arpino, R.; Mosca, L.; Cacciapuoti, G.; Porcelli, M. S-Adenosylmethionine Inhibits Colorectal Cancer Cell Migration through Mirna-Mediated Targeting of Notch Signaling Pathway. Int. J. Mol. Sci. 2022, 23, 7673. [Google Scholar] [CrossRef] [PubMed]
- Phillips, C.M.; Zatarain, J.R.; Nicholls, M.E.; Porter, C.; Widen, S.G.; Thanki, K.; Johnson, P.; Jawad, M.U.; Moyer, M.P.; Randall, J.W.; et al. Upregulation of Cystathionine-β-Synthase in Colonic Epithelia Reprograms Metabolism and Promotes Carcinogenesis. Cancer Res. 2017, 77, 5741–5754. [Google Scholar] [CrossRef]
- Módis, K.; Coletta, C.; Asimakopoulou, A.; Szczesny, B.; Chao, C.; Papapetropoulos, A.; Hellmich, M.R.; Szabo, C. Effect of S-Adenosyl-l-Methionine (SAM), an Allosteric Activator of Cystathionine-β-Synthase (CBS) on Colorectal Cancer Cell Proliferation and Bioenergetics in Vitro. Nitric Oxide 2014, 41, 146–156. [Google Scholar] [CrossRef]
- Liskova, V.; Chovancova, B.; Babula, P.; Rezuchova, I.; Pavlov, K.P.; Matuskova, M.; Krizanova, O. Cystathionine β-Synthase Affects Organization of Cytoskeleton and Modulates Carcinogenesis in Colorectal Carcinoma Cells. Front. Oncol. 2023, 13, 1178021. [Google Scholar] [CrossRef]
- Zhao, L.Y.; Tong, D.D.; Xue, M.; Ma, H.L.; Liu, S.Y.; Yang, J.; Liu, Y.X.; Guo, B.; Ni, L.; Liu, L.Y.; et al. MeCP2, a Target of MiR-638, Facilitates Gastric Cancer Cell Proliferation through Activation of the MEK1/2–ERK1/2 Signaling Pathway by Upregulating GIT1. Oncogenesis 2017, 6, e368. [Google Scholar] [CrossRef]
- Zhao, L.; Liu, Y.; Tong, D.; Qin, Y.; Yang, J.; Xue, M.; Du, N.; Liu, L.; Guo, B.; Hou, N.; et al. MeCP2 Promotes Gastric Cancer Progression Through Regulating FOXF1/Wnt5a/β-Catenin and MYOD1/Caspase-3 Signaling Pathways. EBioMedicine 2017, 16, 87–100. [Google Scholar] [CrossRef]
- Tong, D.; Zhang, J.; Wang, X.; Li, Q.; Liu, L.; Lu, A.; Guo, B.; Yang, J.; Ni, L.; Qin, H.; et al. MiR-22, Regulated by MeCP2, Suppresses Gastric Cancer Cell Proliferation by Inducing a Deficiency in Endogenous S-Adenosylmethionine. Oncogenesis 2020, 9, 99. [Google Scholar] [CrossRef]
- Ma, M.; Kong, P.; Huang, Y.; Wang, J.; Liu, X.; Hu, Y.; Chen, X.; Du, C.; Yang, H. Activation of MAT2A-ACSL3 Pathway Protects Cells from Ferroptosis in Gastric Cancer. Free Radic. Biol. Med. 2022, 181, 288–299. [Google Scholar] [CrossRef]
