Abstract
A total of 265 Salmonella Enteritidis isolates collected from retail markets and children’s hospitals in Shanghai were used to investigate the prevalence and molecular epidemiology of plasmid-mediated fosfomycin resistance genes. Nine of the isolates—7 from the 146 (4.79%) retail chicken-related samples and 2 from the 119 (1.68%) samples from clinical children—were fosfomycin-resistant (FosR). The fosA3 gene was detected in all of the nine FosR isolates, which were located on Inc F-type (8/9, 88.9%) and unknown-type (1/9, 11.1%) transferable plasmids. In total, five plasmid types, namely Inc HI2 (1/9, 11.1%), Inc I1 (3/9, 33.3%), Inc X (8/9, 88.9%), Inc FIIs (9/9, 100%), and Inc FIB (9/9, 100%), were detected in these FosR isolates, which possessed five S1 nuclease pulsed-field gel electrophoresis (S1-PFGE) profiles. The extended-spectrum β-lactamase determinant blaCTX-M-14 subtype was identified in one FosR S. Enteritidis isolate, which was located in a transferable unknown-type plasmid co-carrying fosA3 and tetR genes. Sequence homology analysis showed that this plasmid possessed high sequence similarity to previously reported blaCTX-M-14- and fosA3-positive plasmids from E. coli strains, implying that plasmids carrying the fosA3 gene might be disseminated among Enterobacterales. These findings highlight further challenges in the prevention and treatment of Enterobacteriaceae infections caused by plasmids containing fosA3.
1. Introduction
Because of the increase in the incidence of multidrug-resistant (MDR) bacteria and the depletion of newly developed antibiotics, the reassessment of “older” antibiotics has emerged as an interesting option. Fosfomycin was discovered five decades ago and is a promising candidate for the treatment of various MDR pathogens [1], especially extended-spectrum beta-lactamase (ESBL)-producing Enterobacterales [2,3]. However, previous studies have demonstrated that overall fosfomycin resistance in China is higher than in other parts of the world in both human and animal hosts [4]. Preliminary evidence suggests that fosA3 is the primary gene responsible for fosfomycin resistance in common clinical pathogens in China [4], such as Escherichia coli and Klebsiella pneumoniae isolates [5,6,7], spread through the dissemination of Inc F and Inc N plasmids rather than the clonal expansion of specific strains [8,9]. Therefore, these plasmids may be spread among enterobacteria, including foodborne pathogens.
Foodborne salmonellosis has become a serious public health issue worldwide. Salmonella is a problematic bacterial pathogen that is frequently resistant to multiple antibiotics. Although the prevalence rate of fosfomycin resistance in Salmonella remains unclear, the gene fosA3 has been recently found among fosfomycin-resistant Salmonella isolates from wild birds [10], food animals [11,12,13,14], and hospitalized patients [15,16,17]; it was also found in isolates obtained from food [18] and a healthy catering worker [19]. Further analysis showed that the genetic environment of the fosA3 gene in Salmonella had a structure identical to that in E. coli isolates, indicating that the transferability of fosA3-harboring plasmids in enterobacteria accounts for the further transmission of antimicrobial resistance.
Salmonella enterica subsp. enterica serovar Enteritidis (S. Enteritidis) is the most common serovar identified in chicken meat [20] and is frequently associated with human illness [21]. Information on the occurrence and epidemiological characteristics of fosfomycin-resistant S. Enteritidis isolates in China remains scarce. In this study, we investigated the prevalence and molecular epidemiology of plasmid-mediated fosfomycin resistance genes in S. Enteritidis isolates collected from retail chicken samples and children with gastroenteritis. The transfer mechanism of fosfomycin resistance in these isolates was characterized using plasmid assays and sequencing.
2. Materials and Methods
2.1. S. Enteritidis Isolates and Fosfomycin Susceptibility Testing
S. Enteritidis isolates (n = 265) for this study were obtained from 146 retail chicken-related samples and 119 children under the age of 10 in Shanghai, China from 2010 to 2012. Food sources, patient demographics, and isolation dates of these isolates were reported in our previous studies [22,23]. The minimal inhibitory concentration (MIC) of the isolates against fosfomycin (FOS; Sigma, CA, USA) was determined using an agar dilution method according to the guidelines recommended by the Clinical and Laboratory Standards Institute [24]. The breakpoint for fosfomycin was MIC ≥ 256 μg/mL, according to the interpretive standards of the CLSI [24]. The following 12 antibiotics were tested in fosfomycin resistance (FosR) isolates: tetracycline (TET), sulfisoxazole (SUL), ampicillin (AMP), ceftriaxone (CRO), streptomycin (STR), azithromycin (AZI), trimethoprim-sulfonamides (SXT), amikacin (AMK), ciprofloxacin (CIP), chloramphenicol (CHL), kanamycin (KAN), and gentamicin (GEN). The sensitivity analysis of the aforementioned antibiotics was evaluated using MIC, based on the disk diffusion test. Antibiotics were purchased from Sigma-Aldrich (St. Louis, MO, USA). E. coli ATCC 25922 and Enterococcus faecalis ATCC 29212 were used as quality control organisms.