- Zhang, Y.; Yang, H.; Zhao, J.; Wan, P.; Hu, Y.; Lv, K.; Hu, Y.; Yang, X.; Ma, M. Activation of MAT2A-RIP1 Signaling Axis Reprograms Monocytes in Gastric Cancer. J. Immunother. Cancer 2021, 9, e001364. [Google Scholar] [CrossRef] [PubMed]
- Shukeir, N.; Pakneshan, P.; Chen, G.; Szyf, M.; Rabbani, S.A. Alteration of the Methylation Status of Tumor-Promoting Genes Decreases Prostate Cancer Cell Invasiveness and Tumorigenesis In Vitro and In Vivo. Cancer Res. 2006, 66, 9202–9210. [Google Scholar] [CrossRef] [PubMed]
- Mathes, A.; Duman, M.B.; Neumann, A.; Dobreva, G.; Schmidt, T. S-Adenosylmethionine Treatment Affects Histone Methylation in Prostate Cancer Cells. Gene 2024, 893, 147915. [Google Scholar] [CrossRef] [PubMed]
- Liu, C.; Kelnar, K.; Liu, B.; Chen, X.; Calhoun-Davis, T.; Li, H.; Patrawala, L.; Yan, H.; Jeter, C.; Honorio, S.; et al. Identification of MiR-34a as a Potent Inhibitor of Prostate Cancer Progenitor Cells and Metastasis by Directly Repressing CD44. Nat. Med. 2011, 17, 211. [Google Scholar] [CrossRef]
- Tang, Y.; Tang, Y.; Cheng, Y.S. MiR-34a Inhibits Pancreatic Cancer Progression through Snail1-Mediated Epithelial–Mesenchymal Transition and the Notch Signaling Pathway. Sci. Rep. 2017, 7, 38232. [Google Scholar] [CrossRef]
- Shukla-Dave, A.; Castillo-Martin, M.; Chen, M.; Lobo, J.; Gladoun, N.; Collazo-Lorduy, A.; Khan, F.M.; Ponomarev, V.; Yi, Z.; Zhang, W.; et al. Ornithine Decarboxylase Is Sufficient for Prostate Tumorigenesis via Androgen Receptor Signaling. Am. J. Pathol. 2016, 186, 3131–3145. [Google Scholar] [CrossRef]
- Zabala-Letona, A.; Arruabarrena-Aristorena, A.; Martín-Martín, N.; Fernandez-Ruiz, S.; Sutherland, J.D.; Clasquin, M.; Tomas-Cortazar, J.; Jimenez, J.; Torres, I.; Quang, P.; et al. MTORC1-Dependent AMD1 Regulation Sustains Polyamine Metabolism in Prostate Cancer. Nature 2017, 547, 109–113. [Google Scholar] [CrossRef]
- Bistulfi, G.; Affronti, H.C.; Foster, B.A.; Karasik, E.; Gillard, B.; Morrison, C.; Mohler, J.; Phillips, J.G.; Smiraglia, D.J. The Essential Role of Methylthioadenosine Phosphorylase in Prostate Cancer. Oncotarget 2016, 7, 14380. [Google Scholar] [CrossRef]
- Barve, A.; Vega, A.; Shah, P.P.; Ghare, S.; Casson, L.; Wunderlich, M.; Siskind, L.J.; Beverly, L.J. Perturbation of Methionine/S-Adenosylmethionine Metabolism as a Novel Vulnerability in MLL Rearranged Leukemia. Cells 2019, 8, 1322. [Google Scholar] [CrossRef]