2.2. Plasmid Studies
Plasmid incompatibility (Inc.) groups were assigned by PCR-based replicon typing using the genomic DNA of the FosR isolates as a template. Amplification by PCR was performed with 18 specific primer pairs designed for FIA, FIB, FIC, HI1, HI2, I1, L/M, N, P, W, T, A/C, K, B/O, X, Y, F, and FIIs basic replicons using a previously described corresponding protocol [25]. The presence of plasmid-encoded fosfomycin resistance (fosA3, fosC2, and fosKP96), β-lactams (blaTEM, blaCTX-M, and blaOXA), tetracycline (tetA and tet B), and sulfonamides (sul1 and sul2) were determined by PCR as described previously by Sato et al. [26] and Zhou et al. [22]. All the primers used are listed in Tables S1 and S2. The PCR cycling parameters were as follows: initial denaturation at 94 °C for 10 min; denaturation at 94 °C for 1 min; corresponding annealing temperature conditions (Tables S1 and S2) for 30 s; extension at 72 °C for 1 min and 30 cycles; and extension at 72 °C for 10 min.
Conjugation experiments were performed using E. coli C600 as the recipient [27]. Briefly, each FosR S. Enteritidis isolate as a donor and E. coli C600 after overnight incubation were mixed and then transferred to a filter in Luria–Bertani broth (Oxoid, Cambridge, UK) agar plates for overnight culture. Trans-conjugants were selected on MacConkey agar plates supplemented with fosfomycin (256 μg/mL) and rifampin (200 µg/mL). Plasmids of the parental isolates and trans-conjugants were sized by S1 nuclease pulsed-field gel electrophoresis (S1-PFGE) [28].
2.3. Plasmid Sequencing and Analysis
Genomic DNA was extracted from overnight cultures of S. Enteritidis SJTUF11561 using the QIAamp DNA Mini Kit (Qiagen, CA, USA). The whole genome sequencing (WGS) was performed on the PacBio RS II sequencing platform at the Majorbio Corporation (Shanghai, China). Briefly, a 10 kb DNA library was constructed and sequenced using single-molecule real-time sequencing. The sequence data were assembled using the Canu V1.3 software [29], and the complete sequence was annotated using the RAST, BLASTn, and BLASTp programs on the NCBI platform, followed by manual inspection. Insertion sequences and repetitive elements were identified using IS finder2. A schematic plasmid map was constructed using WinPlas2.7 software. OriTfinder (http://202.120.12.134/oriTfinder/oriTfinder.html, accessed on 12 March 2024) and Plasmidfinder (https://cge.cbs.dtu.dk/services/PlasmidFinder/, accessed on 12 March 2024) were used to identify the origins of transfers and plasmid types in the DNA sequences of bacterial plasmids, respectively.
2.4. Nucleotide Sequence Accession Number
The complete sequences of S. Enteritidis SJTUF11561 and its plasmids (p11561A, p115561B, p11561C, p11561D, and p11561E) were deposited in the NCBI database under PRJNA1136444.
3. Results
3.1. Prevalence of Fosfomycin Resistance and Plasmid-Mediated Determinants
In total, 9 out of 256 (3.51%) isolates, including 7 out of 146 (4.79%) from retail chicken-related samples and 2 out of 119 (1.68%) from children with gastroenteritis, were fosfomycin-resistant (Table 1). The MIC value of two retail chicken-related isolates was 512 μg/mL; the other fosfomycin-resistant isolates had an MIC value of 256 μg/mL (Table 1 and Table 2). All FosR isolates were positive for fosA3 (Table 2), and none possessed fosKP96 and fosC2 genes. The incidence of sul2 was 100%, and blaTEM was present in six out of nine (66.7%) FosR isolates. Two FosR isolates from chicken manure and both the isolates from clinical children carried tetA (four out of nine, 44.4%). Only one FosR isolate (SJTUF 11561) carried blaCTX-M (Table 2).

Table 1.
Minimal inhibitory concentration (MIC) of fosfomycin among 265 Salmonella Enteritidis isolates.

Table 2.
The antibiotic resistance phenotypes, resistance genes, and plasmid incompatibility group profiles of the fosfomycin-resistant strains.
3.2. Characteristics of FosA3-Carrying S. Enteritidis Isolates and Plasmids
All of the nine FosR isolates were resistant to two or more drugs (Table 2). The incidence of sulfisoxazole (SUL) and fosfomycin (FOS) was 100% in the FosR isolates. Specifically, strain SJTUF 11346 only possessed SUL-FOS. Except for SJTUF 11346, all of the FosR isolates from chicken-related samples possessed ampicillin (AMP), streptomycin (STR), and trimethoprim-sulphonamide (SXT) resistance, while two clinical isolates did not. Additionally, four isolates from chicken manure and clinical children possessed tetracycline (TET) resistance.
A total of five types of Inc groups were detected in these fosA3-positive isolates, including Inc HI2 (1/9, 11.1%), Inc I1 (3/9, 33.3%), Inc X (8/9, 88.9%), Inc FIIs (9/9, 100%), and Inc FIB (9/9, 100%) (Table 2). SJTUF 11346 only contained the Inc FIB and FIIs plasmid types, whereas Inc HI2 only existed in SJTUF 11561. The rest of seven isolates were divided into two groups: five possessing Inc FIIs, FIB, and X and two possessing Inc FIIs, FIB, X, and I1. S1-PFGE showed five different plasmid profiles in the nine FosR isolates (Figure 1, lines 1–5). In general, strains with uniform S1-PFGE profiles possessed the same plasmid types (Figure 1, lines 1, 2, and 5), such as SJTUF 10993, SJTUF 10994, SJTUF 10959, SJTUF 10960, SJTUF 11565, SJTUF 11642, and SJTUF 11653. Moreover, the latter five strains with the same plasmid types (IncFIIs, FIB, and X) showed two distinct S1-PFGE profiles (Figure 1, lines 2 and 5).