- Harachi, M.; Masui, K.; Honda, H.; Muragaki, Y.; Kawamata, T.; Cavenee, W.K.; Mischel, P.S.; Shibata, N. Dual Regulation of Histone Methylation by MTOR Complexes Controls Glioblastoma Tumor Cell Growth via EZH2 and SAM. Mol. Cancer Res. 2020, 18, 1142–1152. [Google Scholar] [CrossRef]
- Mosca, L.; Pagano, C.; Tranchese, R.V.; Grillo, R.; Cadoni, F.; Navarra, G.; Coppola, L.; Pagano, M.; Mele, L.; Cacciapuoti, G.; et al. Antitumoral Activity of the Universal Methyl Donor S-Adenosylmethionine in Glioblastoma Cells. Molecules 2024, 29, 1708. [Google Scholar] [CrossRef] [PubMed]
- Liteplo, R.G.; Munro, S. DNA Methylating Capacity in Metastatic Variants of a Human Melanoma Cell Line. Cancer Lett. 1988, 41, 191–198. [Google Scholar] [CrossRef] [PubMed]
- Liteplo, R.G. Altered Methionine Metabolism in Metastatic Variants of a Human Melanoma Cell Line. Cancer Lett. 1989, 44, 23–31. [Google Scholar] [CrossRef] [PubMed]
- Zhou, D.; Shi, H.; Mu, W.; Zhang, B.; Meng, T.; Zhang, S. Potential Role of S-Adenosylmethionine in Osteosarcoma Development. Onco Targets Ther. 2016, 9, 3653. [Google Scholar] [CrossRef]
- Mehdi, A.; Attias, M.; Arakelian, A.; Szyf, M.; Piccirillo, C.A.; Rabbani, S.A. S-Adenosylmethionine Blocks Tumorigenesis and with Immune Checkpoint Inhibitor Enhances Anti-Cancer Efficacy against BRAF Mutant and Wildtype Melanomas. Neoplasia 2023, 36, 100874. [Google Scholar] [CrossRef]
- Ilisso, C.P.; Sapio, L.; Delle Cave, D.; Illiano, M.; Spina, A.; Cacciapuoti, G.; Naviglio, S.; Porcelli, M. S-Adenosylmethionine Affects ERK1/2 and Stat3 Pathways and Induces Apotosis in Osteosarcoma Cells. J. Cell Physiol. 2016, 231, 428–435. [Google Scholar] [CrossRef]
- Parashar, S.; Cheishvili, D.; Arakelian, A.; Hussain, Z.; Tanvir, I.; Khan, H.A.; Szyf, M.; Rabbani, S.A. S-Adenosylmethionine Blocks Osteosarcoma Cells Proliferation and Invasion in Vitro and Tumor Metastasis in Vivo: Therapeutic and Diagnostic Clinical Applications. Cancer Med. 2015, 4, 732–744. [Google Scholar] [CrossRef]
- Ye, F.; Chen, E.R.; Nilsen, T.W. Kaposi’s Sarcoma-Associated Herpesvirus Utilizes and Manipulates RNA N 6-Adenosine Methylation to Promote Lytic Replication. J. Virol. 2017, 91, e00466-17. [Google Scholar] [CrossRef]
- Zhang, X.; Meng, W.; Feng, J.; Gao, X.; Qin, C.; Feng, P.; Huang, Y.; Gao, S.J. METTL16 Controls Kaposi’s Sarcoma-Associated Herpesvirus Replication by Regulating S-Adenosylmethionine Cycle. Cell Death Dis. 2023, 14, 591. [Google Scholar] [CrossRef]
- Pozzi, V.; Molinelli, E.; Campagna, R.; Serritelli, E.N.; Cecati, M.; De Simoni, E.; Sartini, D.; Goteri, G.; Martin, N.I.; van Haren, M.J.; et al. Knockdown of Nicotinamide N-Methyltransferase Suppresses Proliferation, Migration, and Chemoresistance of Merkel Cell Carcinoma Cells in Vitro. Hum. Cell 2024, 37, 729–738. [Google Scholar] [CrossRef]
- Harmankaya, İ.; Akar, S.; Uğraş, S.; Güler, A.H.; Ezveci, H.; Aydoğdu, M.; Çelik, Ç. Nicotinamide N-Methyltransferase Overexpression May Be Associated with Poor Prognosis in Ovarian Cancer. J. Obstet. Gynaecol. 2021, 41, 248–253. [Google Scholar] [CrossRef] [PubMed]
- Sartini, D.; Santarelli, A.; Rossi, V.; Goteri, G.; Rubini, C.; Ciavarella, D.; Lo Muzio, L.; Emanuelli, M. Nicotinamide N-Methyltransferase Upregulation Inversely Correlates with Lymph Node Metastasis in Oral Squamous Cell Carcinoma. Mol. Med. 2007, 13, 415. [Google Scholar] [CrossRef] [PubMed]
- Neelakantan, H.; Wang, H.Y.; Vance, V.; Hommel, J.D.; McHardy, S.F.; Watowich, S.J. Structure-Activity Relationship for Small Molecule Inhibitors of Nicotinamide N-Methyltransferase. J. Med. Chem. 2017, 60, 5015–5028. [Google Scholar] [CrossRef] [PubMed]
- Kannt, A.; Rajagopal, S.; Kadnur, S.V.; Suresh, J.; Bhamidipati, R.K.; Swaminathan, S.; Hallur, M.S.; Kristam, R.; Elvert, R.; Czech, J.; et al. A Small Molecule Inhibitor of Nicotinamide N-Methyltransferase for the Treatment of Metabolic Disorders. Sci. Rep. 2018, 8, 3660. [Google Scholar] [CrossRef]
- Lee, H.Y.; Suciu, R.M.; Horning, B.D.; Vinogradova, E.V.; Ulanovskaya, O.A.; Cravatt, B.F. Covalent Inhibitors of Nicotinamide N-Methyltransferase (NNMT) Provide Evidence for Target Engagement Challenges in Situ. Bioorg. Med. Chem. Lett. 2018, 28, 2682–2687. [Google Scholar] [CrossRef]
- Babault, N.; Allali-Hassani, A.; Li, F.; Fan, J.; Yue, A.; Ju, K.; Liu, F.; Vedadi, M.; Liu, J.; Jin, J. Discovery of Bisubstrate Inhibitors of Nicotinamide N-Methyltransferase (NNMT). J. Med. Chem. 2018, 61, 1541–1551. [Google Scholar] [CrossRef]
- Wang, Y.; Muylaert, C.; Wyns, A.; Vlummens, P.; De Veirman, K.; Vanderkerken, K.; Zaal, E.; Berkers, C.; Moreaux, J.; De Bruyne, E.; et al. S-Adenosylmethionine Biosynthesis Is a Targetable Metabolic Vulnerability in Multiple Myeloma. Haematologica 2023, 109, 256. [Google Scholar] [CrossRef]