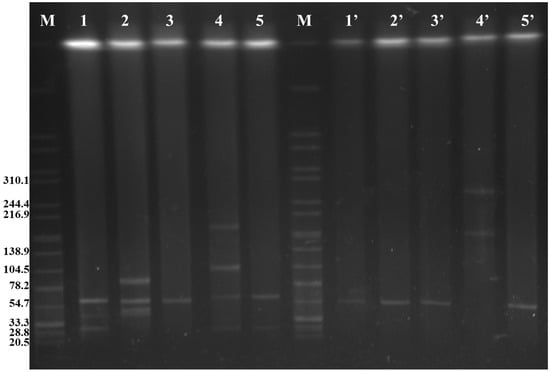
Figure 1.
S1 nuclease pulsed-field gel electrophoresis of FosR S. Enteritidis donors (1–5) and corresponding E. coli trans-conjugants (1′–5′). M: H9812; 1: SJTUF 10993 and SJTUF 10994; 2: SJTUF 10959 and SJTUF 10960; 3: SJTUF 11346; 4: SJTUF 11561; 5: SJTUF 11565, SJTUF 11642, and SJTUF 11653; 1′: trans-conjugants of SJTUF 10993 and SJTUF 10994; 2′: trans-conjugants of SJTUF 10959 and SJTUF 10960; 3′: trans-conjugants of SJTUF 11346; 4′: trans-conjugant of SJTUF 11561; and 5′: trans-conjugants of SJTUF 11565, SJTUF 11642, and SJTUF 11653.
3.3. Conjugation Experiments
Conjugation experiments were performed to confirm the transmissibility of fosfomycin resistance in the recipient E. coli strain in nine FosR isolates at a frequency of 10−4–10−6 per donor cell (Table 2). Fosfomycin MICs for all trans-conjugants were ≥512 μg/mL. In eight of these FosR isolates, a single plasmid with the size of ~60 kb was transferred to the recipients (Figure 1, Lines 1′–3′ and 5′); moreover, two F-type plasmids (Inc FIIs and FIB) and the fosA3 gene were also tested in these trans-conjugants (Table 2). The results showed that the fosfomycin resistance of the eight fosA3-harboring isolates was successfully transferred to recipient cells (Table 2) and that the fosA3 gene was probably located on the Inc F plasmids. Notably, the plasmid type of the SJTUF 11561 trans-conjugant was the same as that of the parental isolate (Table 2). The S1-PFGE profile showed that two larger plasmids with sizes of ~150 kb and ~250 kb were detected in the trans-conjugant (Figure 1, line 4′). These results suggest that pairwise fusion likely occurred in the four tested plasmids (with sizes of ~30, ~60, ~120, and ~190 kb) of the parental strain SJTUF 11561. Unfortunately, these fusion plasmids were not sufficiently stable for further analysis.
3.4. Sequence Analysis of Plasmids in SJTUF 11561
Due to the co-occurrence of the fosA3 gene and blaCTX-M in SJTUF11561, it was selected for further sequencing. Additionally, this strain carried all four types of detected plasmids and had the highest transfer rate. According to WGS, five plasmids were identified in SJTUF 11561: p11561A, p115561B, p11561C, p11561D, and p11561E (Table 3). The molecular weights of the last four larger plasmids corresponded to the S1-PFGE analysis (Figure 1 and Table 3), whereas the smallest plasmid (p11561 A, ~8 kb) was not shown in the S1-PFGE pattern, possibly attributable to the display ability of the detection method. We confirmed that several antimicrobial resistance genes were located in three of these plasmids (p11561A, p115561B, and p11561C) (Table 3). Specifically, the fosA3-, blaCTX-M-14-, and tetR-bearing plasmid p11561A was 8056 bp in length but did not match the plasmid typing. Plasmid p11561B was 24,484 bp in size and belonged to the Inc X1 plasmid type, which harbors aph(6)-Id, aph(3″)-Ib, blaTEM, and sul2 genes. Plasmid p11561C was 64,327 bp in size, comprising the backbone elements of the Inc FIB and Inc FIIs plasmids, indicating that p115561C is a hybrid plasmid bearing both virulence genes (spvA, spvR, spvB, and pagC) and the resistance gene blaTEM. Plasmids p11561D (109,062 bp, Inc I1-I) and p11561E (168,488 bp, Inc HI2) were not responsible for resistance genes. However, three virulence genes (afaD, cia, and terC) were present in the two plasmids (Table 3).

Table 3.
Plasmid analysis of SJTUF 11561 mined from whole genome sequencing.