- Attia, R.R.; Gardner, L.A.; Mahrous, E.; Taxman, D.J.; LeGros, L.; Rowe, S.; Ting, J.P.-Y.; Geller, A.; Kotb, M. Selective Targeting of Leukemic Cell Growth in Vivo and in Vitro Using a Gene Silencing Approach to Diminish S-Adenosylmethionine Synthesis. J. Biol. Chem. 2008, 283, 30788–30795. [Google Scholar] [CrossRef]
- Frau, M.; Tomasi, M.L.; Simile, M.M.; Demartis, M.I.; Salis, F.; Latte, G.; Calvisi, D.F.; Seddaiu, M.A.; Daino, L.; Feo, C.F.; et al. Role of Transcriptional and Posttranscriptional Regulation of Methionine Adenosyltransferases in Liver Cancer Progression. Hepatology 2012, 56, 165–175. [Google Scholar] [CrossRef]
- Liu, Q.; Chen, J.; Liu, L.; Zhang, J.; Wang, D.; Ma, L.; He, Y.; Liu, Y.; Liu, Z.; Wu, J. The X Protein of Hepatitis B Virus Inhibits Apoptosis in Hepatoma Cells through Enhancing the Methionine Adenosyltransferase 2A Gene Expression and Reducing S-Adenosylmethionine Production. J. Biol. Chem. 2011, 286, 17168–17180. [Google Scholar] [CrossRef]
- Sadek, K.M.; Lebda, M.A.; Nasr, N.E.; Nasr, S.M.; EL-Sayed, Y. Role of LncRNAs as Prognostic Markers of Hepatic Cancer and Potential Therapeutic Targeting by S-Adenosylmethionine via Inhibiting PI3K/Akt Signaling Pathways. Environ. Sci. Pollut. Res. Int. 2018, 25, 20057–20070. [Google Scholar] [CrossRef]
- Greenberg, A.K.; Rimal, B.; Felner, K.; Zafar, S.; Hung, J.; Eylers, E.; Phalan, B.; Zhang, M.; Goldberg, J.D.; Crawford, B.; et al. S-Adenosylmethionine as a Biomarker for the Early Detection of Lung Cancer. Chest 2007, 132, 1247. [Google Scholar] [CrossRef] [PubMed]
- Fatemi, N.; Tierling, S.; Es, H.A.; Varkiani, M.; Mojarad, E.N.; Aghdaei, H.A.; Walter, J.; Totonchi, M. DNA Methylation Biomarkers in Colorectal Cancer: Clinical Applications for Precision Medicine. Int. J. Cancer 2022, 151, 2068–2081. [Google Scholar] [CrossRef] [PubMed]
- Pascale, R.M.; Simile, M.M.; Miglio, M.R.D.; Nufris, A.; Daino, L.; Seddaiu, M.A.; Rao, P.M.; Rajalakshmi, S.; Sarma, D.S.R.; Feo, F. Chemoprevention by S-Adenosyl-L-Methionine of Rat Liver Carcinogenesis Initiated by 1,2-Dimethylhydrazine and Promoted by Orotic Acid. Carcinogenesis 1995, 16, 427–430. [Google Scholar] [CrossRef] [PubMed]
- Pascale, R.; Simile, M.M.; Ruggiu, M.E.; Seddaiu, M.A.; Satta, G.; Sequenza, M.J.; Daino, L.; Vannini, M.G.; Lai, P.; Feo, F. Reversal by 5-Azacytidine of the S-Adenosyl-L-Methionine-Induced Inhibition of the Development of Putative Preneoplastic Foci in Rat Liver Carcinogenesis. Cancer Lett. 1991, 56, 259–265. [Google Scholar] [CrossRef]
- Feo, F.; Frau, M.; Feo, C.F.; Pascale, R.M. New Insights on the Role of Epigenetic Alterations in Hepatocellular Carcinoma. J. Hepatocell. Carcinoma 2014, 1, 65–83. [Google Scholar] [CrossRef]
- Stoyanov, E.; Mizrahi, L.; Olam, D.; Schnitzer-Perlman, T.; Galun, E.; Goldenberg, D.S.; Stoyanov, E.; Mizrahi, L.; Olam, D.; Schnitzer-Perlman, T.; et al. Tumor-Suppressive Effect of S-Adenosylmethionine Supplementation in a Murine Model of Inflammation-Mediated Hepatocarcinogenesis Is Dependent on Treatment Longevity. Oncotarget 2017, 8, 104772–104784. [Google Scholar] [CrossRef]
- Yan, L.; Liang, X.; Huang, H.; Zhang, G.; Liu, T.; Zhang, J.; Chen, Z.; Zhang, Z.; Chen, Y. S-Adenosylmethionine Affects Cell Cycle Pathways and Suppresses Proliferation in Liver Cells. J. Cancer 2019, 10, 4368–4379. [Google Scholar] [CrossRef]
- Morgan, T.R.; Osann, K.; Bottiglieri, T.; Pimstone, N.; Hoefs, J.C.; Hu, K.Q.; Hassanein, T.; Boyer, T.D.; Kong, L.; Chen, W.P.; et al. A Phase II Randomized, Controlled Trial of S-Adenosylmethionine in Reducing Serum α-Fetoprotein in Patients with Hepatitis C Cirrhosis and Elevated AFP. Cancer Prev. Res. 2015, 8, 864–872. [Google Scholar] [CrossRef]
- Li, J.; Ramani, K.; Sun, Z.; Zee, C.; Grant, E.G.; Yang, H.; Xia, M.; Oh, P.; Ko, K.; Mato, J.M.; et al. Forced Expression of Methionine Adenosyltransferase 1A in Human Hepatoma Cells Suppresses in Vivo Tumorigenicity in Mice. Am. J. Pathol. 2010, 176, 2456–2466. [Google Scholar] [CrossRef]
- Ilisso, C.P.; Delle Cave, D.; Mosca, L.; Pagano, M.; Coppola, A.; Mele, L.; Caraglia, M.; Cacciapuoti, G.; Porcelli, M. S-Adenosylmethionine Regulates Apoptosis and Autophagy in MCF-7 Breast Cancer Cells through the Modulation of Specific MicroRNAs. Cancer Cell Int. 2018, 18, 197. [Google Scholar] [CrossRef] [PubMed]
- Delle Cave, D.; Ilisso, C.P.; Mosca, L.; Pagano, M.; Martino, E.; Porcelli, M.; Cacciapuoti, G. The Anticancer Effects of S-Adenosylmethionine on Breast Cancer Cells. JSM Chem. 2018, 5, 1049–1056. [Google Scholar]
- Chik, F.; Machnes, Z.; Szyf, M. Synergistic Anti-Breast Cancer Effect of a Combined Treatment with the Methyl Donor S-Adenosyl Methionine and the DNA Methylation Inhibitor 5-Aza-2’-Deoxycytidine. Carcinogenesis 2014, 35, 138–144. [Google Scholar] [CrossRef]