The full-length fragment of pSJTUF11561(1–8056 bp) bearing fosA3 and blaCTX-M-14 was compared with other plasmids identified using BLASTn. It showed similarity to a partial-length fragment of pC0121T (GenBank accession no. JX442753; 600–8000 bp; 91% coverage; 99.99% identity), which is an Inc FII resistance plasmid from E. coli (Figure 2). Sequence comparisons showed that all three resistance genes (blaCTX-M-14, fosA3, tetR) from both plasmids were surrounded by the transposase gene (tnpA); the difference was that IS26 copies were inserted downstream of the resistance gene (tetR) in plasmid pSJTUF11561 but not in pC0121T. Moreover, the gene tnpA in the IS26 copies was inverted, compared to that of pC0121T. The second plasmid pN0863T (accession no. JQ823170; 1–4731 bp; 50% coverage; 100.00% identity) showed a high similarity to pSJTUF11561 (Figure 2). Plasmid pN0863T was recovered from an E. coli isolated from the fecal specimens of a stray dog. The resistance gene tetR was not detected in pN0863T, and there was a pair of reversed tnpA genes at both ends of the selected fragment, in which the tnpA gene located downstream of the resistance gene (fosA3) was inverted, compared to the tnpA located on the IS26 copies of plasmid pSJTUF11561. Moreover, between the two resistance genes (blaCTX-M-14 and fosA3) located in pSJTUF11561, there was more than one hypothetical protein when compared with those located in pC0121T and pN0863T.
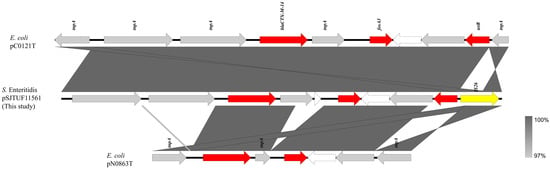
Figure 2.
Genetic environment comparison of fosA3 and blaCTX-M-14 genes in plasmids p11561A (this study), pC0121T (JX442753), and pN0863T (JQ823170). Gray shading indicates shared regions with a high degree of homology. Genes are represented by arrows and colored depending on gene function as depicted: red, antimicrobial resistance; yellow, mobile element; white, hypothetical protein; and gray, other protein (or genes).
4. Discussion
Previous studies have shown that the resistance rates of fosfomycin are significantly different among different Gram-negative bacteria isolated from clinical samples, being 9.6% (5/52) in E. coli [30], 36.1% (13/36) in Pseudomonas aeruginosa [30], 74.2% (23/31) in Klebsiella pneumoniae [30], 64.3% (9/14) in Acinetobacter baumannii [30], 9.5% (25/263) in Shigella [31], and 2.8% (14/501) in S. Enteritidis [16]. In the current study, the resistance rate of 119 strains of S. Enteritidis isolated from clinical children was 1.68% (2/119), which was significantly lower than the aforementioned Gram-negative bacteria. We speculate that there might be two possible reasons for this: (1) These selected isolates were from childhood cases, and fosfomycin is currently rarely used in children; therefore, it is less likely to acquire drug resistance, and (2) the current clinical application of fosfomycin is in combination with cephalosporins, aminoglycosides, quinolones, and other antibacterial drugs for the treatment of methicillin-resistant Staphylococcus aureus [32], MDR Pseudomonas aeruginosa (MDRP) [33], and Shigella dysenteriae [34], but the frequency of its use in the treatment of Salmonella infection is relatively low. In addition, compared with the recent reports by Gu et al. [11] and Fang et al. [12], in which the fosfomycin resistance rates of isolates from farm chickens and other food animals were 2.1% (6/288) and 2.6% (8/310), respectively, the resistance rate was a little higher (4.79%, 7/146) in retail chicken in this study, indicating the cumulative risk of fosfomycin resistance from broiler farming to retail. To the best of our knowledge, although Lin et al. [18] isolated two fosA3-harboring Salmonella isolates from retail chicken meat, this is the first report on the prevalence of fosfomycin resistance in Salmonella isolated from retail foods. We would like to expand the types of food and serotypes of Salmonella to enrich the survey data in future research.
FosA3 is the most frequently identified fosfomycin-modifying enzyme worldwide [4]. Inc FII plasmids play a predominant role in the dissemination of fosfomycin-modifying enzymes in Enterobacterales among clinical isolates and food animals in Asian countries [3,35,36]. Recent reports from China have described fosA3 on transferable Inc FII plasmids in S. Enteritidis from chickens purchased from a market in Hong Kong [18] and from clinical isolates in Shanghai [16]. Similarly, the results from the plasmid typing of the parental isolate and its trans-conjugants in this study showed that the fosA3 gene in eight FosR S. Enteritidis strains was also related to the Inc F-type plasmid.
The dissemination of the fosA3 gene is closely associated with that of the ESBL gene blaCTX-M [37,38,39]. Previous studies have reported that some plasmids from Salmonella carrying the fosA3 gene also carried blaCTX-M genes [17,40,41,42], whereas some fosA3 genes in plasmids from Salmonella did not coexist with blaCTX-M genes [12]. Both of the above situations were observed in this study; specifically, eight out of nine plasmids carried the fosA3 gene, but no blaCTX-M genes were detected (Table 2), whereas the fosA3 gene was co-transferred with blaCTX-M in the one remaining plasmid from SJTUF 11561 (Table 2). In previous studies, blaCTX-M-14 genes, which co-transferred with the fosA3 gene in Salmonella, were usually detected in the Inc HI2 plasmid [12,16,19,43], and blaCTX-M-55 genes were detected in the Inc FII [12,16,18] or unknown-type plasmids [16]. Unlike previous studies, we found the coexistence of fosA3 and blaCTX-M-14 genes on an 8 kb plasmid of unknown type (Figure 2), suggesting that plasmids associated with the co-transfer of fosA3 and blaCTX-M might be of diverse types.