- Mahmood, N.; Arakelian, A.; Cheishvili, D.; Szyf, M.; Rabbani, S.A. S-Adenosylmethionine in Combination with Decitabine Shows Enhanced Anti-Cancer Effects in Repressing Breast Cancer Growth and Metastasis. J. Cell. Mol. Med. 2020, 24, 10322–10337. [Google Scholar] [CrossRef] [PubMed]
- Ilisso, C.P.; Castellano, M.; Zappavigna, S.; Lombardi, A.; Vitale, G.; Dicitore, A.; Cacciapuoti, G.; Caraglia, M.; Porcelli, M. The Methyl Donor S-Adenosylmethionine Potentiates Doxorubicin Effects on Apoptosis of Hormone-Dependent Breast Cancer Cell Lines. Endocrine 2015, 50, 212–222. [Google Scholar] [CrossRef]
- Cave, D.D.; Desiderio, V.; Mosca, L.; Ilisso, C.P.; Mele, L.; Caraglia, M.; Cacciapuoti, G.; Porcelli, M. S-Adenosylmethionine-Mediated Apoptosis Is Potentiated by Autophagy Inhibition Induced by Chloroquine in Human Breast Cancer Cells. J. Cell. Physiol. 2018, 233, 1370–1383. [Google Scholar] [CrossRef]
- Mosca, L.; Pagano, M.; Ilisso, C.P.; Cave, D.D.; Desiderio, V.; Mele, L.; Caraglia, M.; Cacciapuoti, G.; Porcelli, M. AdoMet Triggers Apoptosis in Head and Neck Squamous Cancer by Inducing ER Stress and Potentiates Cell Sensitivity to Cisplatin. J. Cell. Physiol. 2019, 234, 13277–13291. [Google Scholar] [CrossRef]
- Mosca, L.; Minopoli, M.; Pagano, M.; Vitiello, F.; Carriero, M.; Cacciapuoti, G.; Porcelli, M. Effects of S-adenosyl-L-methionine on the Invasion and Migration of Head and Neck Squamous Cancer Cells and Analysis of the Underlying Mechanisms. Int. J. Oncol. 2020, 56, 1212–1224. [Google Scholar] [CrossRef]
- Bedard, G.T.; Gilaj, N.; Peregrina, K.; Brew, I.; Tosti, E.; Shaffer, K.; Tyler, P.C.; Edelmann, W.; Augenlicht, L.H.; Schramm, V.L. Combined Inhibition of MTAP and MAT2a Mimics Synthetic Lethality in Tumor Models via PRMT5 Inhibition. J. Biol. Chem. 2024, 300, 105492. [Google Scholar] [CrossRef]
- Mosca, L.; Pagano, M.; Pecoraro, A.; Borzacchiello, L.; Mele, L.; Cacciapuoti, G.; Porcelli, M.; Russo, G.; Russo, A. S-Adenosyl-l-Methionine Overcomes UL3-Mediated Drug Resistance in P53 Deleted Colon Cancer Cells. Int. J. Mol. Sci. 2020, 22, 103. [Google Scholar] [CrossRef]
- Zhao, Y.; Li, J.S.; Guo, M.Z.; Feng, B.S.; Zhang, J.P. Inhibitory Effect of S-Adenosylmethionine on the Growth of Human Gastric Cancer Cells in Vivo and in Vitro. Chin. J. Cancer 2010, 29, 752–760. [Google Scholar] [CrossRef] [PubMed]
- Schmidt, T. S-Adenosylmethionine Affects ERK1/2 and STAT3 Pathway in Androgen-Independent Prostate Cancer Cells. Mol. Biol. Rep. 2022, 49, 4805–4817. [Google Scholar] [CrossRef] [PubMed]
- Liu, M.; Mobet, Y.; Shen, H. S-Adenosylmethionine Inhibits the Proliferation of Retinoblastoma Cell Y79, Induces Apoptosis and Cell Cycle Arrest of Y79 Cells by Inhibiting the Wnt2/β-Catenin Pathway. Arch. Immunol. Ther. Exp. 2024, 72, 20. [Google Scholar] [CrossRef]
- Santini, D.; Vincenzi, B.; Massacesi, C.; Picardi, A.; Gentilucci, U.V.; Esposito, V.; Liuzzi, G.; La Cesa, A.; Rocci, L.; Marcucci, F.; et al. S-Adenosylmethionine (AdoMet) Supplementation for Treatment of Chemotherapy-Induced Liver Injury. Anticancer Res. 2003, 23, 5173–5179. [Google Scholar] [PubMed]
- Vincenzi, B.; Santini, D.; Frezza, A.M.; Berti, P.; Vespasiani, U.; Picardi, A.; Tonini, G. The Role of S-Adenosyl Methionine in Preventing FOLFOX-Induced Liver Toxicity: A Retrospective Analysis in Patients Affected by Resected Colorectal Cancer Treated with Adjuvant FOLFOX Regimen. Expert Opin. Drug Saf. 2011, 10, 345–349. [Google Scholar] [CrossRef]
- Onorato, A.; Napolitano, A.; Spoto, S.; Incorvaia, L.; Russo, A.; Santini, D.; Tonini, G.; Vincenzi, B. S-Adenosylmethionine Supplementation May Reduce Cancer-Related Fatigue: A Prospective Evaluation Using the FACIT-F Questionnaire in Colon Cancer Patients Undergoing Oxaliplatin-Based Chemotherapy Regimens. Chemotherapy 2021, 66, 161–168. [Google Scholar] [CrossRef]
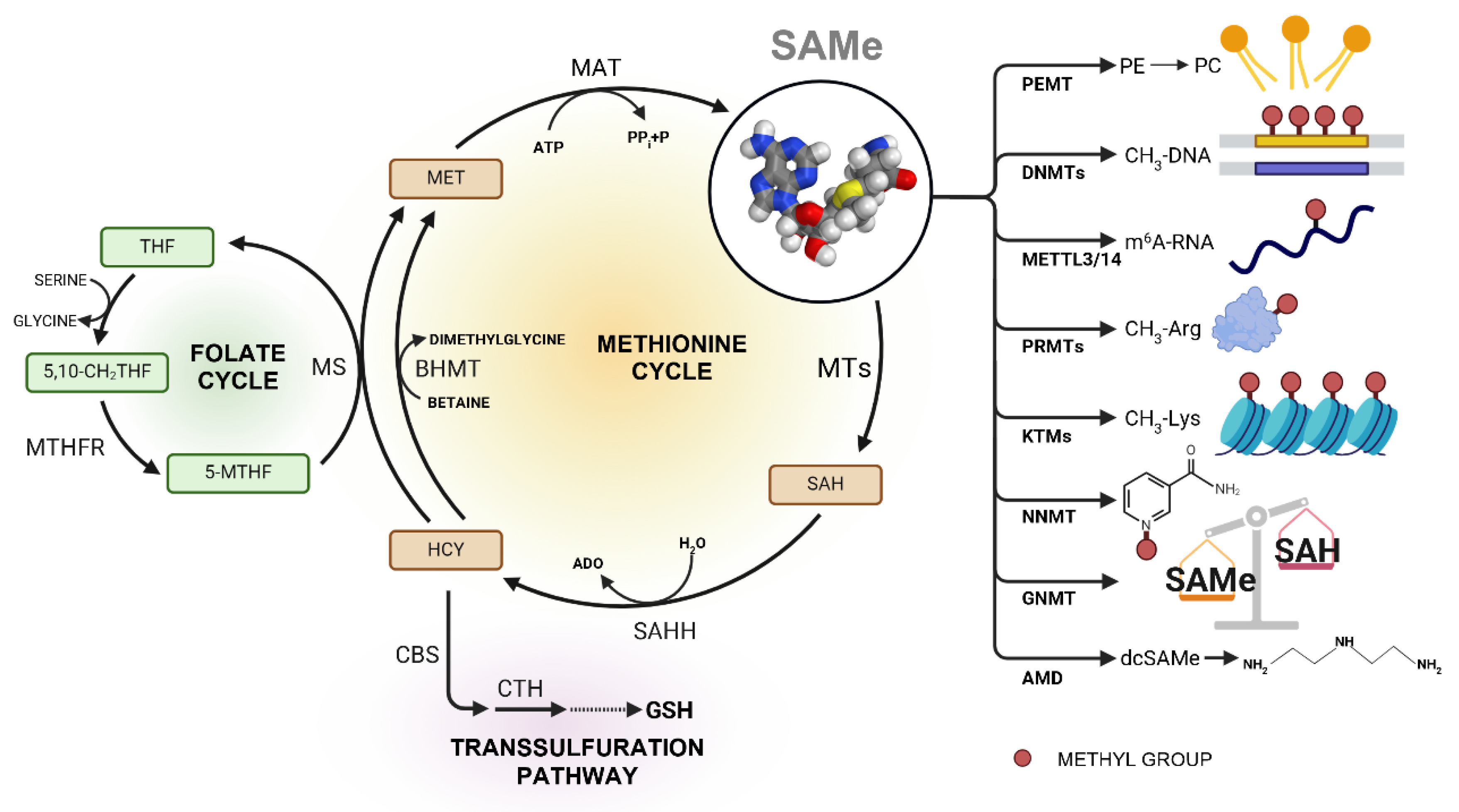

Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Fernández-Ramos, D.; Lopitz-Otsoa, F.; Lu, S.C.; Mato, J.M. S-Adenosylmethionine: A Multifaceted Regulator in Cancer Pathogenesis and Therapy. Cancers 2025, 17, 535. https://doi.org/10.3390/cancers17030535
Fernández-Ramos D, Lopitz-Otsoa F, Lu SC, Mato JM. S-Adenosylmethionine: A Multifaceted Regulator in Cancer Pathogenesis and Therapy. Cancers. 2025; 17(3):535. https://doi.org/10.3390/cancers17030535
Chicago/Turabian StyleFernández-Ramos, David, Fernando Lopitz-Otsoa, Shelly C. Lu, and José M. Mato. 2025. "S-Adenosylmethionine: A Multifaceted Regulator in Cancer Pathogenesis and Therapy" Cancers 17, no. 3: 535. https://doi.org/10.3390/cancers17030535
APA StyleFernández-Ramos, D., Lopitz-Otsoa, F., Lu, S. C., & Mato, J. M. (2025). S-Adenosylmethionine: A Multifaceted Regulator in Cancer Pathogenesis and Therapy. Cancers, 17(3), 535. https://doi.org/10.3390/cancers17030535