A typical fosA3 cassette (IS26-fosA3-orf1-orf2-orf3-IS26) can be randomly inserted into the area adjacent to the blaCTX-M cassette in Enterobacterales to form different genetic structures [13,18,36,40]. In our study, the fosA3 gene detected in the pSJTUF11561 plasmid was located in the genetic environment of IS26-ISEcp1-IS10-blaCTX-M-14-ΔIS903D-fosA3-orf1-orf2-Δorf3-IS26, which was also the typical fosA3 cassette inserted downstream of a blaCTX-M-14 cassette (Figure 2). Sequence comparison analysis in this current study (Figure 2) showed that the genetic environment of fosA3 of pSJTUF11561 was practically identical to two published plasmids isolated from E. coli, including pC0121T (Inc FII, IS26-ISEcp1-IS10-blaCTX-M-14-ΔIS903D-fosA3-orf1-orf2-Δorf3-IS26) [44] and pN0863T (Inc N, IS26-ΔISEcp1-blaCTX-M-14-ΔIS903D-fosA3-orf1-Δorf2-IS26) [44]. It was surprising that the genetic environment of fosA3 detected in pSJTUF11561 was different, with some plasmids in Salmonella carrying both fosA3 and blaCTX-M. Specifically, the fosA3 gene of Salmonella pGDD27–24 from a duck isolate had the common genetic background of IS26-tetR-orf2-ofr1-fosA3-IS26-blaTEM-1-orf477-blaCTX-M-55-ΔISEcp1 [12], and the fosA3 gene of Salmonella pYZU1189 from a healthy woman had the common genetic background of ΔISEcp1-blaCTX-M-14-IS903B-fosA3-orf [19]. Interestingly, the common genetic backgrounds of these two fosA3 genes in these Salmonella strains were similar to that reported in E. coli [12,19]. These results suggest that plasmids carrying fosA3 might be disseminated among Enterobacterales, not just among genera such as Salmonella or E. coli.
Furthermore, the S1-PFGE and PCR results of SJTUF 11561 showed that the number of plasmids decreased (Figure 1, lines 4 and 4′) and the size of plasmids increased (Figure 1, lines 4 and 4′), but the replication types did not change (Table 2) in the trans-conjugant, compared to the parental isolate, which revealed that the fusion occurred in the conjugation process. A similar phenomenon was reported previously, where both fosA3 and blaCTX-M genes located in E. coli plasmids isolated from chickens were successfully transferred to recipients by conjugation experiments, and the plasmid size in these trans-conjugants was much larger than that in the donor strains [45]. Multiple replicons and translocases were detected in the other four plasmids, which may be used to assist the transfer of fosA3 and blaCTX-M genes through the fusion process.
5. Conclusions
In summary, this is the first report on the prevalence rate of fosfomycin resistance in Salmonella Enteritidis isolated from retail chicken; moreover, this study demonstrated that FosR Salmonella Enteritidis can be detected in children with gastroenteritis who are currently rarely treated with fosfomycin. The plasmid-mediated fosA3 gene played a predominant role in fosfomycin resistance because all of the FOSR Salmonella Enteritidis isolates were positive for the fosA3 gene and could be transferred by plasmids, including Inc F and the unknown type. Furthermore, the fosA3 gene was co-distributed with other important antibiotic resistance genes, including blaTEM, sul2, tetA, and blaCTX-M. Our sequencing results support the idea that fosA3 and blaCTX-M might be disseminated among Enterobacterales but not limited to the Salmonella genus; therefore, further spread of these plasmids at the Enterobacterales level is a serious public health concern.
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/pathogens13090816/s1, Table S1. Primers used in PCR-based plasmid replicon typing; Table S2. Primers used for antimicrobial resistance genes.
Author Contributions
Data curation, H.L.; formal analysis, L.Z.; funding acquisition, X.Z.; investigation, S.Y.; methodology, L.L. and X.X.; supervision, X.Z.; writing—original draft, L.L. and S.Y.; writing—review and editing, H.L. and X.Z. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by the National Natural Science Foundation of China, grant no. 32072320, and Climbing Plan for Excellent Young Staffs at Shanghai University of Medicine & Health Sciences, grant no. A3-2601-24-311001.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
The complete sequences of S. Enteritidis SJTUF11561 and its plasmids (p11561A, p115561B, p11561C, p11561D, and p11561E) have been deposited in the NCBI database under PRJNA1136444.
Acknowledgments
The authors thank Xianming Shi in the Shanghai Jiao Tong University for kindly offering the Salmonella strains.
Conflicts of Interest
The authors declare no conflicts of interest.
References
- Silver, L.L. Fosfomycin: Mechanism and resistance. Cold Spring Harb. Perspect. Med. 2017, 7, a025262. [Google Scholar] [CrossRef] [PubMed]
- Falagas, M.E.; Vouloumanou, E.K.; Samonis, G.; Vardakas, K.Z. Fosfomycin. Clin. Microbiol. Rev. 2016, 29, 321–347. [Google Scholar] [CrossRef] [PubMed]
- Zurfluh, K.; Treier, A.; Schmitt, K.; Stephan, R. Mobile fosfomycin resistance genes in Enterobacteriaceae-An increasing threat. Microbiologyopen 2020, 9, e1135. [Google Scholar] [CrossRef] [PubMed]
- Yang, T.Y.; Lu, P.L.; Tseng, S.P. Update on fosfomycin-modified genes in Enterobacteriaceae. J. Microbiol. Immunol. Infect. 2019, 52, 9–21. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Zheng, B.; Li, Y.; Zhu, S.; Xue, F.; Liu, J. Antimicrobial susceptibility and molecular mechanisms of fosfomycin resistance in clinical Escherichia coli isolates in mainland China. PLoS ONE 2015, 10, e0135269. [Google Scholar] [CrossRef][Green Version]
- Bartoloni, A.; Sennati, S.; Di Maggio, T.; Mantella, A.; Riccobono, E.; Strohmeyer, M.; Revollo, C.; Villagran, A.L.; Pallecchi, L.; Rossolini, G.M. Antimicrobial susceptibility and emerging resistance determinants (blaCTX-M, rmtB, fosA3) in clinical isolates from urinary tract infections in the Bolivian Chaco. Int. J. Infect. Dis. 2016, 43, 1–6. [Google Scholar] [CrossRef][Green Version]
- Mattioni, M.V.; Hrabak, J.; Bitar, I. Fosfomycin resistance mechanisms in Enterobacterales: An increasing threat. Front. Cell. Infect. Microbiol. 2023, 13, 1178547. [Google Scholar] [CrossRef]
- Lee, S.Y.; Park, Y.J.; Yu, J.K.; Jung, S.; Kim, Y.; Jeong, S.H.; Arakawa, Y. Prevalence of acquired fosfomycin resistance among extended-spectrum β-lactamase-producing Escherichia coli and Klebsiella pneumoniae clinical isolates in Korea and IS26-composite transposon surrounding fosA3. J. Antimicrob. Chemother. 2012, 67, 2843–2847. [Google Scholar] [CrossRef]
- Cao, X.L.; Shen, H.; Xu, Y.Y.; Xu, X.J.; Zhang, Z.F.; Cheng, L.; Chen, J.H.; Arakawa, Y. High prevalence of fosfomycin resistance gene fosA3 in blaCTX-M-harbouring Escherichia coli from urine in a Chinese tertiary hospital during 2010–2014. Epidemiol. Infect. 2017, 145, 818–824. [Google Scholar] [CrossRef]
- Villa, L.; Guerra, B.; Schmoger, S.; Fischer, J.; Helmuth, R.; Zong, Z.; García-Fernández, A.; Carattoli, A. IncA/C plasmid carrying blaNDM-1, blaCMY-16, and fosA3 in a Salmonella enterica serovar corvallis strain isolated from a migratory wild bird in Germany. Antimicrob. Agents Chemother. 2015, 59, 6597–6600. [Google Scholar] [CrossRef]
- Gu, X.X.; Zhang, W.H.; Zhang, L.J.; Yang, L.; Fu, J.L.; Jiang, H.X. Prevalence and dissemination of fosfomycin resistance gene fosA3 among Salmonella isolates from food-producing animals. Vet. Sci. China 2017, 47, 514–522. [Google Scholar]
- Fang, L.X.; Jiang, Q.; Deng, G.H.; He, B.; Sun, R.Y.; Zhang, J.F.; Cen, D.J.; Miao, Y.Y.; Wang, D.; Guo, W.Y.; et al. Diverse and flexible transmission of fosA3 associated with heterogeneous multidrug resistance regions in Salmonella enterica Serovar Typhimurium and Indiana isolates. Antimicrob. Agents Chemother. 2020, 64, e02001-19. [Google Scholar] [CrossRef]
- Zhang, L.J.; Gu, X.X.; Zhang, J.; Yang, L.; Lu, Y.W.; Fang, L.X.; Jiang, H.X. Characterization of a fosA3 carrying IncC-IncN plasmid from a multidrug-resistant ST17 Salmonella Indiana isolate. Front. Microbiol. 2020, 11, 1582. [Google Scholar] [CrossRef]
- Wang, D.; Fang, L.X.; Jiang, Y.W.; Wu, D.S.; Jiang, Q.; Sun, R.Y.; Wang, M.G.; Sun, J.; Liu, Y.H.; Liao, X.P. Comparison of the prevalence and molecular characteristics of fosA3 and fosA7 among Salmonella isolates from food animals in China. J. Antimicrob. Chemother. 2022, 77, 1286–1295. [Google Scholar] [CrossRef]
- Wong, M.H.; Chan, E.W.; Chen, S. IS26-mediated formation of a virulence and resistance plasmid in Salmonella Enteritidis. J. Antimicrob. Chemother. 2017, 72, 2750–2754. [Google Scholar] [CrossRef]
- Zhang, Z.; Chang, J.; Xu, X.; Zhou, M.; Shi, C.; Liu, Y.; Shi, X. Dissemination of IncFII plasmids carrying fosA3 and blaCTX-M-55 in clinical isolates of Salmonella enteritidis. Zoonoses Public Health 2021, 68, 760–768. [Google Scholar] [CrossRef] [PubMed]
- Vázquez, X.; Fernández, J.; Rodríguez-Lozano, J.; Calvo, J.; Rodicio, R.; Rodicio, M.R. Genomic analysis of two MDR isolates of Salmonella enterica serovar infantis from a Spanish hospital bearing the blaCTX-M-65 gene with or without fosA3 in pESI-like Plasmids. Antibiotics 2022, 11, 786. [Google Scholar] [CrossRef]
- Lin, D.C.; Chen, S. First detection of conjugative plasmid-borne fosfomycin resistance gene fosA3 in Salmonella isolates of food origin. Antimicrob. Agents Chemother. 2015, 59, 1381–1383. [Google Scholar] [CrossRef] [PubMed]
- Wang, Z.; Xu, H.; Tang, Y.; Li, Q.; Jiao, X. A multidrug-resistant monophasic Salmonella Typhimurium co-harboring mcr-1, fosA3, blaCTX-M-14 in a transferable IncHI2 plasmid from a healthy catering worker in China. Infect. Drug Resist. 2020, 13, 3569–3574. [Google Scholar] [CrossRef]
- Yang, J.; Zhang, Z.; Zhou, X.; Cui, Y.; Shi, C.; Shi, X. Prevalence and characterization of antimicrobial resistance in Salmonella enterica isolates from retail foods in Shanghai, China. Foodborne Pathog. Dis. 2020, 17, 35–43. [Google Scholar] [CrossRef]
- Moreau, M.R.; Wijetunge, D.S.; Bailey, M.L.; Gongati, S.R.; Goodfield, L.L.; Hewage, E.M.; Kennett, M.J.; Fedorchuk, C.; Ivanov, Y.V.; Linder, J.E.; et al. Growth in egg yolk enhances Salmonella Enteritidis colonization and virulence in a mouse model of human colitis. PLoS ONE 2016, 11, e0150258. [Google Scholar] [CrossRef] [PubMed]
- Zhou, X.; Xu, L.; Xu, X.; Zhu, Y.; Suo, Y.; Shi, C.; Shi, X. Antimicrobial resistance and molecular characterization of Salmonella enterica serovar Enteritidis from retail chicken products in Shanghai, China. Foodborne Pathog. Dis. 2018, 15, 346–352. [Google Scholar] [CrossRef] [PubMed]
- Xu, L.; Zhou, X.; Xu, X.; Matthews, K.R.; Liu, Y.; Kuang, D.; Shi, X. Antimicrobial resistance, virulence genes and molecular subtypes of S. Enteritidis isolated from children in Shanghai. J. Infect. Dev. Ctries. 2018, 12, 573–580. [Google Scholar] [CrossRef] [PubMed]
- CLSI. CLSI Supplement M100; Performance Standards for Antimicrobial Susceptibility Testing, 28th ed.; Clinical Laboratory Standards Institute: Wayne, PA, USA, 2018. [Google Scholar]
- Carattoli, A.; Bertini, A.; Villa, L.; Falbo, V.; Hopkins, K.L.; Threlfall, E.J. Identification of plasmids by PCR-based replicon typing. J. Microbiol. Methods 2005, 63, 219–228. [Google Scholar] [CrossRef] [PubMed]
- Sato, N.; Kawamura, K.; Nakane, K.; Wachino, J.; Arakawa, Y. First detection of fosfomycin resistance gene fosA3 in CTX-M-producing Escherichia coli isolates from healthy individuals in Japan. Microb. Drug Resist. 2013, 19, 477–482. [Google Scholar] [CrossRef]
- Chen, L.; Chen, Z.L.; Liu, J.H.; Zeng, Z.L.; Ma, J.Y.; Jiang, H.X. Emergence of RmtB methylase producing Escherichia coli and Enterobacter cloacae isolates from pigs in China. J. Antimicrob. Chemother. 2007, 59, 880–885. [Google Scholar] [CrossRef]
- Wong, M.H.; Yan, M.; Chan, E.W.; Biao, K.; Chen, S. Emergence of clinical Salmonella enterica serovar Typhimurium isolates with concurrent resistance to ciprofloxacin, ceftriaxone, and azithromycin. Antimicrob. Agents Chemother. 2014, 58, 3752–3756. [Google Scholar] [CrossRef]
- Koren, S.; Walenz, B.P.; Berlin, K.; Miller, J.R.; Bergman, N.H.; Phillippy, A.M. Canu: Scalable and accurate long-read assembly via adaptive k-mer weighting and repeat separation. Genome Res. 2017, 27, 722–736. [Google Scholar] [CrossRef]
- Cai, H.M.; Tang, Y.; Jiang, X.F. Plasmid-mediated fosfomycin resistance genes in Gram-negative bacilli. Lab. Med. 2015, 30, 622–625. [Google Scholar]
- Cheng, Y.; Mei, Q.; Liu, Y.Y.; Cheng, J.; Xiong, Z.Z.; Li, J.B. Detection plasmid-mediated fosfomycin resistance genes in clinical isolated shigella. Acta. Univ. Med. Anhui 2015, 50, 441–445. [Google Scholar]
- Omori, K.; Kitagawa, H.; Takada, M.; Maeda, R.; Nomura, T.; Kubo, Y.; Shigemoto, N.; Ohge, H. Fosfomycin as salvage therapy for persistent methicillin-resistant Staphylococcus aureus bacteremia: A case series and review of the literature. J. Infect. Chemother. 2024, 30, 352–356. [Google Scholar] [CrossRef] [PubMed]
- Okazaki, M.; Suzuki, K.; Asano, N.; Araki, K.; Shukuya, N.; Egami, T.; Higurashi, Y.; Morita, K.; Uchimura, H.; Watanabe, T. Effectiveness of fosfomycin combined with other antimicrobial agents against multidrug-resistant Pseudomonas aeruginosa isolates using the efficacy time index assay. J. Infect. Chemother. 2002, 8, 37–42. [Google Scholar] [CrossRef] [PubMed]
- Mikhail, S.; Singh, N.B.; Kebriaei, R.; Rice, S.A.; Stamper, K.C.; Castanheira, M.; Rybak, M.J. Evaluation of the synergy of ceftazidime-avibactam in combination with meropenem, amikacin, aztreonam, colistin, or fosfomycin against well-characterized multidrug-resistant Klebsiella pneumoniae and Pseudomonas aeruginosa. Antimicrob. Agents Chemother. 2019, 63, e00779-19. [Google Scholar] [CrossRef] [PubMed]
- Ho, P.L.; Chan, J.; Lo, W.U.; Law, P.Y.; Chow, K.H. Plasmid-mediated fosfomycin resistance in Escherichia coli isolated from pig. Vet. Microbiol. 2013, 162, 964–967. [Google Scholar] [CrossRef] [PubMed]
- Yang, X.; Liu, W.; Liu, Y.; Wang, J.; Lv, L.; Chen, X.; He, D.; Yang, T.; Hou, J.; Tan, Y.; et al. F33: A-: B-, IncHI2/ST3, and IncI1/ST71 plasmids drive the dissemination of fosA3 and blaCTX-M-55/-14/-65 in Escherichia coli from chickens in China. Front. Microbiol. 2014, 5, 688. [Google Scholar] [CrossRef]
- Freitag, C.; Michael, G.B.; Li, J.; Kadlec, K.; Wang, Y.; Hassel, M.; Schwarz, S. Occurrence and characterisation of ESBL-encoding plasmids among Escherichia coli isolates from fresh vegetables. Vet. Microbiol. 2018, 219, 63–69. [Google Scholar] [CrossRef]
- Cheng, K.; Fang, L.X.; Ge, Q.W.; Wang, D.; He, B.; Lu, J.Q.; Zhong, Z.X.; Wang, X.R.; Yu, Y.; Lian, X.L.; et al. Emergence of fosA3 and blaCTX-M-14 in multidrug-resistant Citrobacter freundii isolates from flowers and the retail environment in China. Front. Microbiol. 2021, 12, 586504. [Google Scholar] [CrossRef]
- Nigiz, Ş.; Hazırolan, G.; Köseoglu Eser, Ö.; Gür, D. First detection of Klebsiella pneumoniae isolate co-harboring fosfomycin resistance gene fosA3 and blaCTX-M among gram negative urine isolates in a Turkish hospital. Microb. Drug Resist. 2022, 28, 317–321. [Google Scholar]
- Wong, M.H.; Chan, E.W.; Xie, L.; Li, R.; Chen, S. IncHI2 plasmids are the key vectors responsible for oqxAB transmission among Salmonella species. Antimicrob. Agents Chemother. 2016, 60, 6911–6915. [Google Scholar] [CrossRef]
- He, D.; Liu, L.; Guo, B.; Wu, S.; Chen, X.; Wang, J.; Zeng, Z.; Liu, J.H. Chromosomal location of the fosA3 and blaCTX-M genes in Proteus mirabilis and clonal spread of Escherichia coli ST117 carrying fosA3-positive IncHI2/ST3 or F2:A-:B- plasmids in a chicken farm. Int. J. Antimicrob. Agents 2017, 49, 443–448. [Google Scholar] [CrossRef]
- Jiang, Y.; Wang, Z.Y.; Li, Q.C.; Lu, M.J.; Wu, H.; Mei, C.Y.; Shen, P.C.; Jiao, X.; Wang, J. Characterization of extensively drug-resistant Salmonella enterica Serovar Kentucky sequence Type 198 isolates from chicken meat products in xuancheng, China. Microbiol. Spectr. 2023, 11, e0321922. [Google Scholar] [CrossRef] [PubMed]
- Sun, R.Y.; Fang, L.X.; Ke, B.X.; Sun, J.; Wu, Z.W.; Feng, Y.J.; Liu, Y.H.; Ke, C.W.; Liao, X.P. Carriage and transmission of mcr-1 in Salmonella Typhimurium and its monophasic 1,4,[5],12:i:- variants from diarrheal outpatients: A 10-year genomic epidemiology in Guangdong, southern China. Microbiol. Spectr. 2023, 11, e0311922. [Google Scholar] [CrossRef] [PubMed]
- Ho, P.L.; Chan, J.; Lo, W.U.; Law, P.Y.; Li, Z.; Lai, E.L.; Chow, K.H. Dissemination of plasmid-mediated fosfomycin resistance fosA3 among multidrug-resistant Escherichia coli from livestock and other animals. J. Appl. Microbiol. 2013, 114, 695–702. [Google Scholar] [CrossRef] [PubMed]
- Jiang, W.; Men, S.; Kong, L.; Ma, S.; Yang, Y.; Wang, Y.; Yuan, Q.; Cheng, G.; Zou, W.; Wang, H. Prevalence of plasmid-mediated fosfomycin resistance gene fosA3 Among CTX-M-producing Escherichia coli Isolates from Chickens in China. Foodborne Pathog. Dis. 2017, 14, 210–218. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).